Abstract
Purpose
We performed a case-control genome-wide association study (GWAS) to identify single nucleotide polymorphisms (SNPs) associated with musculoskeletal adverse events (MS-AEs) in women treated with aromatase inhibitors (AIs) for early breast cancer.
Patients and Methods
A nested case-control design was used to select patients enrolled onto the MA.27 phase III trial comparing anastrozole with exemestane. Cases were matched to two controls and were defined as patients with grade 3 or 4 MS-AEs (according to the National Cancer Institute's Common Terminology Criteria for Adverse Events v3.0) or those who discontinued treatment for any grade of MS-AE within the first 2 years. Genotyping was performed with the Illumina Human610-Quad BeadChip.
Results
The GWAS included 293 cases and 585 controls. A total of 551,358 SNPs were analyzed, followed by imputation and fine mapping of a region of interest on chromosome 14. Four SNPs on chromosome 14 had the lowest P values (2.23E-06 to 6.67E-07). T-cell leukemia 1A (TCL1A) was the gene closest (926-7000 bp) to the four SNPs. Functional genomic studies revealed that one of these SNPs (rs11849538) created an estrogen response element and that TCL1A expression was estrogen dependent, was associated with the variant SNP genotypes in estradiol-treated lymphoblastoid cells transfected with estrogen receptor alpha and was directly related to interleukin 17 receptor A (IL17RA) expression.
Conclusion
This GWAS identified SNPs associated with MS-AEs in women treated with AIs and with a gene (TCL1A) which, in turn, was related to a cytokine (IL17). These findings provide a focus for further research to identify patients at risk for MS-AEs and to explore the mechanisms for these adverse events.
INTRODUCTION
The third-generation aromatase inhibitors (AIs) anastrozole, exemestane, and letrozole are established adjuvant therapies for postmenopausal women with early-stage breast cancer. This is based on multiple large, randomized clinical trials that have been conducted in the initial therapy setting1,2 after 2 to 3 years of tamoxifen3–6 and in the extended adjuvant therapy setting after about 5 years of tamoxifen.7,8 An American Society of Clinical Oncology (ASCO) panel concluded that optimal adjuvant therapy for postmenopausal women with receptor-positive breast cancer includes an AI, either as initial therapy or after treatment with tamoxifen.9 However, a substantial proportion of women are suboptimally adherent to anastrozole therapy,10 and about half of patients treated with AIs have joint-related complaints,11,12 which likely contributes to decreased compliance.
MA.27 is a phase III trial comparing the nonsteroidal AI anastrozole with the steroidal AI exemestane as adjuvant therapy for early breast cancer. Musculoskeletal complaints were the most frequent reason given by patients on this trial for discontinuing therapy. We used a genome-wide association study (GWAS)13 to identify any SNP (single nucleotide polymorphism) associated with musculoskeletal adverse events (MS-AEs) in women receiving AI adjuvant therapy for early breast cancer, followed by studies of the possible functional basis for the associations.
PATIENTS AND METHODS
Source of Patients
Cases and controls were obtained from the MA.27 trial conducted by the NCIC Clinical Trials Group (coordinating group), Cancer and Leukemia Group B (CALGB), Eastern Cooperative Oncology Group (ECOG), North Central Cancer Treatment Group, Southwest Oncology Group, and International Breast Cancer Study Group (IBCSG). MA.27 included postmenopausal women with completely resected stages I to III breast cancer (American Joint Committee on Cancer [AJCC] Version 6) that was estrogen receptor (ER) positive and/or progesterone receptor positive. Patients were randomly assigned to 5 years of anastrozole or exemestane. This research was performed after approval by local institutional review boards in accordance with assurances filed with and approved by the Department of Health and Human Services.
Accrual of 6,827 North American patients occurred between May 2003 and July 2008, with the majority providing DNA and consent for genetic testing. Non–North American patients (n = 693) entered by the IBCSG did not contribute DNA. MA.27 initially included a second random assignment to celecoxib or placebo, but this was discontinued in December 2004 after the entry of 1,622 patients because of reports of cardiovascular toxicity associated with celecoxib.14
Case Definition for MS-AEs
Cases had at least one of the following six MS-AEs: joint pain, muscle pain, bone pain, arthritis, diminished joint function, or other musculoskeletal problems. Cases were required to either (1) have at least grade 3 toxicity, according to the National Cancer Institute's (NCI's) Common Terminology Criteria for Adverse Events v3.0, or (2) go off treatment for any grade of MS-AE within the first 2 years (ie, an MS-AE occurring after 2 years was not considered a case). Participants who fulfilled the case definition while on celecoxib or within 3 months after stopping celecoxib were excluded as cases.
Control Definition
Controls did not experience any of the MS-AEs, were followed for at least 2 years, and had at least 6 months longer follow-up than a case to which they were matched. This meant that all controls were off celecoxib for at least 6 months.
Study Design
A nested, matched case-control design was used, with matching on the following factors: treatment arm (exemestane, anastrozole), prior adjuvant chemotherapy (yes, no), age at start of AI treatment (± 5 years), celecoxib (yes, no), and time on study. When possible, each case was matched exactly with two controls. Otherwise, we used close matching based on a distance between each case and all potential controls determined with an optimal matching algorithm.15 The majority of MA.27 patients were white (94%), and this GWAS was restricted to white patients. Additional covariates evaluated were body mass index, bisphosphonate use (yes, no), fractures in past 10 years (yes, no), baseline ECOG performance status, prior hormone replacement therapy (HRT; yes, no), prior adjuvant radiotherapy (yes, no), and prior taxane (yes, no).
Genotyping and Quality Control
Two cases and two controls were randomly chosen as duplicates for quality control of genotype concordance. A white parent-child Centre d'Etude du Polymorphisme Humain (CEPH) trio from the HapMaP was included to check for Mendelian transmission of alleles. Genotypes were determined by the RIKEN Center for Genomic Medicine with the Illumina Human610-Quad BeadChip platform (Illumina, San Diego, CA).
Statistical Analyses
Primary analyses were based on conditional logistic regression to account for the matched design. SNP genotypes were coded as additive effects on the log odds ratio by coding as 0, 1, or 2 for the count of the minor allele. This resulted in a likelihood ratio test with 1 df for each SNP. The primary covariates used to match cases and controls were implicitly controlled in conditional logistic regression.
