Abstract
Decoding of UGA selenocysteine codons in eubacteria is mediated by the specialized elongation factor SelB, which conveys the charged tRNASec to the A site of the ribosome, through binding to the SECIS mRNA hairpin. In an attempt to isolate the eukaryotic homolog of SelB, a database search in this work identified a mouse expressed sequence tag containing the complete cDNA encoding a novel protein of 583 amino acids, which we called mSelB. Several lines of evidence enabled us to establish that mSelB is the bona fide mammalian elongation factor for selenoprotein translation: it binds GTP, recognizes the Sec-tRNASec in vitro and in vivo, and is required for efficient selenoprotein translation in vivo. In contrast to the eubacterial SelB, the recombinant mSelB alone is unable to bind specifically the eukaryotic SECIS RNA hairpin. However, complementation with HeLa cell extracts led to the formation of a SECIS-dependent complex containing mSelB and at least another factor. Therefore, the role carried out by a single elongation factor in eubacterial selenoprotein translation is devoted to two or more specialized proteins in eukaryotes.
Keywords: elongation factor/SelB/selenocysteine/translation/tRNASec
Introduction
Selenocysteine is the major biological form of selenium. Only eubacteria, archaea and animals can incorporate it co-translationally into selenoproteins, a class of enzymes instrumental in oxidation–reduction reactions. Selenocysteine is part of the active site in these proteins, which comprise various prokaryotic enzymes but also the animal glutathione peroxidases and thioredoxin reductases that are of prime importance for maintaining the redox status of the cell (reviewed in Burk and Hill, 1999). The conservation of a complex molecular machinery for designating an internal UGA triplet as the selenocysteine and not the stop codon attests to the crucial role borne by this amino acid (reviewed in Atkins et al., 1999). The mechanism leading to selenoprotein translation has been unraveled in eubacteria (reviewed in Atkins et al., 1999; Commans and Böck, 1999). The tRNASec is charged with serine by the conventional seryl-tRNA synthetase, and the seryl residue is converted to selenocysteine on the tRNASec by selenocysteine synthase. The general translation factor EF-Tu does not participate in this process where it is replaced by the specialized elongation factor SelB (Forchhammer et al., 1989). The SelB–GTP–Sec-tRNASec complex binds a hairpin adjacent to the UGA selenocysteine codon in the mRNA, forming a quaternary complex that directs the charged tRNASec to the A site of the ribosome. Regarding archaea, the isolation of the Methanococcus jannaschii SelB homolog was reported very recently (Rother et al., 2000).
Much less is known about selenocysteine incorporation into animal selenoproteins. Two RNA partners are well characterized, the tRNASec and a hairpin structure called SECIS, which resides in the 3′-untranslated region (3′-UTR) of selenoprotein mRNAs. The SECIS element is mandatory for recognition of UGA as a selenocysteine codon (Berry et al., 1991; reviewed in Hubert et al., 1996a; Low and Berry, 1996). With structure–function studies, we have proposed an experimental model for the structure of the SECIS hairpin, which established the pivotal role of non-Watson–Crick base pairs in mediating selenoprotein translation (Walczak et al., 1996, 1998). More recently, it was discovered that SECIS hairpins can adopt two different apical structures, depending on the size of the apical loop in different SECIS RNAs (Grundner-Culeman et al., 1999; Fagegaltier et al., 2000a). Several SECIS-binding proteins have been described (Shen et al., 1995, 1998; Hubert et al., 1996b; Fujiwara et al., 1999; Copeland et al., 2000; Fagegaltier et al., 2000b). Among these, only the 120 kDa SBP2 was shown to be required effectively for mammalian selenoprotein translation (Copeland et al., 2000). Its sequence does not show any characteristic feature of elongation factors, in contrast to the eubacterial system where a single protein, SelB, possesses two functions: one as an elongation factor that binds the charged tRNASec, the other one as the mRNA hairpin-binding factor. The latter function is mediated by the C-terminal domain of SelB, which is not found in EF-Tu (Kromayer et al., 1996).
Several proteins have been described to be associated with the tRNASec. Partially purified fractions containing SePF, a putative bovine SelB factor, protected the Sec-tRNASec ester bond against mild hydrolysis (Yamada et al., 1994). Autoantibodies from patients with a severe chronic active hepatitis precipitated a 48 kDa polypeptide associated with the tRNASec (Gelpi et al., 1992). Recently, it was shown that the mammalian protein SECp43 specifically binds the tRNASec (Ding and Grabowski, 1999). However, none of these proteins was reported to bear the SelB function, and the absence of a known mammalian homolog of the bacterial SelB prompted us to undertake experiments to isolate its cDNA. Here, we report the identification of the cDNA and the functional characterization of the elongation factor mSelB, a novel mouse protein of 583 amino acids specialized for selenoprotein translation.
Results
Identification of a partial human SelB sequence and obtaining the complete mouse SelB cDNA
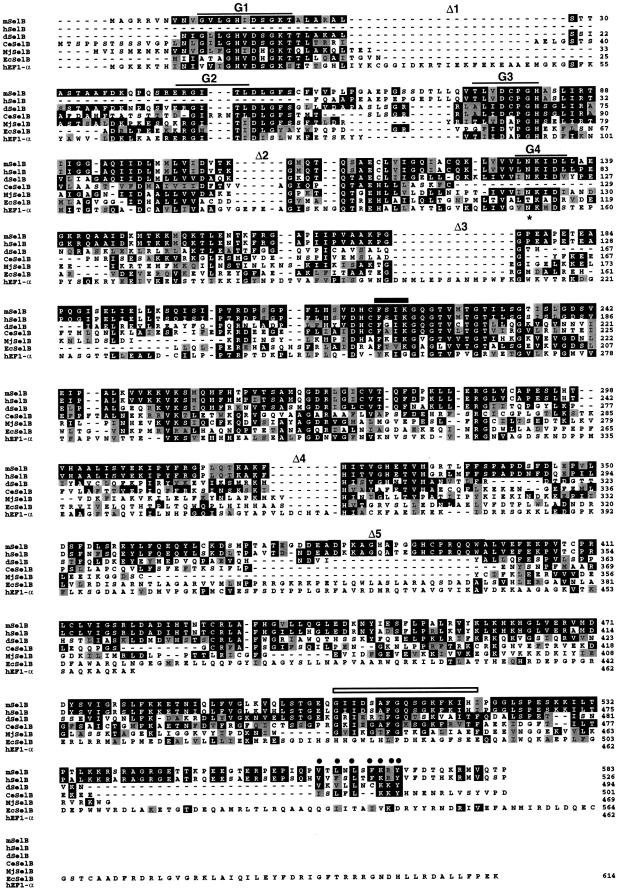
Two characteristic features of the Escherichia coli (Ec) and M.jannaschii (Mj) SelB sequences are: (i) the presence of deletions Δ1–Δ4 (Figure 1) with respect to the EF-Tu or EF1-α elongation factors (Hilgenfeld et al., 1996; Rother et al., 2000); and (ii) the FXI/VK sequence (the solid horizontal bar in Figure 1), which we found to constitute one of the hallmarks of the EcSelB and MjSelB sequences since the terminal lysine of the motif is replaced by threonine and glycine residues in EF-Tu and EF1-α, respectively (Figure 1). The tBLASTn program was used to identify the mammalian SelB homolog by querying expressed sequence tag (EST) databases at NCBI with the MjSelB amino acid sequence. Among the hits, one human EST (DDBJ/EMBL/GenBank accession No. R53658) showed the best match and was selected because its translation indicated that it indeed contained the FSIK sequence and the Δ4 deletion. Complete sequencing of this EST indicated the presence of a 1095 bp open reading frame (ORF), but sequence alignments with EcSelB, MjSelB, EF-Tu and EF1-α led to the conclusion that the EST lacked the region lying upstream of the FSIK sequence. 5′ RACE PCR and library screening provided information for a new 1991 bp human sequence harboring an ORF with a coding capacity of 526 amino acids for the putative human SelB (hSelB). As the ORF was still incomplete, its sequence was used for a second round of EST database searches, leading to the identification of a mouse EST (DDBJ/EMBL/Genbank accession No. AI317100). After complete sequencing, its translation and sequence comparisons with MjSelB, EcSelB and EF1-α indicated that it contained the complete cDNA for the putative mouse SelB (mSelB), with a 1749 bp ORF encoding a 583 amino acid protein of 63.5 kDa predicted molecular mass. The deduced amino acid sequences of the putative hSelB and mSelB are shown in Figure 1. Analysis of their sequences over the common region indicated that they share 88% amino acid identity.