To avoid biases that might arise from differences in genetic ancestry (ie, population stratification), EIGENSTRAT software was used to determine eigenvalues for the SNP correlation matrix that statistically differed from zero on the basis of Tracy-Widom P values.16,17 The corresponding eigenvectors were used as covariates in logistic regression models. We performed additional analyses to evaluate the robustness of our findings that are described in the Appendix (online only). Statistical analyses were conducted with the R statistical computing package, and SAS (SAS Institute, Cary, NC) and PLINK software.18
Imputation and Fine Mapping
SNPs were imputed within 300 kb on either side of the region containing the three SNPs with smallest P values on chromosome 14 using MACH 1.0 software,19 with the white CEPH European Ancestry (CEU) as the reference panel. A region (± 200 kb) around these same three SNPs was fine mapped at the RIKEN Center for Genomic Medicine. First, 29 SNPs were genotyped that were registered in the HapMaP database. After considering linkage disequilibrium (LD) among the SNPs, the strongest associated region of 20 kb (range, 95.23 to 95.25 Mb) was resequenced in 94 samples using an ABI3730 Genetic Analyzer (Applied Biosystems, Carlsbad, CA). From the 119 SNPs that were identified by resequencing, 16 additional SNPs with a minor allele frequency of 0.05 or more were genotyped using multiplex polymerase chain reaction–based Invader assay.
Functional Genomic Studies
The three genotyped SNPs on chromosome 14 with the smallest P values, as well as an imputed SNP with a small P value that was validated by fine mapping, were studied functionally using electrophoretic motility shift (EMS) assays, chromatin immunoprecipitation (ChIP) assays, determination of their relationship to TCLIA expression after estrogen exposure, and transfection studies. Details of the methods used to perform these functional assays are described in the Appendix.
RESULTS
Cases and Controls
This analysis involved 293 cases and 585 controls which, including the duplicate samples and CEPH trios, had call rates of 0.982 to 0.999. Additional details are provided in the Appendix.
Patient Characteristics
Table 1 data show that the cases and controls were well balanced for most factors except prior HRT, which was significantly higher in cases (66% v 44%; P < .001), and fractures within the past 10 years, which was also slightly higher in cases (13% v 9%; P = .06).
Table 1.
Patient Characteristics
| Characteristic | Cases(n = 293) |
Controls(n = 585) |
Wilcoxon Rank Sum P | Fisher's Exact P | ||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Age, years | ||||||
| Median | 63.3 | 64.1 | .61 | |||
| Q1 | 57.8 | 58.1 | ||||
| Q3 | 70.2 | 70.2 | ||||
| Range | 46.1-86.9 | 45.1-84.8 | ||||
| Treatment arm (blinded) | ||||||
| A | 163 | 56 | 326 | 56 | 1.00 | |
| B | 130 | 44 | 259 | 44 | ||
| Celecoxib (blinded) | ||||||
| C | 221 | 75 | 426 | 73 | .42 | |
| D | 72 | 25 | 159 | 27 | ||
| Prior chemotherapy | ||||||
| No | 200 | 68 | 405 | 69 | .82 | |
| Yes | 93 | 32 | 180 | 31 | ||
| Prior taxane | ||||||
| No | 244 | 84 | 490 | 84 | .92 | |
| Yes | 48 | 16 | 94 | 16 | ||
| Unknown/missing | 1 | 1 | ||||
| Prior radiation therapy | ||||||
| No | 100 | 34 | 175 | 30 | .22 | |
| Yes | 192 | 66 | 407 | 70 | ||
| Unknown/missing | 1 | 3 | ||||
| Prior HRT | ||||||
| No | 94 | 35 | 289 | 53 | < .001 | |
| Yes | 178 | 65 | 258 | 47 | ||
| Unknown/missing | 21 | 38 | ||||
| Fracture in past 10 years | ||||||
| No | 255 | 87 | 534 | 91 | .06 | |
| Yes | 38 | 13 | 51 | 9 | ||
| BMI at baseline | ||||||
| Missing | 2 | 0.7 | 8 | 1 | ||
| Known | 291 | 577 | ||||
| Median | 28.2 | 27.9 | .51 | |||
| Q1 | 25.0 | 24.4 | ||||
| Q3 | 33.1 | 32.4 | ||||
| Range | 17.7-56.8 | 16.9-50.8 | ||||
| ECOG PS at baseline | ||||||
| 0 | 237 | 80.9 | 491 | 84 | .24 | |
| 1 | 55 | 18.8 | 88 | 15 | ||
| 2 | 1 | 0.3 | 6 | 1 | ||
| Bisphosphonate use | ||||||
| No | 249 | 95 | 473 | 87 | .66 | |
| Yes | 12 | 5 | 72 | 13 | ||
| Unknown/missing | 10 | 40 | ||||
| MS-AEs withdrew from therapy | ||||||
| Yes | ||||||
| Grade 1 | 17 | 6 | 0 | |||
| Grade 2 | 108 | 37 | 0 | |||
| Grade 3 | 101 | 34 | 0 | |||
| Grade 4 | 6 | 2 | 0 | |||
| No | ||||||
| Grade 3 | 58 | 20 | 0 | |||
| Grade 4 | 3 | 1 | 0 | |||
Abbreviations: Q1, first quartile; Q3, third quartile; HRT, hormone replacement therapy; BMI, body mass index; ECOG, Eastern Cooperative Oncology Group; PS, performance status; MS-AE, musculoskeletal adverse event.
MS-AEs
The maximum grade MS-AEs are presented in Table 1 according to whether the patients discontinued AI therapy. Among the 293 cases, the number of days until the first MS-AE ranged from 10 to 726 (median, 223 days; mean, 276 days). The majority of cases had joint pain as their only MS-AE (184 cases; 62.8%) or in combination with other MS-AEs (56 cases; 19.1%).
Genotyping Results
In all, 592,236 SNPs were genotyped, but 11,281 (1.9%) were considered failures by the laboratory. Of these, 29,478 SNPs with a minor allele frequency (MAF) < 0.01 were excluded because of limited power for association analyses. The exact test for Hardy-Weinberg equilibrium was performed in the controls. The quantile-quantile plot of these P values (Appendix Fig A1, online only) illustrates SNPs with a departure from Hardy-Weinberg equilibrium, and we excluded 82 SNPs with a P value < 1E-06; sensitivity analyses were conducted with differing P value thresholds, and they did not affect the analysis (data not shown). Therefore 551,395 SNPs were used for the association analyses.
Control for Potential Population Stratification
To control for potential population stratification, SNPs were chosen that were uncorrelated with each other to avoid local genomic LD having undesirable impact on global genomic estimates of population stratification. SNPs were considered uncorrelated when the absolute value of the Pearson correlation was < 0.063.20–22 This resulted in 7,077 SNPs being used in the EIGENSTRAT analyses. From these analyses, eight eigenvalues were identified with Tracy-Widom P values < .05. None of the corresponding eight eigenvectors differed significantly (ie, all P > .05) between cases and controls.
GWAS Analyses: Cases Versus Controls
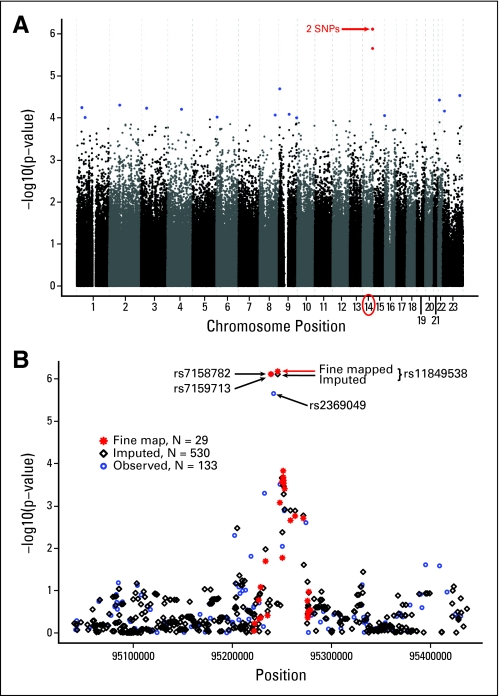
By the conditional logistic regression analyses adjusted for population stratification, the smallest P value was 7.74E-07, close to the commonly accepted threshold for genome-wide significance of 1E-07. Adjusting for the eigenvectors had little influence on the results (see quantile-quantile plot for the conditional logistic regression results, both adjusted and unadjusted for the eigenvectors, in Appendix Fig A2, online only), which also illustrates that the variation inflation factor lambda in Devlin and Roeder23 is close to 1.0. The distribution of P values across the genome is illustrated in the Manhattan plot (Fig 1A). The most striking P values (< 1E-06) were for three SNPs on chromosome 14 (Table 2). Adjusting for the eight eigenvectors, or additionally for prior history of fractures and HRT use, did not substantially alter our findings, nor did the results change substantially according to the unadjusted and unmatched Armitage P values (Table 2). Exploratory analyses for possible SNP-SNP interactions and per allele differences between the two blinded treatment arms were all nonsignificant after adjusting for multiple testing.
Fig 1.
(A) Manhattan plot of –log10 (P values) from conditional logistic regression adjusted for eight eigenvectors versus chromosomal position of single nucleotide polymorphisms (SNPs). (B) Chromosome 14 region of interest: Manhattan plot of –log10 (P values) from conditional logistic regression adjusted for eight eigenvectors for observed (blue circle), imputed (black diamond), and fine mapped (red asterisk) SNP genotypes.
Table 2.
SNPs With Smallest P Values Identified by Genotyping
| SNP | Chromosome | Position (bp) | Minor Allele Frequency |
Unadjusted* |
Armitage |
Adjusted for Eight Eigenvectors* |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | P | P | OR | 95% CI | P | P† | |||
| rs7158782 | 14 | 95238884 | 0.189 | 0.109 | 3.48E-06 | 3.34E-06 | 2.13 | 1.58 to 2.87 | 7.74E-07 | 4.73E-07 |
| rs7159713 | 14 | 95239330 | 0.189 | 0.109 | 3.48E-06 | 3.34E-06 | 2.13 | 1.58 to 2.87 | 7.74E-07 | 4.73E-07 |
| rs2369049 | 14 | 95241604 | 0.176 | 0.100 | 9.17E-06 | 6.98E-06 | 2.08 | 1.54 to 2.83 | 2.23E-06 | 1.96E-06 |
| rs4742490 | 9 | 8361609 | 0.375 | 0.277 | 4.41E-05 | 2.79E-05 | 1.65 | 1.31 to 2.08 | 2.04E-05 | 1.05E-03 |
| rs6637820 | 23 | 130227989 | 0.123 | 0.062 | 1.35E-05 | 8.21E-06 | 2.28 | 1.55 to 3.37 | 2.93E-05 | 3.61E-05 |
| rs1207405 | 22 | 24970849 | 0.111 | 0.058 | 6.84E-05 | 9.12E-05 | 2.28 | 1.54 to 3.38 | 3.76E-05 | 4.47E-04 |
| rs17017756 | 2 | 79821583 | 0.140 | 0.218 | 1.28E-04 | 9.99E-05 | 0.55 | 0.42 to 0.74 | 4.97E-05 | 2.98E-05 |
| rs260964 | 1 | 39330359 | 0.352 | 0.259 | 7.97E-05 | 5.85E-05 | 1.60 | 1.27 to 2.01 | 5.70E-05 | 8.32E-06 |
| rs409228 | 3 | 41040417 | 0.230 | 0.321 | 6.25E-05 | 5.59E-05 | 0.61 | 0.48 to 0.78 | 5.89E-05 | 5.44E-05 |
| rs12186280 | 4 | 108046724 | 0.102 | 0.052 | 8.87E-05 | 8.63E-05 | 2.24 | 1.51 to 3.32 | 6.22E-05 | 3.68E-04 |
| rs6633380 | 23 | 13756099 | 0.296 | 0.212 | 1.28E-04 | 6.63E-05 | 1.65 | 1.29 to 2.10 | 6.88E-05 | 3.93E-05 |
| rs2515034 | 8 | 119565108 | 0.092 | 0.044 | 1.04E-04 | 7.31E-05 | 2.34 | 1.54 to 3.57 | 8.53E-05 | 5.04E-04 |
| rs11145462 | 9 | 79332930 | 0.399 | 0.491 | 1.80E-04 | 2.10E-04 | 0.65 | 0.53 to 0.81 | 8.19E-05 | 1.99E-04 |
| rs4246309 | 15 | 98584524 | 0.447 | 0.347 | 6.66E-05 | 5.07E-05 | 1.52 | 1.23 to 1.88 | 8.81E-05 | 1.39E-04 |
Abbreviations: SNP, single nucleotide polymorphism; bp, base pair; OR, odds ratio.
Conditional logistic regression.
Prior history of fractures and hormone usage.
Imputation and Fine Mapping
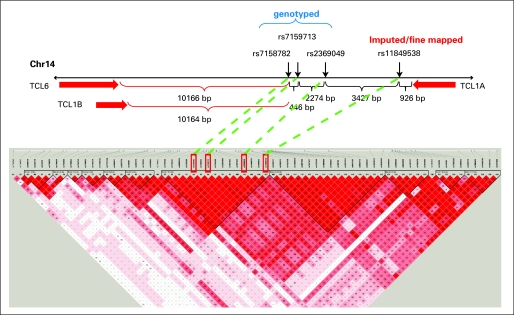
Imputing SNPs within 300 kb of the smallest P value SNPs on chromosome 14, illustrated in Figure 1B, showed that rs7159713 and rs2369049 were in LD with rs7158782 (Pearson correlation of minor allele dosage > 0.8) and that an additional imputed SNP (rs11849538) also showed an association with MS-AEs (MAF cases/controls: 0.172/0.091; odds ratio, 2.21; P = 6.67E-07). Using the imputed data, we focused on a 200-kb region and genotyped 29 SNPs, including the imputed SNP (rs11849538), which was verified by this genotyping and the DNA sequencing. We examined the LD block of the candidate region and focused on the strongest associated region of 20 kb (95.23 to 95.25 Mb), which included four SNPs (rs7158782, rs7159713, rs2369049, and rs11849538). We resequenced this region and identified a total of 119 SNPs that included 49 novel SNPs and 70 SNPs already registered in the dbSNP database. Hence, we genotyped 16 additional SNPs with MAFs of 0.05 or greater, but no SNP showed a stronger association than rs11849538. Therefore, we concluded that rs11849538 or the other three highly linked SNPs (rs7158782, rs7159713, and rs2369049) might have functional significance.