Fig. 1. Alignment of SelB sequences from mouse (mSelB), human (hSelB), C.elegans (CeSelB), Drosophila (dSelB), M.jannaschii (MjSelB), E.coli (EcSelB) and of the human hEF1-α sequence. The alignment was made with ClustalW (Thompson et al., 1994) and manually refined with MegAlign (DNASTAR). Identical amino acids are in reverse, similar residues are shaded in gray. The G1–G4 GTP-binding domains are indicated, as well as the Δ1–Δ5 deletions mentioned in the text. The open bar depicts the mSelB G503–I519 block of homology, and the solid bar maps the hSelB FSIK sequence (positions 162–165). The closed circles and the asterisk position residues that are mentioned in the Discussion.
Sequence features of the human and mouse SelB
Figure 1 shows the presence of significant blocks of homology between amino acid positions V8 and V290 in mSelB, and I8 and V325 in hEF1-α, yielding 23% amino acid identity. Sequence alignments between the mammalian SelB, MjSelB and EcSelB indicated that the blocks of homology extend to positions F337 in mSelB, Y319 in MjSelB and L307 in EcSelB. In the latter region, the amino acid identity between mSelB and EcSelB is 24% but reaches 36% between mSelB and MjSelB.
The eubacterial and MjSelB proteins contain the GTP-binding domains but possess two features that distinguish them from EF-Tu and EF1-α, respectively: the presence of a C-terminal extension and the lack of most of the contact domains (Δ1–Δ4 in Figure 1) with the guanine nucleotide exchange factor(s) (Hilgenfeld et al., 1996; Rother et al., 2000). Four blocks of sequence similarity to the G1–G4 GTP-binding domains of EcSelB, MjSelB and hEF1-α were detected in mSelB (only G3 and G4 in hSelB since the sequence is incomplete). Other stretches of sequence identity or similarity with EcSelB and hEF1-α were found flanking the G2 and G3 domains, or corresponding to amino acid positions D215–I236, V253–M256, L272–I274 and R286–G287 in the mSelB sequence. Amino acids at these positions in EF-Tu were described to contact the T or acceptor stems, or the aminoacyl group, in the crystal structures of the Thermus aquaticus tRNAPhe–EF-Tu and E.coli tRNACys–EF-Tu complexes (Nissen et al., 1995, 1999). Also showing up in mSelB were the four deletion regions Δ1–Δ4 (Δ2–Δ4 in hSelB) that occur in EcSelB and MjSelB.
Observation of the C-terminal parts of the mSelB, hSelB, MjSelB and EcSelB SelB sequences revealed the presence of sequence similarities corresponding to mSelB positions F403–P583, creating a C-terminal extension with respect to hEF1-α (Figure 1). The mammalian extension is longer than that of MjSelB, but shorter than in EcSelB.
mSelB is a guanine nucleotide-binding protein
The amino acid sequence conservation in the G1–G4 domains (Figure 1) predicted that mSelB is a guanine nucleotide-binding protein. To investigate whether this correlates with GTP binding in vitro, binding experiments were performed with a His-tagged recombinant protein that was overexpressed in E.coli and purified in two chromatographic steps as shown in Figure 2. The binding activities were characterized by adding a fixed amount of mSelB to various concentrations of [3H]GTP or the non-hydrolyzable [γ-35S]GTP analog. The experiments in Figure 3 established that mSelB is actually a guanine nucleotide-binding protein. From the saturation curve in Figure 3A, we calculated that the apparent dissociation constant Kd was 0.3 µM for GTP. Similar experiments with [3H]GDP gave an apparent Kd of 0.6 µM for the binding of GDP (Figure 3B).
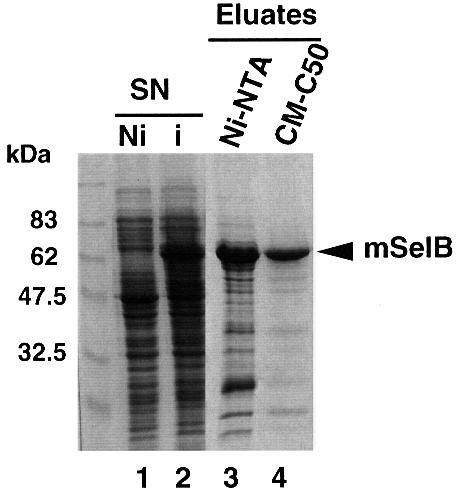
Fig. 2. Purification steps of the recombinant mouse SelB. Supernatants (SN) of E.coli transformed with the His-tagged mSelB expression vector, induced (i in lane 2) or non-induced (Ni in lane 1) by IPTG, were fractionated by affinity chromatography on an Ni-NTA column (lane 3). The eluted fractions were pooled and loaded onto a carboxymethyl-Sephadex C50 column (lane 4). Samples were run on a 10% SDS–polyacrylamide gel and the proteins revealed by Coomassie staining.
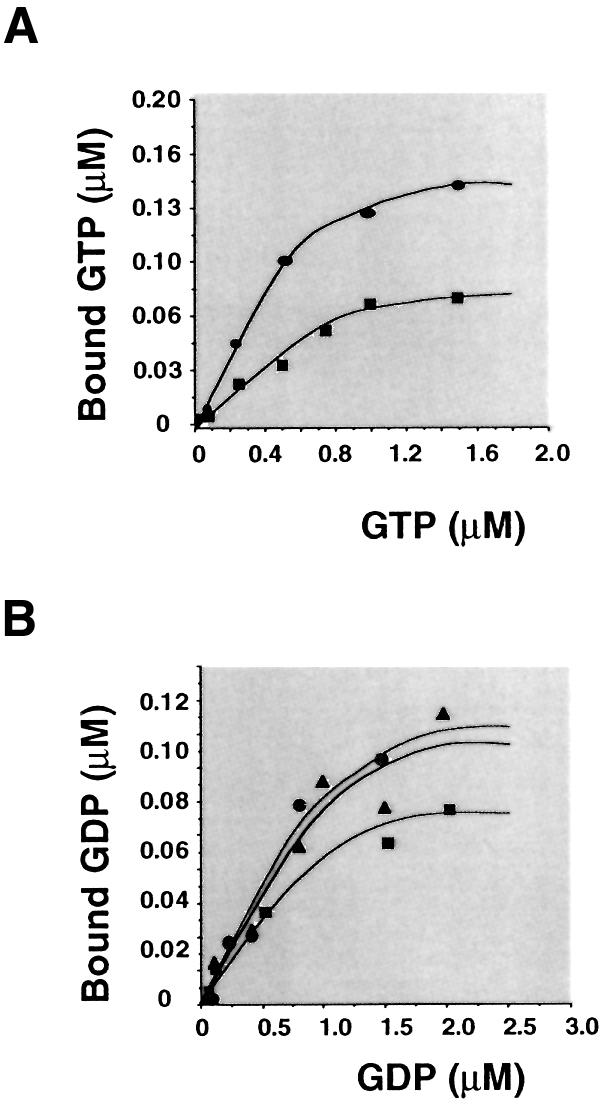
Fig. 3. Determination of the apparent dissociation constants of mSelB for GTP and GDP. Binding assays were performed as described in Materials and methods. (A) The concentration-dependent binding of [3H]GTP to 0.5 (circles) or 0.3 µM (squares) mSelB. (B) Concentration- dependent binding of [3H]GDP to 0.3 (squares), 0.6 (triangles) or 0.9 µM (circles) mSelB.