Functional Genomic Studies of SNPs on Chromosome 14
The three genotyped SNPs (rs7158782, rs7159713, and rs2369049) and the imputed SNP (rs11849538) were all close to the T-cell leukemia 1A (TCL1A) gene (Fig 2). All four of these SNPs were in LD (R2 > 0.85). We attempted to determine whether any of these SNPs might be functional on the basis of EMS or ChIP assays, and if they were, whether they might be associated with variation in the expression of the closest gene, TCL1A; whether estrogens might play a role in their functional effects; and, finally, whether TCL1A might influence the expression of receptors or cytokines known to play a role in arthritis.
Fig 2.
Single nucleotide polymorphisms identified on chromosome 14 (Chr 14), their relationship to T-cell leukemia (TCL) genes, and linkage disequilibrium relationships. bp, base pair.
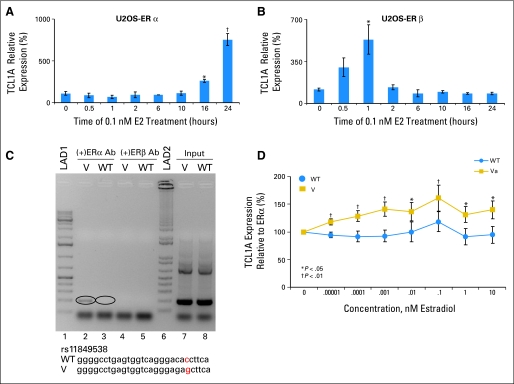
We first determined that TCL1A is highly and variably expressed in 288 lymphoblastoid cell lines from three different ethnic groups for which we have expression array data as well as genome-wide SNP data. EMS assays performed with lymphoblastoid cell nuclear extract showed a shift (ie, protein binding) for all but the rs2369049 SNP, with less binding by the variant than by the wild type (WT) sequences in all cases (Appendix Figs 3A to 3C, online only). The TRANSFAC database predicted that rs7158782 would disrupt a GATA-1 binding motif, and this prediction was supported by a ChIP assay (Appendix Fig 3C). However, of particular importance for this study, the TRANSFAC database also predicted that the SNP closest to the 3′ end of TCL1A—rs11849538—would create an estrogen response element (ERE), and this prediction was supported by the results of a ChIP assay (Fig 3C) performed using ERα-transfected lymphoblastoid cells with known genotype for the rs11849538 SNP. We then determined whether TCL1A expression might be estrogen dependent by exposing U20S cells stably transfected with ERα or ERβ to 0.1 nmol/L 17-β-estradiol (E2) and found eight- and six-fold increases in TCL1A mRNA expression after 18 hours and 1 hour, respectively (Figs 3A and 3B), linking TCL1A expression to estrogens.
Fig 3.
(A) Relative T-cell leukemia 1A (TCL1A) expression in U2OS cells transfected with estrogen receptor alpha (ERα) exposed to 0.1 nmol (nM)/L 17-β-estradiol over 24 hours. (B) Relative TCL1A expression in U2OS cells transfected with ER beta (ERβ) exposed to 0.1 nmol/L 17-β-estradiol over 24 hours. (C) Chromatin immunoprecipitation assay using ERα-transfected lymphoblastoid cells with known genotype for the rs11849538 single nucleotide polymorphism (SNP). DNA ladder 1 (LAD1) and DNA ladder 2 (LAD2) are both Invitrogen (Carlsbad, CA) 1-kb DNA ladders, with LAD2 being a 1-kb Plus DNA ladder. Lanes 2 to 5 are polymerase chain reaction (PCR) products from DNA that was bound to human ERα antibody (Ab). The inputs for the variant (V) and wild type (WT) were PCR amplification products of pools of sheared DNA from the entire genome. (D) SNP-related differences in TCL1A expression and estrogen response in nine V (rs11849538) and nine WT lymphoblastoid cell lines transfected with ERα. (*) P < .05. (†) P < .01.
We then determined the effect of different genotypes at these four SNPs on the estrogen-dependent TCL1A expression. To do that, we transiently transfected lymphoblastoid cell lines with known genotypes for the four SNPs with ERα, exposed the cell lines to various concentrations of E2, and determined the relationship of the SNPs to TCL1A expression (Fig 3D). In all three ethnic groups, the cells with the variant sequences—sequences that created an ERE at rs11849538—showed greater TCL1A expression than did those with the WT sequence.
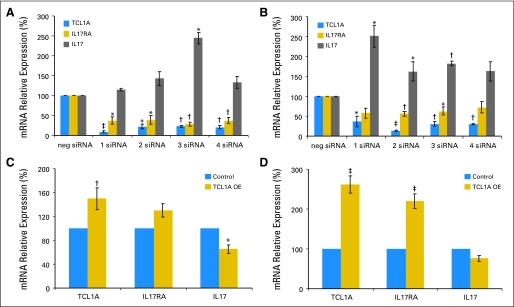
Finally, we knew that interleukin 17 (IL17) and the IL17 receptor A (IL17RA) were both therapeutic targets in patients with rheumatoid arthritis,24 so we determined whether the expression of TCL1A was correlated with the expression of either IL17 or IL17RA in the same 288 lymphoblastoid cell lines. Expression of TCL1A and IL7RA were correlated (r = 0.36; P < 1.9E-10). We then demonstrated in U2OS cells that small interfering RNA knockdown of TCL1A resulted in decreased expression of IL17RA but increased expression of IL17 mRNA (Figs 4A and 4B), while overexpression of TCL1A resulted in increased IL17RA expression and decreased expression of mRNA for the ligand IL17 (Figs 4C and 4D).
Fig 4.
T-cell leukemia 1A (TCL1A), interleukin 17 receptor A (IL17RA), and interleukin 17 (IL17) mRNA levels after TCL1A small interfering RNA (siRNA) knockdown in (A) U2OS–estrogen receptor alpha (ERα) cells and (B) U2OS–ER beta (ERβ) after transient overexpression (OE) of TCL1A in (C) U2OS-ERα cells and (D) U2OS-ERβ cells. (*) P < .05. (†) P < .01. (‡) P < .001.
DISCUSSION
This genome-wide nested case-control study identified four SNPs in tight LD on chromosome 14 that were associated with MS-AEs in women receiving AIs for resected early-stage breast cancer, with P values that ranged from 2.23E-06 to 6.67E-07, close to the Bonferroni threshold of 1E-07. The closest gene to these SNPs was TCL1A with the SNP having the smallest P value (rs11849538) located only 926 bp from the 3′ end of that gene.
The significant advantages of our study design are that it avoids selection biases and exposure recall biases among cases, maximizes representativeness of controls, optimizes measurement of exposure (randomized treatment allocation), and ensures unbiased follow-up of all participants in a protocol-specified manner. In fact, our study design can be viewed as being strong as a cohort study but much more efficient.25 There are, however, general limitations of all GWASs,26 including the potential for false-positive associations, that underscore the requirement for replication. There were imbalances between cases and controls in terms of HRT and prior fractures, but these did not confound our findings. We recognize that the NCI criteria used to measure MS-AEs can be somewhat subjective, possibly with heterogeneous causes that could reduce power, but this does not have an impact on our findings. Additionally, our functional genomic studies are sufficiently compelling to justify further research.
The purpose of this study was both to identify genetic markers for MS-AEs and to explore mechanisms that might be related to this drug-related AE in women exposed to AI-dependent decreased estrogen levels. Therefore, we examined functional characteristics of the SNPs as they might relate to estrogen action. It was striking that the SNP with the smallest P value (rs11849538) created an ERE shown by ChIP assay to be functional (Fig 3C). We determined whether estrogens and/or the SNPs might be functionally related to TCL1A and we demonstrated an eight-fold induction of TCL1A expression by 24 hours in ERα-transfected cells (Fig 3A) and significantly higher TCL1A expression after exposure to varying concentrations of E2 in lymphoblastoid cell lines containing the variant SNPs when compared with cells having the WT sequence after transient transfection with ERα (Fig 3D).
TCL1A expression has previously been associated with a number of hematopoietic malignancies, including T-cell and B-cell lymphomas,27 and has been shown to enhance Akt serine threonine kinase activity, thus functioning as an Akt coactivator.28 TCL1A expression is thought to be restricted to early developmental cells of the immune system, including CD4−, CD8−, and CD3− thymocytes.28 However, there were no previous reports of the regulation of TCL1A by estrogen or of an association of TCL1A expression with cytokine receptor expression. Patients who carry the SNP variant identified in our GWAS that creates an ERE (rs11849538) might be more responsive to a given level of estrogen and thus display higher levels of TCL1A expression for any given level of estrogen (Fig 3D). A reduction in estrogen levels during AI therapy might result in proportionally greater reductions in TCL1A expression in women with these SNPs than in women with the WT sequence. The mechanism by which differential changes in TCL1A expression might induce MS-AEs remains to be determined, but our observations with regard to its relationship to IL17RA expression indicate that the association of TCL1A expression with cytokine function is worthy of further exploration in the course of future studies.
Finally, it is intriguing to speculate that our findings in women receiving AIs who develop MS-AEs may provide insight into the “arthritis of the menopause” described by Cecil and Archer 85 years ago.29 AI therapy might be considered an estrogen-deprivation stress test that could provide novel insights into symptoms related to estrogen deprivation that occur during menopause—74% of women without breast cancer in the Women's Health Initiative clinical trials reported joint pain.30
In summary, this GWAS identified four SNPs on chromosome 14 that were related to MS-AEs in patients receiving AI adjuvant therapy. The combination of four strong SNP signals and the equally strong functional linkage of these SNPs to AI effect focused our attention on these polymorphisms as possible biomarkers for risk for this important adverse drug reaction, on TCL1A as the potential link, and on cytokines as potential mechanistic factors. The determination of the mechanism of these MS-AEs would enable a focused approach to amelioration of symptoms, thus facilitating compliance and improving the benefits of AIs for women with early breast cancer.
Acknowledgment
We acknowledge the women who participated in the MA.27 clinical trial and provided DNA and consent for its use in genetic studies.
Glossary Terms
- Estrogen response element (ERE):
Specific DNA sequences with high affinity for the estrogen receptor that are involved in gene expression in response to estradiol.
- Genome-wide association study (GWAS):
Hypothesis-free studies that evaluate the association of genetic variations throughout the entire genome with traits, using high throughput genotyping technologies to assay SNPs.
- Genotyping:
The process used for obtaining the genotype of a given gene or a genetic marker. Typically, polymerase chain reaction-based methods are used. However, in the case of single nucleotide polymorphism genotyping, microarray platforms are used routinely. Genotyping data serves several purposes, including a means to determine genetic diversity, to identify important genetic traits and in forensic and population studies. It is used increasingly in determining paternity of offspring. From a somatic point of view (within a tumor), genotyping is used to determine loss of heterozygosity.
- HapMaP:
An international project that created a publically available genome-wide database of common human sequence variations, http://hapmap.ncbi.nlm.nih.gov.
- Hardy-Weinberg equilibrium:
A state in which genotype frequencies and ratios remain constant from generation to generation and in which genotype frequencies are a product of allele frequencies. A randomly mating population tends toward a Hardy-Weinberg equilibrium state if there are no mutations, migrations, or environmental factors favoring particular genotypes.
- Imputation:
In a GWAS, the use of a reference data set (eg, HapMaP) and linkage disequilibrium in a region to infer the alleles of SNPs not directly genotyped.
- Linkage disequilibrium:
Nonrandom association of linked genes. This is the tendency of the alleles of two separate but already linked loci to be found together more frequently than would be expected by chance alone.
- Manhattan plot:
In a GWAS, the display of negative log (P values) on the Y-axis for SNPs across the 22 autosomes and sex chromosomes on the X-axis. The higher the point lies on the Y-axis, the lower the P value and the greater the significance.
- Population stratification:
Differences in the allele frequencies in populations due to differences in ancestry.
- SNP (single nucleotide polymorphism):
Genetic polymorphisms are natural variations in the genomic DNA sequence present in greater than 1% of the population, with SNP representing DNA variations in a single nucleotide. SNPs are being widely used to better understand disease processes, thereby paving the way for genetic-based diagnostics and therapeutics.
Appendix
Methods
Identification of cases and controls.
A total of 297 cases were identified. Matching resulted in 291 cases with two perfectly matched controls, four cases with one perfectly matched control, and one close control (allowing ± 10 years in three controls and eliminating the need to match on prior chemotherapy in one control). One case had two close controls after permitting age matching ± 10 years and one case had no matched controls. In addition, one control was excluded after review revealed that her control status was not valid. This resulted in 888 eligible patients (297 cases and 591 controls). However, four controls were not plated and, after genotyping, six samples (four cases and two controls) had call rates < 98% and were excluded from analyses. This resulted in 293 cases and 585 controls.