mSelB binds specifically the selenocysteyl-tRNASec in vitro
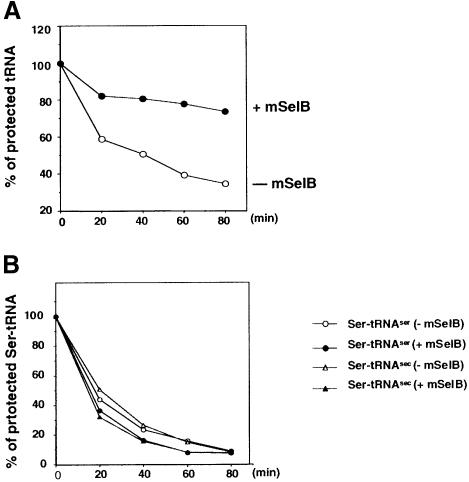
To provide additional evidence that mSelB is indeed the selenoprotein translation factor, we used a protection assay to ask whether it could specifically recognize the tRNASec. This experiment takes advantage of the ability of elongation factors to protect the aminoacyl-tRNA ester bond against mild alkaline hydrolysis when bound to the charged tRNA (Forchhammer et al., 1989; Yamada et al., 1994). A T7 tRNASec transcript was selenocysteylated in vitro with a 75Se selenol group and then submitted to hydrolysis. Figure 4A shows that an 80 min incubation led to 40% of residual selenocysteyl-tRNASec in the absence of mSelB. In contrast, the presence of the recombinant mSelB prevented substantial hydrolysis from occurring since the percentage of protected Sec-tRNASec did not fall below 78%. To attest that the protection was afforded only to the tRNASec, the seryl-tRNASer was submitted to the same treatment. Under similar conditions, mSelB did not preclude hydrolysis of that ester bond (Figure 4B). Furthermore, and most importantly, the ester bond of the seryl-tRNASec, the precursor in the selenocysteine biosynthesis pathway, was clearly susceptible to mild alkaline hydrolysis (Figure 4B). This established that the protection was ascribed to the selenocysteyl moiety.
Fig. 4. mSelB specifically protects the selenocysteyl-tRNASec ester bond against mild alkaline hydrolysis. (A) Hydrolysis of the selenocysteyl-tRNASec in the presence (closed circles) or absence (open circles) of mSelB. (B) Hydrolysis of the seryl-tRNASec in the absence (open triangles) or presence (closed triangles) of mSelB. Hydrolysis of the seryl-tRNASer was performed likewise in the absence (open circles) or presence (closed circles) of mSelB.
From this set of experiments, we conclude that mSelB specifically recognizes the selenocysteyl-tRNASec, but neither the seryl-tRNASec nor the seryl-tRNASer.
The tRNASec is associated with mSelB in vivo
We wished to determine whether the protection detected in vitro reflects an association between mSelB and the tRNASec in vivo. To this end, a hemagglutinin (HA)-tagged mSelB cDNA was transiently transfected into COS-7 cells and the total cell extracts (Figure 5A, lane 3) were submitted to an immunoprecipitation with anti-HA antibodies to recover the tagged mSelB present in the total extract. In the first place, anti-HA antibodies verified that the HA-tagged mSelB was immunoprecipitated effectively from the extract (Figure 5A, lane 4) and that the signal did not show up in immunoprecipitated mock-transfected cell extracts (lane 2). The band observed in the control lane 1 (but not in the immunoprecipitate in lane 2) results from cross-reaction of the polyclonal anti-HA antibody with a cellular protein, as previously reported (Lescure et al., 1999).
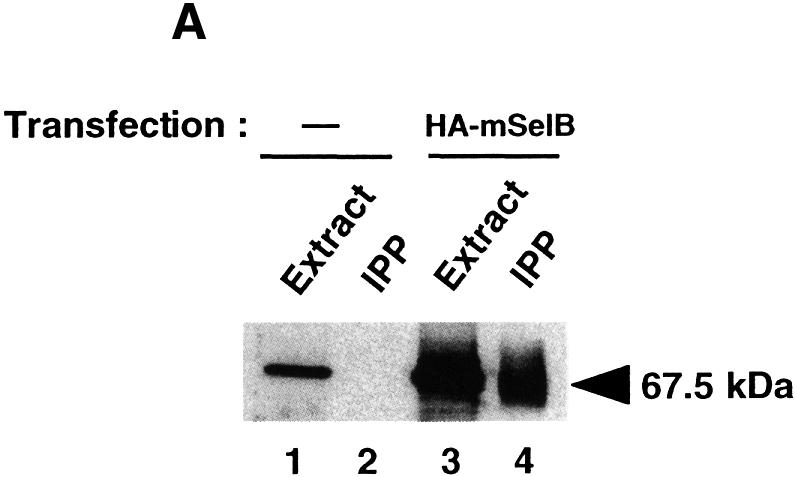
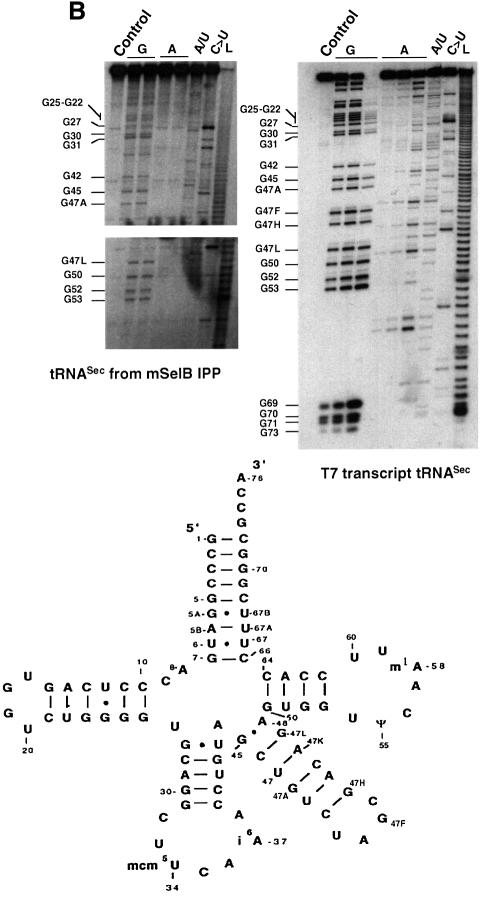
Fig. 5. The tRNASec is associated with mSelB in vivo. (A) Extracts of COS-7 cells transfected with the HA-tagged mSelB expression vector, or mock-transfected, were blotted with the anti-HA antibody before (lanes 3 and 1, respectively) or after immunoprecipitation (IPP in lanes 4 and 2, respectively). The signal in lane 1 results from cross-reaction of the anti-HA with a cellular protein. Samples were run on a 10% SDS–polyacrylamide gel and revealed by chemiluminescence. (B) Enzymatic determination of the tRNASec sequence with RNase T1 (G), RNase U2 (A), RNase PhyM (A/U) and RNase CL3 (C>U). L is an alkaline ladder; control is a lane that received no enzyme. Shown on the left are sequencing gels (two separate migrations) for the immunoprecipitated tRNASec arising from the experiment in (A). The T7 tRNASec transcript was sequenced in parallel for band assignments. The two-dimensional structure of the mammalian tRNASec (Sturchler et al., 1993) is represented with the modified nucleotides.
In a second step, we tested whether the tRNASec was associated with mSelB. A fraction of the immunoprecipitated mSelB was phenol extracted. After mild treatment of the RNA moiety with phosphodiesterase and phosphatase to remove the 3′-terminal CCA, the 3′ end was labeled by action of the tRNA nucleotidyl transferase in the presence of [α-32P]ATP. The sequencing gel in Figure 5B identified the tRNASec, indicating that it was co-immunoprecipitated with the HA-tagged mSelB. In contrast to the T7 tRNASec, the CUUCAAA sequence (positions 32–38) in the anticodon of the immunoprecipitated tRNASec could not be resolved because of missing bands and aberrant migration on the gel, presumably provoked by the mcm5U and i6A modified bases. The same phenomenon was reported by Gelpi et al. (1992) in the course of sequencing the tRNASec isolated from an immunoprecipitated ribonucleoparticle in HeLa cells extracts. Collectively, these experiments showed that the tRNASec is associated with mSelB in vivo.
mSelB is required for efficient selenoprotein translation
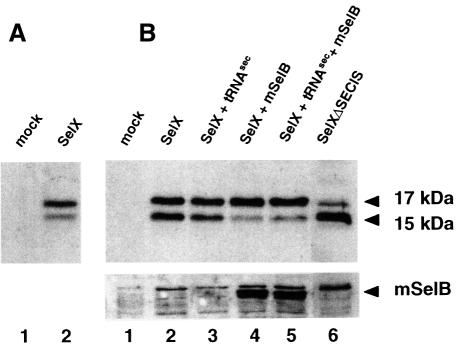
To investigate whether mSelB is involved in the translation of the selenocysteine codon in vivo, we used the following strategy. COS-7 cells were transfected with the cDNA encoding the HA-tagged selenoprotein SelX (Lescure et al., 1999) under experimental conditions (co-transfection of the tRNASec gene) allowing predominant translation of the 17 kDa full-length protein (Figure 6A, lane 2). The weaker intensity signal at 15 kDa arose from premature termination of translation at the selenocysteine codon read as a stop (Lescure et al., 1999). This is supported by the control using the SelX cDNA lacking the SECIS element, which should not lead to the 17 kDa protein (Figure 6B, lane 6). In that lane, the residual full-length SelX very probably arose from unspecific UGA suppression, as previously reported (Walczak et al., 1998; Lescure et al., 1999). If cells were transfected with an excess of SelX cDNA, the intensity of the 15 kDa band was on a par with that of the full-length protein (Figure 6B, lane 2). Co-transfection of the tRNASec gene did not significantly alter the pattern (Figure 6B, compare lanes 2 and 3). This could be interpreted to mean that one (or several) component(s) of the selenoprotein translation machinery, other than the tRNASec, was limiting in the cells to obtain efficient selenoprotein translation when higher amounts of SelX cDNA were transfected. Assuming that the specialized translation factor SelB was one such limiting component, the cells were co-transfected with the mSelB-encoding cDNA. Figure 6B, lane 4 shows that the translation pattern was affected dramatically since the intensity of the 15 kDa product dropped to the level observed in Figure 6A, lane 2, with a concomitant increase in the intensity of the 17 kDa protein (compare lanes 2 and 3 with lane 4 in Figure 6B). The effect was observed regardless of the presence or absence of the tRNASec gene (Figure 6B, lanes 5 and 4, respectively). A control with an anti-mSelB anti-peptide antibody established that mSelB was actually translated in the cells (lanes 4 and 5 in the bottom panel of Figure 6B). This experiment established that mSelB was limiting under our conditions and that its subsequent expression in transfected cells led to stimulation of selenoprotein translation by efficient readthrough of the selenocysteine codon.
Fig. 6. mSelB is required for efficient selenoprotein translation. (A) Separation of the 17 and 15 kDa SelX polypeptides (lane 2, with 3 µg of SelX expression vector). (B) Fractionation of the SelX polypeptides arising from cells transfected with an excess (10 µg) of SelX expression vector (lane 2), and co-transfected with the tRNASec gene (lane 3). Under the same conditions, co-transfection of the mSelB expression vector is shown, in the absence (lane 4) or presence (lane 5) of the tRNASec gene. Lane 6: expression of SelX in cells transfected with a cDNA lacking the SECIS element. Lanes 1 in (A) and (B) are controls with mock-transfected cells. Proteins were fractionated by 12% SDS–PAGE, blotted with the anti-HA antibody and revealed with the ECL kit. Bottom panel: same extracts as above on a separate (10%) gel where mSelB in lanes 4 and 5 was revealed with the anti-mSelB anti-peptide antibody.
In contrast to eubacteria, the eukaryotic SelB does not bind the SECIS element specifically
Eubacterial SelB factors recognize and bind specifically the bacterial SECIS RNA, even in the absence of Sec-tRNASec (Kromayer et al., 1996). To investigate whether mSelB possesses the same property, bandshift assays were performed with the SECIS element of the glutathione peroxidase (GPx) mRNA. Figure 7A shows that a retarded complex (marked by the arrow) appeared with 0.4 µg (lane 2) of recombinant mSelB. The intensity of the complex did not increase with higher amounts of mSelB (Figure 7A, 1 and 2 µg in lanes 3 and 4, respectively), and it even disappeared in the presence of a 2-fold molar excess (versus mSelB) of bulk yeast tRNA (lane 5). With the G24 SECIS mutant, which carries four Watson–Crick base pairs instead of the non-Watson–Crick base pair quartet (Walczak et al., 1998), a faint but nevertheless observable retarded complex was still obtained at high mSelB concentrations (Figure 7A, lanes 7–9). Collectively, the data showed that the complex formed between mSelB and the SECIS element was unspecific. We next wished to determine whether an ancillary factor, contained in fractionated HeLa whole-cell extracts, could promote the binding of mSelB to the SECIS element. Figure 7B, lane 3 showed that incubation of the wild-type SECIS with the extract led to three retarded bands: complex B marked by the arrow, and the other two denoted by the asterisks. Of the three, only complex B was specific since it was totally abrogated by the G24 SECIS mutant (Figure 7B, lane 8). Since the SECIS-binding protein SBP2 also responded to point mutations in the non-Watson–Crick base pair quartet of the SECIS element (Copeland and Driscoll, 1999), it is very likely that complex B contains SBP2. Surprisingly, complementation of 1 µg of the recombinant mSelB with the extract led to formation of the retarded complex A (Figure 7B, lane 4), with a much lower electrophoretic mobility than the unspecific complex formed with mSelB alone (Figure 7B, lane 2). Formation of complex A is dependent on mSelB since it appeared only in its presence (compare lanes 4 and 3, which contained whole-cell extract in the presence or absence of mSelB, respectively). Pre-incubation of mSelB with the anti-peptide anti-mSelB antibody was inhibitory to complex A formation (Figure 7B, lane 5), whereas this effect was not observed with the pre-immune IgGs (Figure 7B, lane 6), arguing that mSelB is actually contained in complex A. Finally, we could determine that the retarded complex A contained at least one component binding the SECIS element, because utilization of the G24 SECIS mutant abolished its formation (Figure 7B, lane 9).