Functional Genomic Studies
Electrophoretic motility shift (EMS) assays.
Nuclear extracts were prepared from cultured lymphoblastoid cells. The cells were pelleted by centrifugation at 1,500 × g for 2 minutes. The pellets were suspended in 400 μL of ice-cold hypotonic buffer A (10 mmol/L HEPES pH 7.9, 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.1 mmol/L ethyleneglycoltetracetic acid [EGTA], 0.4% NP-40, 0.5 mmol/L phenylmethylsulfonylfluoride [(PMSF], and 0.5 mmol/L dithiothreitol [DTT]) and were incubated on ice for 15 minutes. The cell suspensions were then precipitated by centrifugation at 14,000 × g for 30 seconds, and the pellets were resuspended in 200 μL of buffer B (20 mmol/L HEPES pH 7.9, 400 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 0.5 mmol/L PMSF, and 1 mmol/L DTT), followed by shaking for 15 minutes at 4°C. After centrifugation, the supernatants were stored in aliquots at −70°C. The quantity of protein in the nuclear extracts was determined using the micro bicinchoninic acid protein assay (Pierce, Rockford, IL).
DNA/protein reactions and EMS assays were performed by using 10 μg of lymphoblastoid cell nuclear extract protein incubated with 1× binding buffer, 2.5% glycerol, 5 mmol/L MgCl2, 50 μg/μL Poly (dI·dT), 0.05% NP-40 (Pierce), and 10 fmol biotin 5′-labeled probes (IDT Integrated DNA Technologies, Coralville, IA) at room temperature for 20 minutes. In the same preparation, 2 pmol of unlabeled double-stranded probe was added as competitor before addition of the biotin-labeled probe for the competition studies. Samples were loaded on 6% Tris-borate-EDTA (TBE) gels and were run at 100 V for 1 hour in 0.5× TBE buffer.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed using the ChampionChIP one-day kit (SABiosciences, Frederick, MD). Polymerase chain reaction (PCR) was used to monitor the ChIP assay results. PCR reactions contained 1.25 units of Taq polymerase enzyme (Promega, Madison, WI), 1× reaction buffer, 1.5 mmol/L MgCl2, 0.5 mmol/L deoxyribonucleotide triphosphate (dNTP; Invitrogen, Carlsbad, CA), and 357 nmol/L each primer; a 2-μL amount of DNA from the ChIP assay was added in a final volume of 50 μL. In all, 20 μL of the PCR products were loaded on 1.2% agarose gel and were run at 100 V for 1.5 hours in 1× TAE buffer (Tris-acetic acid-EDTA).
Cell culture studies.
The human U2OS–estrogen receptor alpha (ERα) and U2OS–ER beta (ERβ) cell lines were engineered by using the U2OS human osteosarcoma cell line with the T-REx System (Invitrogen), which allows control of the expression of the transgene by using doxycline (Dox) as an inducer. These cells were stably transfected with ERα and ERβ constructs in the laboratory of Thomas Spelsberg, PhD, at the Mayo Clinic (he generously provided the cells used in these studies). John Hawse, PhD, of the Mayo Clinic, provided consultative input.
For the experiments described in this study, human U2OS-ERα and U2OS-ERβ cell lines were cultured in six-well plates at a cell density of 60% in DMEM containing 10% (vol/vol) fetal bovine serum (FBS). Twenty-four hours later, the cells were transferred to DMEM containing 5% charcoal stripped serum for an additional 24 hours, followed by incubation in DMEM containing 100 ng/mL Dox. The cells were then cultured in DMEM with 100 ng/mL Dox plus 0.1 nmol/L 17-β-estradiol (E2) for 0, 0.5, 1, 2, 6, 10, 16, and 24 hours. Total RNA was then isolated from the cells with the RNeasy mini kit (Qiagen, Chatsworth, CA). One hundred nanograms of total RNA was used to perform quantitative reverse transcriptase PCR (qRT-PCR) with TCL1A primers (Qiagen) and β-actin primers (Qiagen) as controls.
Lymphoblastoid cell line transfection and estrogen exposure.
Three cell lines with variant genotypes for the four chromosome 14 single nucleotide polymorphisms (SNPs) and three cell lines with the wild-type (WT) sequence from each of three ethnic groups (African American, European American, and Han Chinese American; a total of 18 individual cell lines), were cultured in RPMI-1640 media containing 15% (vol/vol) FBS. The genotypes for all four SNPs in each cell line were confirmed by PCR.
The 18 cell lines were transiently transfected with a pcDNA4-ERα construct that was kindly provided by Dr. Spelsberg. For each cell line, 2 × 106 cells were suspended in the Cell Line Nucleofector Kit V (Lonza, Cologne, Germany) with 2 μg purified pcDNA4-ERα, and the cells were transfected with the Amaxa apparatus (Amaxa Biosystem, Cologne, Germany) using the T-030 program. For each cell line, cells from six electroporation procedures were pooled to obtain an adequate number of cells for use in the proposed experiments (ie, 12 × 106 cells per experiment).
Electroporated cells were then plated in RPMI-1640 medium supplemented with 15% FBS and were allowed to recover from the electroporation process for 48 hours. The transiently transfected cells were then cultured in six-well plates with RPMI-1640 media containing 5% (vol/vol) charcoal stripped serum for an additional 24 hours, followed by incubation in the same media containing 0, 0.00001, 0.0001, 0.001, 0.01, 0.1, 1, or 10 nmol/L E2 for 24 hours. Total RNA was then isolated from the cells with the RNeasy mini kit (Qiagen). Two hundred nanograms of total RNA was used to perform qRT-PCR with TCL1A and with ERα primers (Qiagen). The expression of TCL1A was corrected on the basis of ERα expression.
TCL1A transfection.
U2OS-ERα and U2OS-ERβ cells were plated in a six-well tissue culture plate at a density of 500,000 cells per well in DMEM with 15% FBS for 24 hours at 37°C. Cells were transfected with 50 nmol/L TCL1A small interfering RNA (siRNA) or negative control siRNA (Qiagen) for TCL1A knockdown using Lipofectamine RNAiMAX (Invitrogen). Overexpression studies used FuGENE 6 (Roche, Indianapolis, IN). After 24 hours at 37°C, the medium was removed from the cells and replaced for an additional 24 hours with DMEM containing 10% FBS. mRNA was extracted using RNA Mini Kit (Qiagen) for qRT-PCR assay.
Supplemental Statistical Analyses
Additional analyses were conducted to evaluate the robustness of our findings with respect to population stratification. First, we evaluated whether any of the nonmatching investigational covariates were significantly associated with musculoskeletal adverse events (MS-AEs) by using forward stepwise conditional logistic regression. Any factors that were statistically associated with an MS-AE (P < .05) were also evaluated as adjusting covariates. Second, we performed conditional logistic regression but without any adjusting covariates to determine the influence of covariates. Third, we performed the usual Armitage test for trend, which ignores the matching and the covariates. Because several cases did not have matched controls and vice versa, the Armitage test for trend included participants who were excluded from conditional logistic regression analyses. As stated in the main article, these analyses had little impact on the ranking of P values.
Additional exploratory analyses of the genome-wide assay genotype data were run for the SNPs listed in Table 3 plus the SNP with the smallest P value (rs11849538). Using conditional logistic regression and adjustment for the eight eigenvectors, we tested SNP-SNP interactions for SNPs residing on different chromosomes (a total of 128 interaction tests). None of these results approached statistical significance (smallest uncorrected P = .02). We also tested whether the per-allele odds ratio differed between the two blinded treatment arms (smallest P = .11), or differed between the two blinded celecoxib groups (smallest P = .02). We also tested whether the SNPs reported were associated with the grade of MS-AEs. After adjusting for the eight eigenvectors, the smallest P value over all the SNPs was .09, suggesting that the grade of MS-AE was not associated with any of the SNPs. Finally, to evaluate whether the time to the first MS-AE was associated with any of the SNPs, we analyzed cases only, using time to first MS-AE as the outcome variable in a Cox regression model. After adjusting for the eight eigenvectors, the smallest P value over all the SNPs was .04. In summary, none of these exploratory analyses provided small enough P values to be robustly statistically significant after adjusting for the post hoc multiple testing.
Fig A1.
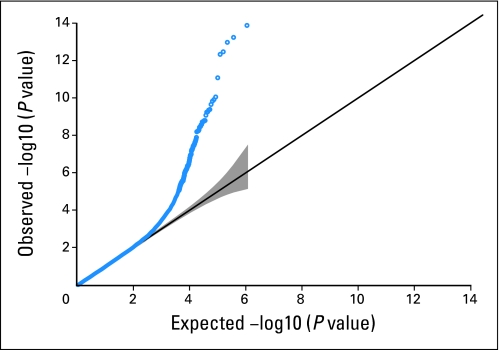
Quantile-quantile plot of Hardy-Weinberg equilibrium (HWE) exact P values among controls. The gray area represents point-wise 95% CIs for the null distribution of HWE.
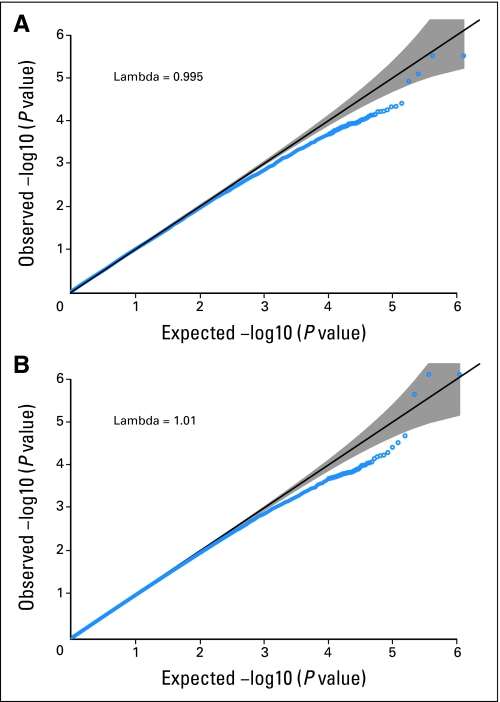
Fig A2.
Quantile-quantile plots of P values from conditional logistic regression analyses, unadjusted (A) and adjusted (B) for eight eigenvectors. The gray area represents point-wise CIs for the null distribution of no association.
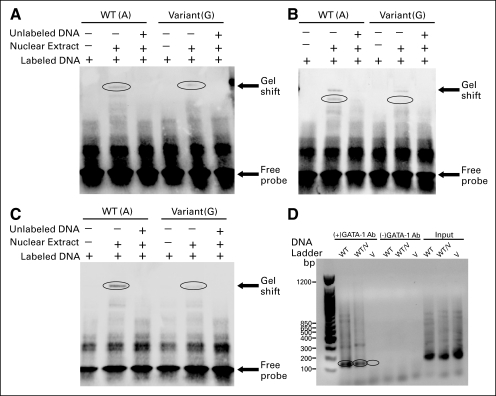
Fig A3.
Electrophoretic mobility shift assays using nuclear extract from lymphoblastoid cell lines for (A) rs7158782, (B) rs7159713, (C) rs11849538, and (D) chromatin immunoprecipitation assays for rs7158782 using GATA-1 antibody (Ab). The circled areas highlight the band associated with GATA-1 binding (wild-type [WT] and WT/variant [V]) as well as the lack of that band for the V sequence.
Footnotes
See accompanying editorial on page 4665
Supported in part by National Institutes of Health (NIH) Grants No. U01GM61388, U01GM63173, P50CA116201, and U10CA77202; by Grant No. CCS 015469 from the Canadian Cancer Society; and by the Biobank Japan Project funded by the Japanese Ministry of Education, Culture, Sports, Science and Technology; the Breast Cancer Research Foundation, New York, NY; and the NIH Pharmacogenomics Research Network–RIKEN Center for Genomic Medicine Global Alliance, Pfizer supported the clinical trial from which the patients in this study were obtained.