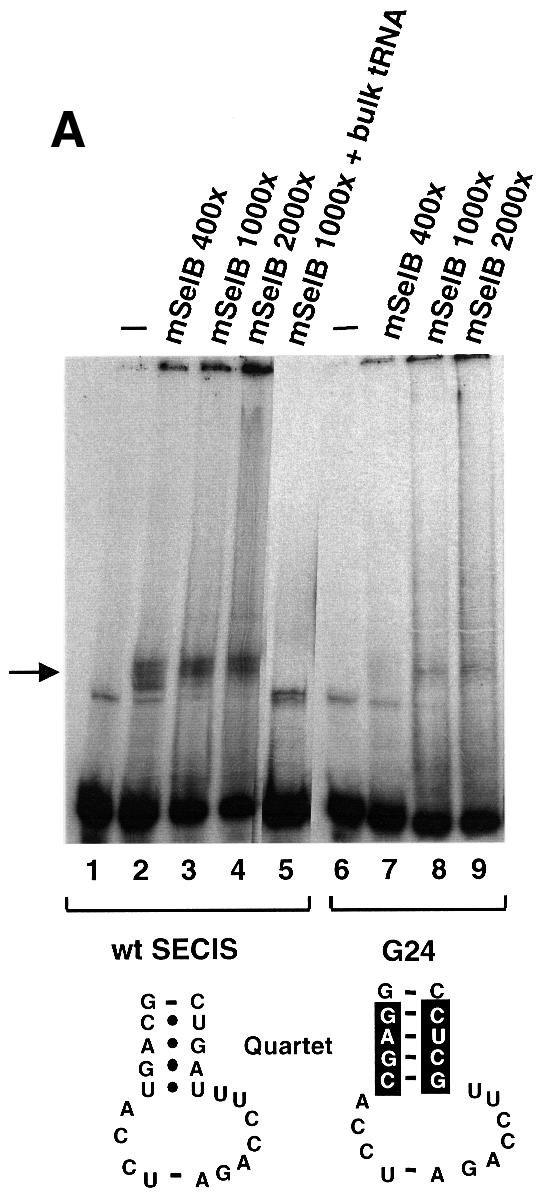
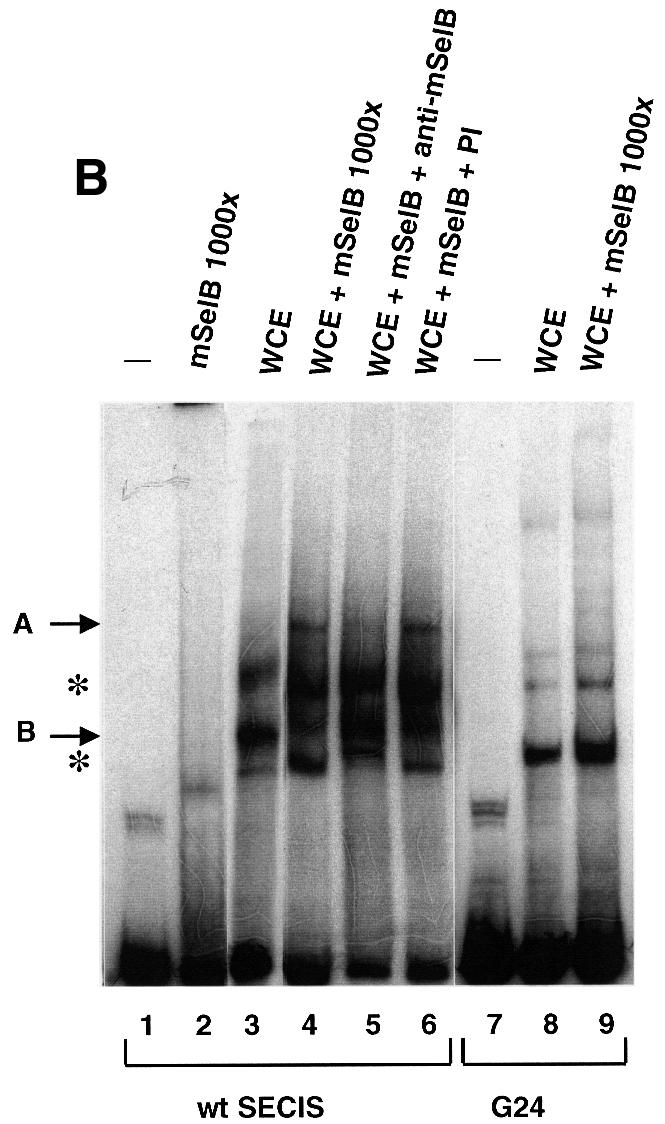
Fig. 7. A protein from HeLa whole-cell extracts forms with mSelB a SECIS-dependent complex. (A) Gel retardation assays obtained with the wild-type GPx SECIS RNA alone (lane 1) or in the presence of increasing amounts of recombinant mSelB protein (400-, 1000- and 2000-fold molar excess mSelB/SECIS RNA, corresponding to 0.4, 1 and 2 µg in lanes 2–4, respectively). Lane 5 contained 700 ng of total yeast tRNA (2-fold molar excess/mSelB). Lanes 6–9 used the G24 GPx SECIS mutant depicted at the bottom. The non-Watson–Crick base pair quartet of the SECIS element is displayed. (B) The wild-type SECIS was incubated alone (lane 1), or in the presence of a 1000-fold molar excess of mSelB alone or complemented with 600 ng of fractionated whole-cell extract (lanes 3 and 4, respectively). The IgG fraction containing the anti-mSelB anti-peptide antiboby or the pre-immune IgGs (PI) were added in lanes 5 and 6, respectively. The G24 SECIS mutant was used in lanes 7–9 under the same conditions as in lanes 1, 3 and 4. Asterisks denote unspecific complexes. WCE, whole-cell extract.
Therefore, the recombinant mSelB alone is unable to recognize the SECIS element specifically. However, mSelB forms a SECIS-dependent complex with at least one component contained in the HeLa whole-cell extract, likely to be SBP2.
Putative SelB homologs in the Caenorhabditis elegans and Drosophila genomes
A query of the C.elegans and Drosophila genomes with the mSelB amino acid sequence identified two genes (DDBJ/EMBL/GenBank accession Nos Z99709 and AC004434, respectively) that yielded 501 (C.elegans) and 494 (Drosophila) amino acid polypeptides bearing 26 and 38% sequence identity with mSelB, respectively (Figure 1). The sequence alignment in Figure 1 showed that both sequences carry the G1–G4 homology blocks and the Δ1–Δ4 deletions. It is worth noting the presence of the other deletion Δ5 in the C.elegans and Drosophila sequences that partially amputates the region corresponding to amino acid positions T373–Q397 in mSelB and A317–Q340 in hSelB. This deletion also occurs in MjSelB where it extends towards the N-terminus.
The C.elegans and Drosophila proteins also carry C-terminal extensions with respect to EF1-α. Although shorter, they harbor significant blocks of homology with the mammalian and MjSelB. Taking into account the occurrence of the GTP-binding domain sequence similarities, the SelB-specific Δ1–Δ4 deletion domains and the presence of sequence similarities in the C-terminal extensions, it is very likely that the C.elegans and Drosophila genes encode the CeSelB and dSelB functional homologs.
Discussion
By combining database search, sequence alignments and cDNA screening, we obtained a 525 partial amino acid sequence for the human SelB. This information was used to identify the complete cDNA encoding the 583 amino acid mouse SelB. Sequence alignments, the guanine nucleotide-binding activity of this protein, its specific association with the tRNASec both in vitro and in vivo, and finally its requirement for efficient selenoprotein translation in vivo altogether constitute several converging lines of evidence establishing that mSelB is the elongation factor specialized for selenoprotein translation.
The predicted molecular mass of mSelB is 63.5 kDa, to be compared with the 50 kDa of EF1-α. The C-terminal extension of mSelB accounts predominantly for the increased molecular mass. The N-terminal part of mSelB has 23% sequence identity with the general elongation factor hEF1-α. The majority of the homology blocks correspond to the G1–G4 domains that are directly involved in GTP binding in EF-Tu (LaCour et al., 1985). The other conserved amino acids were shown in EF-Tu to contact the tRNA arms (Nissen et al., 1995, 1999), suggesting that the mSelB N-terminal domain binds the charged tRNASec. The 36% amino acid identity in the N-terminal domains between the mammalian SelB and MjSelB sequences is also noteworthy, whereas this percentage drops to 24% in the three couples mSelB–EF1-α, EcSelB–EF-Tu (Hilgenfeld et al., 1996) and mSelB–EcSelB. Among G proteins, the G2 domain is only present in elongation factors (Forchhammer et al., 1989). In this regard, its occurrence in the mammalian SelB strengthens the evidence for their being part of this class of translation factors. In the G4 domain, all G proteins possess the conserved asparagine corresponding to N112 in mSelB (positioned by the asterisk in Figure 1). Remarkably, EcSelB is an exception to the rule since it has a threonine instead (Forchhammer et al., 1989).
The affinity of mSelB for GTP is 2-fold higher than for GDP, in keeping with the 4- and 5-fold higher affinities for GTP determined for MjSelB and EcSelB, respectively (Forchhammer et al., 1989; Rother et al., 2000). Whereas the affinities for GDP are either similar (EF1-α) or much higher (EF-Tu) than for GTP (reviewed in Negrutskii and Elskaya, 1998), it turns out that the SelB proteins are the only class of elongation factors possessing higher affinities for GTP. This corroborates and extends the observation reported earlier by Hilgenfeld et al. (1996). In this regard, the lower affinity of the SelB factors for GDP should obviate the need for the guanine nucleotide exchange factor(s), an assumption supported by the presence of the deletion regions Δ1–Δ4 that correspond precisely in EF-Tu to amino acid residues contacting EF-Ts (Hilgenfeld et al., 1996).
It is noticeable that, in the C-terminal extensions, there is a significant block of homology between the mammalian SelB, CeSelB, dSelB and MjSelB in the region corresponding to positions G503–I519 in mSelB (the open horizontal bar in Figure 1). Remarkably, residue L566 in mSelB is invariant between hSelB, CeSelB and dSelB, and the hydrophobicity of amino acids 562 and 564 is maintained between these proteins (closed circles in Figure 1). Moreover, the striking conservation of the KR/KY triplet (positions 569–571 in mSelB, depicted by closed circles in Figure 1) should provide some clue toward understanding the function of the C-terminal extension in the animal SelB.
A major difference between the eubacterial and eukaryal SelB emerged from our data. Indeed, we could show that mSelB alone was unable to bind the SECIS element specifically, contrasting with EcSelB, which forms a specific complex with the bacterial SECIS hairpin (Kromayer et al., 1996), but in keeping with the archaeal MjSelB, which is unable to do so (Rother et al., 2000). Remarkably, complementation of the recombinant mSelB with HeLa whole-cell extracts enabled formation of a complex with the ability to bind the SECIS element specifically. However, we observed that this complex disappeared with the GPx SECIS mutant carrying four Watson–Crick base pairs instead of the non-Watson–Crick base pair quartet, essential to mediate selenoprotein translation (Walczak et al., 1998). Since it was shown that formation of a complex between the SECIS-binding protein SBP2 and the PHGPx SECIS element was abolished by a single point mutation in the non-Watson–Crick base pair quartet (Copeland and Driscoll, 1999; Copeland et al., 2000), it is very likely that SBP2 contained in the extract leads to formation of the mSelB-containing SECIS-dependent complex. The contacts could be direct or indirect, perhaps mediated by the mSelB C-terminal domain. In any event, we can conclude unambiguously that the selenocysteine UGA decoding process in mammals necessitates at least two distinct proteins, mSelB and SBP2, to ensure elongation of translation and the SECIS-binding function, whereas this task is accomplished by SelB alone in eubacteria. Since selenocysteine decoding in the archae M.jannaschii appears to follow similar lines (Rother et al., 2000), this and our work bring one more piece of evidence underpinning the close relationship existing between the archaeal and eukaryal translation apparatus.
Obviously, eukaryotic selenoprotein synthesis represents a higher degree of complexity than regular translation, but also higher than anticipated. The evolutionary conservation of such a molecular machinery for selenocysteine biosynthesis and UGA decoding, composed of four gene products in eubacteria and at least five in eukaryotes, speaks in favor of the importance that selenoproteins take on. Consistent with this view is the observation that the machinery exists in C.elegans only for translation of the single thioredoxin reductase selenoprotein (Buettner et al., 1999). Mechanistically, several issues are raised, one of which asks how the SECIS element in the 3′-UTR can orchestrate the readthrough of the UGA selenocysteine codon far upstream in the coding region. One hurdle that impeded deciphering of this molecular mechanism was precisely the lack of a known elongation factor for selenoprotein translation. As of now, the identification of the eukaryotic SelB will undoubtedly help hasten the pace of future discoveries.
Materials and methods
Database search
The human and mouse dbEST and the Drosophila genome were queried using tBlastn at NCBI. The C.elegans genome was searched at the Sanger Centre. ESTs R53658 and AI317100 were purchased at Incyte Genomics (formerly Genome Systems) and sequenced.
Obtaining the complete mouse SelB cDNA: plasmid constructs
To obtain the region upstream of the FSIK sequence in hSelB, iterative rounds of Marathon 5′ RACE PCR were performed using the Marathon cDNA amplification kit with human prostate Marathon Ready cDNAs (Clontech). This produced sequences either carrying stop codons or bringing little additional information. A HeLa λ-Zap random library (a gift of J.M.Garnier, IGBMC, Strasbourg) was screened with a 174 bp EcoRI–XmaI fragment arising from one clone of the last 5′ RACE PCR step and that provided a 35 bp complementarity to the region upstream of FSIK. Four independent clones were obtained with sequences overlapping R53658 and containing variable 5′-extended ends. The clone with the longest 5′ end added 484 bp of 5′ sequences to R53658. Compilation of the sequence of this clone with that of R53658 yielded the 1991 bp cDNA putatively encoding the human SelB. It was used for identifying the complete mouse cDNA by database search. The pBluescript SK(–) plasmid (EST AI317100) containing the 2130 bp EcoRI–XhoI cDNA insert with the 1749 bp ORF encoding the mouse SelB protein was called pSK(–)-mSelB.
To construct mSelB tagged with six histidines adjacent to the methionine at the N-terminus, the 2168 bp BamHI–XhoI fragment from pSK(–)-mSelB was ligated to the BamHI–SalI-digested pQE-30 vector (Qiagen) to yield pQE-mSelB.
The HA-tagged mSelB was constructed as follows. NotI digestion of the pXJ(HA)3 vector (Walczak et al., 1998) yielded pXJ(ΔHA)3 and a 111 bp NotI fragment containing the triple HA tag and Kozak sequences. The 2186 bp mSelB-containing fragment arising from NotI–XhoI digestion of pSK(–)-mSelB was ligated to the NotI–XhoI-digested pXJ(ΔHA)3 vector, giving pXJ-mSelB. Ligation of the NotI fragment into the unique NotI site of pXJ-mSelB resulted in construct pXJ(HA)3-mSelB carrying Kozak sequences followed by the triple HA tag in-frame with the N-terminus of the mSelB-coding sequence.
mSelB purification
Overexpression of the His6-tagged mSelB was obtained by growing transformed E.coli TG1 for 8 h at 25°C with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cleared lysates were prepared under native conditions in the lysis buffer containing 50 mM sodium phosphate buffer pH 8, 300 mM NaCl, 10 mM imidazole, 1 µM bestatin, 1 µM pepstatin A, 1 µM E-64, 100 µM pefablock and 1 mg/ml lysozyme. Affinity chromatography on Ni-NTA–agarose (Qiagen) was done at 4°C according to the manufacturer, the washing and elution steps being performed in the lysis buffer containing 20 and 250 mM imidazole, respectively. Fractions were dialyzed against 100 mM sodium phosphate buffer pH 7, 10 mM KCl, 5 mM dithiothreitol (DTT), 5% glycerol and protease inhibitors. Fractions from three Ni-NTA chromatographies were pooled and loaded onto a carboxymethyl-Sephadex C50 column equilibrated in the dialysis buffer. Mouse SelB eluted at 600 mM KCl. The fractions were dialyzed against 100 mM phosphate buffer pH 7, 10 mM KCl, 5 mM DTT, 5% glycerol, and stored at –20°C in 40% glycerol.
Guanine nucleotide-binding assays
Binding experiments were carried out at 25°C in 100 mM potassium phosphate buffer pH 7, 10 mM KCl, 2 mM MgCl2, 1 mM DTT. [8,5′-3H]GTP and [8,5′-3H]GDP (NEN Life Science) were diluted to 4000 d.p.m./pmol. The non-hydrolyzable [γ-35S]GTP (Amersham-Pharmacia) was used at 555 d.p.m./pmol. Incubation with 0.3–0.9 µM mSelB was done for 20 min at 25°C in 200 µl. Aliquots of 30 µl were withdrawn for scintillation counting and 150 µl were transferred to the upper reservoir of Ultrafree-MC units (30 000 PTTK membranes, Millipore), as described in Ormö and Sjöberg (1990). After a 45 s centrifugation at 6500 r.p.m., 30 µl aliquots were withdrawn from the upper and lower reservoirs for scintillation counting and calculation of the free nucleotide concentration. The bound nucleotide concentration was calculated as the difference between the total and filtrate nucleotide concentrations.
Protection assays
[75Se]HSe– was prepared from sodium [75Se]selenite (2400–2800 Ci/g, the Missouri University Research Reactor), to serve as the selenocysteine precursor according to Yamada et al. (1994). Aminoacylations of tRNASec and tRNASer were performed with 0.2 µg of T7 transcripts. The tRNASec was serylated and subsequently selenocysteylated in a medium containing partially purified bovine SerRS and selenocysteine synthase fractions (Yamada et al., 1994). The [75Se]Sec-tRNASec was phenol extracted under acidic conditions. Aminoacylation was verified after hydrolysis of the ester bond to release the [75Se]selenocysteine that was fractionated on TLC plates in n-butanol:acetic acid:H2O (4:1:1). [14C]Ser-tRNASec and [14C]Ser-tRNASer were prepared with partially purified bovine SerRS and l-[14C]serine (150 mCi/mmol).
The saturating amount of mSelB necessary for the protection assay was determined with protein amounts in the range 0–40 µg. The standard assay mixture consisted of 30 µl containing 100 mM Tris–HCl pH 7.5, 75 mM KCl, 0.2 mM DTT, 5 mM of the non-hydrolyzable GTP analog GMP-P(CH2)-P, 5 µg of bovine serum albumin (BSA), with either 3 µl (80 fmol) of the [75Se]Sec-tRNASec preparation and 16–40 µg of mSelB, or 3000 d.p.m. of [14C]Ser-tRNASec or [14C]Ser-tRNASer (5 pmol) with 40 µg of mSelB. Samples were incubated for 30 min at 37°C. Aliquots were ethanol precipitated under acidic conditions. The pellet containing the protected aminoacyl-tRNA was hydrolyzed in 2 M NH4OH, 1 mM β-mercaptoethanol for 1 h at 37°C, and precipitated. The free aminoacyl residue liberated from the protected tRNA was recovered from the supernatant and spotted directly on TLC plates. The amount of [75Se]selenocysteine or [14C]serine in the supernatant was measured with the Fuji Bioimage Analyzer BAS2000 to calculate the ratios of the Sec-tRNASec, Ser-tRNASec and Ser-tRNASer protected by mSelB against mild alkaline hydrolysis.
Generation of an anti-peptide antibody
The synthetic peptide KKMQKTLENTKFRG derived from the mSelB amino acids 151–164 was coupled to keyhole limpet hemocyanin and injected into rabbits to generate a polyclonal anti-peptide antibody. It was purified by precipitation with a saturating solution of ammonium sulfate at 4°C or by protein A–Sepharose affinity chromatography.
COS-7 cells transfections and immunoprecipitations
Calcium phosphate transfections of COS-7 cells were carried out according to Walczak et al. (1998) and Lescure et al. (1999) with 10 µg of total DNA, containing 1 µg of CMV-LacZ and 2 µg of the tRNASec gene, in the presence of 10 nM sodium selenite. For SelX translation, cells were transfected with either 3 (Figure 6A, lane 2) or 10 µg (Figure 6B) of pXJ(HA)3SelX or pXJ(HA)3SelXΔSECIS DNAs (Lescure et al., 1999). Four micrograms of pXJ-mSelB DNA was used in Figure 6B, lanes 4 and 5. Supernatants of the crude cell extracts were analyzed by western blot after normalizing with the β-gal assay. Transfections were done in triplicate in three independent experiments.
For mSelB immunoprecipitation purposes, 8 µg of pXJ(HA)3-mSelB DNA were used but no tRNASec gene added. Each supernatant from 15 extracts of COS-7 transfected cells was added to 350 µl of 20 mM HEPES–NaOH pH 7.9, 150 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5% Tween and protease inhibitors, and incubated for 1 h at 20°C with 40 µl of anti-HA antibodies coupled to protein A–Sepharose. Samples were washed five times in 400 µl of the same buffer. A fraction of each sample was withdrawn for western blot analysis. To analyze the tRNASec, the remainder was resuspended in 100 µl of 50 mM Tris–HCl pH 7.5, 10 mM EDTA, 1% SDS, and submitted to proteinase K digestion for 30 min at 37°C. The tRNA moiety was recovered by ethanol precipitation.
3′ end labeling and sequencing of the tRNASec
The tRNASec was taken up in 20 µl of 50 mM Tris–HCl pH 8, 10 mM MgCl2 and digested with 0.5 µg of phosphodiesterase and 0.5 U of calf intestine phosphatase for 15 min at 20°C. It was resuspended subsequently in the same buffer containing 20 µM CTP, 50 µCi of [α-32P]ATP, 8 mM DTT, and treated with 1.5 µg of purified E.coli tRNA nucleotidyl transferase for 30 min at 37°C. The 3′ end-labeled tRNASec was purified on a 12% sequencing gel. The in vitro transcribed bovine T7 tRNASec was treated similarly to be used as a sequence marker. The tRNASecs were sequenced enzymatically with 1–5 U of RNase T1 (G), 10–25 U of RNase U2 (A), 3 U of RNase PhyM (A/U) and 0.01 U of RNase CL3 (C>U). The digests were run on 12% sequencing gels.
RNA-binding assays
HeLa cells were grown by the cell culture group at IGBMC-Strasbourg. HeLa whole-cell extracts, prepared according to Dignam et al. (1983) in the presence of 10 mM DTT, were fractionated on S-Sepharose columns as described by Copeland and Driscoll (1999). Prior to adding the wild-type or all-Watson–Crick G24 mutant GPx SECIS RNAs (Walczak et al., 1998), the appropriate amounts of recombinant mSelB were pre-incubated in 16 µl of the phosphate saline buffer of Copeland and Driscoll (1999), for 15 min at 20°C. The 20 µl final mixture, containing 105 c.p.m. of radiolabeled RNA and 12 mM DTT, was incubated for a further 30 min at 20°C. Incubation of mSelB in the presence of 600 ng of fractionated whole-cell extract, or 700 ng of competitor yeast tRNA, was performed similarly with pre-incubation of the recombinant mSelB and fractionated whole-cell extract, or mSelB and tRNA, in the absence of SECIS RNA. The IgGs containing the anti-mSelB anti-peptide antibody, or the pre-immune IgGs, obtained by protein A–Sepharose chromatography of the sera, were pre-incubated with the recombinant mSelB for 10 min at 20°C prior to the treatment described above. Samples were loaded onto 4% native polyacrylamide gels.
DDBJ/EMBL/GenBank data
The accession Nos for the mouse and human SelB cDNAs are AF268871 and AF268872, respectively.
Acknowledgments
Acknowledgements
We are indebted to A.Böck for his invaluable help in communicating his results on the M.jannaschii SelB homolog prior to publication. We are grateful to J.M.Garnier and P.Chambon, J.M.Egly and the cell culture group at IGBMC, and to G.Keith for the HeLa λ-Zap library, HeLa cells and tRNA nucleotidyl transferase, respectively. We thank L.Jaeger, A.Lescure and E.Myslinski for fruitful discussions and reading of the manuscript, and C.Loegler for excellent technical assistance. This work was supported by grants from the Ligue contre le Cancer du Haut-Rhin and the Association pour la Recherche contre le Cancer. D.F. was awarded a fellowship from the Ministère de l’Education Nationale, de la Recherche et de la Technologie, and K.Y. was a postdoctoral fellow of the Fondation pour la Recherche Médicale.
References
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673. [Google Scholar]
- Berry M.J., Banu,L., Chen,Y.Y., Mandel,S.J., Kieffer,J.D., Harney,J.W. and Larsen,P.R. (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ UTR. Nature, 353, 273–276. [DOI] [PubMed] [Google Scholar]
- Buettner C., Harney,J.W. and Berry,M.J. (1999) The Caenorhabditis elegans homologue of thioredoxin reductase contains a SECIS element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem., 274, 21598–21602. [DOI] [PubMed] [Google Scholar]
- Burk R.F. and Hill,K.E. (1999) Orphan selenoproteins. BioEssays, 21, 231–237. [DOI] [PubMed] [Google Scholar]
- Commans S. and Böck,A. (1999) Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev., 23, 335–351. [DOI] [PubMed] [Google Scholar]
- Copeland P.R. and Driscoll,D.M. (1999) Purification, redox sensitivity and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem., 274, 25447–25454. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Fletcher,J.E., Carlson,B.A., Hatfield,D.L. and Driscoll,D.M. (2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J., 19, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Martin,P.L., Shastry,B.S. and Roeder,R.G. (1983) Eukaryotic gene transcription with purified components. Methods Enzymol., 101, 582–600. [DOI] [PubMed] [Google Scholar]
- Ding F. and Grabowski,P.J. (1999) Identification of a protein component of a mammalian tRNASec complex implicated in the decoding of UGA as selenocysteine. RNA, 5, 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D., Lescure,A., Walczak,R., Carbon,P. and Krol,A. (2000a) Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res., 28, 2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D., Hubert,N., Carbon,P. and Krol,A. (2000b) The selenocysteine insertion sequence binding protein SBP is different from the Y-box protein dbpB. Biochimie, 82, 117–122. [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Leinfelder,W. and Böck,A. (1989) Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature, 342, 453–456. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Busch,K., Gross,H.J. and Mizutani,T. (1999) A SECIS binding protein (SBP) is distinct from selenocysteyl-tRNA protecting factor (SePF). Biochimie, 81, 213–218. [DOI] [PubMed] [Google Scholar]
- Gelpi C., Sontheimer,E.J. and Rodriguez-Sanchez,J.-L. (1992) Autoantibodies against a serine tRNA–protein complex implicated in cotranslational selenocysteine insertion. Proc. Natl Acad. Sci. USA, 89, 9739–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner-Culeman E., Martin,G.W., Harney,J.W. and Berry,M.J. (1999) Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA, 5, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Böck,A. and Wilting,R. (1996) Structural model for the selenocysteine-specific elongation factor SelB. Biochimie, 78, 971–978. [DOI] [PubMed] [Google Scholar]
- Hubert N., Walczak,R., Sturchler,C., Myslinski,E., Schuster,C, Westhof,E., Carbon,P. and Krol,A. (1996a) RNAs mediating cotranslational insertion in eukaryotic selenoproteins. Biochimie, 78, 590–596. [DOI] [PubMed] [Google Scholar]
- Hubert N., Walczak,R., Carbon,P. and Krol,A. (1996b) A protein binds the selenocysteine insertion element in the 3′ UTR of mammalian selenoprotein mRNAs. Nucleic Acids Res., 24, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromayer M., Wilting,R., Tormay,P. and Böck,A. (1996) Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol., 262, 413–420. [DOI] [PubMed] [Google Scholar]
- LaCour T.M.F., Nyborg,J., Thirup,S. and Clark,B.F.C. (1985) Structural details of the binding of guanosine diphosphate to EF-Tu from E.coli as studied by X-ray crystallography. EMBO J., 4, 2385–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure A., Gautheret,D., Carbon,P. and Krol,A. (1999) Novel selenoproteins indentified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem., 274, 38147–38154. [DOI] [PubMed] [Google Scholar]
- Low S.C. and Berry,M.J. (1996) Knowing when not to stop. Trends Biochem. Sci., 21, 203–208. [PubMed] [Google Scholar]
- Negrutskii B.S. and Elskaya,A.V. (1998) Eukaryotic translation elongation factor 1α: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acids Res. Mol. Biol., 60, 47–78. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F.C. and Nyborg,J. (1995) Crystal structure of the ternary complex of the Phe-tRNAPhe, EF-Tu, and GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen P., Thirup,S., Kjeldgaard,M. and Nyborg,J. (1999) The crystal structure of Cys-tRNACys–EF-Tu–GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure, 7, 143–156. [DOI] [PubMed] [Google Scholar]
- Ormö M. and Sjöberg,B.-M. (1990) An ultrafiltration assay for nucleotide binding to ribonucleotide reductase. Anal. Biochem., 189, 138–141. [DOI] [PubMed] [Google Scholar]
- Rother M., Wilting,R., Commans,S. and Böck,A. (2000) Identification and characterisation of the selenocysteine-specific translation factor SelB from the archaeon Methanococcus jannaschii.J. Mol. Biol., 299, 351–358. [DOI] [PubMed] [Google Scholar]
- Shen Q., McQuilkin,P.A. and Newburger,P.E. (1995) RNA binding proteins that specifically recognize the selenocysteine insertion sequence of human cellular glutathione peroxidase mRNA. J. Biol. Chem., 270, 30448–30452. [DOI] [PubMed] [Google Scholar]
- Shen Q., Wu,R., Leonard,J.L. and Newburger,P.E. (1998) Identification and molecular cloning of a human selenocysteine insertion sequence-binding protein. A bifunctional role for DNA-binding protein B. J. Biol. Chem., 273, 5443–5446. [DOI] [PubMed] [Google Scholar]
- Sturchler C., Westhof,E., Carbon,P. and Krol,A. (1993) Unique secondary and tertiary structural features of the eukaryotic selenocysteine tRNASec. Nucleic Acids Res., 21, 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak R., Westhof,E., Carbon,P. and Krol,A. (1996) A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA, 2, 367–379. [PMC free article] [PubMed] [Google Scholar]
- Walczak R., Carbon,P. and Krol,A. (1998) An essential non-Watson–Crick base pair motif in 3′ UTR to mediate selenoprotein translation. RNA, 4, 74–84. [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Mizutani,T., Ejiri,S.-I. and Totsuka,T. (1994) A factor protecting mammalian (75Se)Secys-tRNA is different from EF1-α. FEBS Lett., 347, 137–142. [DOI] [PubMed] [Google Scholar]