Presented in part at the 32nd Annual San Antonio Breast Cancer Symposium, December 9-13, 2009, San Antonio, TX.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: James N. Ingle, Pfizer (U); Vered Stearns, Otsuka America Pharmaceuticals (C); Kathleen I. Pritchard, Novartis (C), Pfizer (C), Roche (C) Stock Ownership: None Honoraria: Matthew J. Ellis, AstraZeneca, Pfizer; Vered Stearns, AstraZeneca; Kathleen I. Pritchard, Novartis, Pfizer, Roche Research Funding: Matthew J. Ellis, AstraZeneca; Vered Stearns, Novartis, Pfizer Expert Testimony: Kathleen I. Pritchard, AstraZeneca (C), Novartis (C) Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: James N. Ingle, Daniel J. Schaid, Paul E. Goss, Mohan Liu, Taisei Mushiroda, Judy-Anne W. Chapman, Michiaki Kubo, Joseph Pater, Liewei Wang, Vered Stearns, David A. Flockhart, Yusuke Nakamura, Richard M. Weinshilboum
Financial support: James N. Ingle, Yusuke Nakamura, Richard M. Weinshilboum
Administrative support: James N. Ingle, Taisei Mushiroda, Joseph Pater, Richard M. Weinshilboum
Provision of study materials or patients: James N. Ingle, Paul E. Goss, Mohan Liu, Judy-Anne W. Chapman, Michiaki Kubo, Lois Shepherd, Daniel C. Rohrer, Matthew P. Goetz, Yusuke Nakamura, Richard M. Weinshilboum
Collection and assembly of data: James N. Ingle, Paul E. Goss, Taisei Mushiroda, Judy-Anne W. Chapman, Michiaki Kubo, Gregory D. Jenkins, Anthony Batzler, Lois Shepherd, Joseph Pater, Matthew J. Ellis, Yusuke Nakamura, Richard M. Weinshilboum
Data analysis and interpretation: James N. Ingle, Daniel J. Schaid, Paul E. Goss, Mohan Liu, Taisei Mushiroda, Judy-Anne W. Chapman, Michiaki Kubo, Gregory D. Jenkins, Anthony Batzler, Liewei Wang, Matthew P. Goetz, Kathleen I. Pritchard, David A. Flockhart, Yusuke Nakamura, Richard M. Weinshilboum
Manuscript writing: James N. Ingle, Daniel J. Schaid, Paul E. Goss, Mohan Liu, Taisei Mushiroda, Judy-Anne W. Chapman, Michiaki Kubo, Gregory D. Jenkins, Anthony Batzler, Lois Shepherd, Joseph Pater, Liewei Wang, Matthew J. Ellis, Vered Stearns, Daniel C. Rohrer, Matthew P. Goetz, Kathleen I. Pritchard, David A. Flockhart, Yusuke Nakamura, Richard M. Weinshilboum
Final approval of manuscript: James N. Ingle, Daniel J. Schaid, Paul E. Goss, Mohan Liu, Taisei Mushiroda, Judy-Anne W. Chapman, Michiaki Kubo, Gregory D. Jenkins, Anthony Batzler, Lois Shepherd, Joseph Pater, Liewei Wang, Matthew J. Ellis, Vered Stearns, Daniel C. Rohrer, Matthew P. Goetz, Kathleen I. Pritchard, David A. Flockhart, Yusuke Nakamura, Richard M. Weinshilboum
REFERENCES
- 1.Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists' Group. Forbes JF, Cuzick J, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 2.Coates AS, Keshaviah A, Thürlimann B, et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: Update of study BIG 1-98. J Clin Oncol. 2007;25:486–492. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
- 3.Coombes RC, Kilburn LS, Snowdon CF, et al. Survival and safety of exemestane versus tamoxifen after 2-3 years' tamoxifen treatment (Intergroup Exemestane Study): A randomised controlled trial. Lancet. 2007;369:559–570. doi: 10.1016/S0140-6736(07)60200-1. [DOI] [PubMed] [Google Scholar]
- 4.Jakesz R, Jonat W, Gnant M, et al. Switching of postmenopausal women with endocrine-responsive early breast cancer to anastrozole after 2 years' adjuvant tamoxifen: Combined results of ABCSG trial 8 and ARNO 95 trial. Lancet. 2005;366:455–462. doi: 10.1016/S0140-6736(05)67059-6. [DOI] [PubMed] [Google Scholar]
- 5.Boccardo F, Rubagotti A, Guglielmini P, et al. Switching to anastrozole versus continued tamoxifen treatment of early breast cancer: Updated results of the Italian tamoxifen anastrozole (ITA) trial. Ann Oncol. 2006;17(suppl 7):vii10–vii14. doi: 10.1093/annonc/mdl941. [DOI] [PubMed] [Google Scholar]
- 6.Kaufmann M, Jonat W, Hilfrich J, et al. Improved overall survival in postmenopausal women with early breast cancer after anastrozole initiated after treatment with tamoxifen compared with continued tamoxifen: The ARNO 95 study. J Clin Oncol. 2007;25:2664–2670. doi: 10.1200/JCO.2006.08.8054. [DOI] [PubMed] [Google Scholar]
- 7.Goss PE, Ingle JN, Martino S, et al. Randomized trial of letrozole following tamoxifen as extended adjuvant therapy in receptor-positive breast cancer: Updated findings from NCIC CTG MA.17. J Natl Cancer Inst. 2005;97:1262–1271. doi: 10.1093/jnci/dji250. [DOI] [PubMed] [Google Scholar]
- 8.Mamounas EP, Jeong JH, Wickerham DL, et al. Benefit from exemestane as extended adjuvant therapy after 5 years of adjuvant tamoxifen: Intention-to-treat analysis of the National Surgical Adjuvant Breast and Bowel Project B-33 trial. J Clin Oncol. 2008;26:1965–1971. doi: 10.1200/JCO.2007.14.0228. [DOI] [PubMed] [Google Scholar]
- 9.Winer EP, Hudis C, Burstein HJ, et al. American Society of Clinical Oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: Status report 2004. J Clin Oncol. 2005;23:619–629. doi: 10.1200/JCO.2005.09.121. [DOI] [PubMed] [Google Scholar]
- 10.Partridge AH, LaFountain A, Mayer E, et al. Adherence to initial adjuvant anastrozole therapy among women with early-stage breast cancer. J Clin Oncol. 2008;26:556–562. doi: 10.1200/JCO.2007.11.5451. [DOI] [PubMed] [Google Scholar]
- 11.Crew KD, Greenlee H, Capodice J, et al. Prevalence of joint symptoms in postmenopausal women taking aromatase inhibitors for early-stage breast cancer. J Clin Oncol. 2007;25:3877–3883. doi: 10.1200/JCO.2007.10.7573. [DOI] [PubMed] [Google Scholar]
- 12.Henry NL, Giles JT, Ang D, et al. Prospective characterization of musculoskeletal symptoms in early stage breast cancer patients treated with aromatase inhibitors. Breast Cancer Res Treat. 2008;111:365–372. doi: 10.1007/s10549-007-9774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 14.Solomon SD, McMurray JJ, Pfeffer MA, et al. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N Engl J Med. 2005;352:1071–1080. doi: 10.1056/NEJMoa050405. [DOI] [PubMed] [Google Scholar]
- 15.Rosenbaum PR. Optimal matching for observational studies. J Am Stat Assoc. 1989;84:1024–1032. [Google Scholar]
- 16.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Willer CJ, Sanna S, et al. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: Population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian C, Plenge R, Ransom M, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genetics. 2008;4:29–39. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu K, Wang Z, Li Q, et al. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devlin B, Roeder K. 1999. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 24.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 25.Kopec JA, Esdaile JM. Bias in case-control studies: A review. J Epidemiol Community Health. 1990;44:179–186. doi: 10.1136/jech.44.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manolio TA, Collins FS. The HapMap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443–456. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekarsky Y, Hallas C, Croce CM. The role of TCL1 in human T-cell leukemia. Oncogene. 2001;20:5638–5643. doi: 10.1038/sj.onc.1204596. [DOI] [PubMed] [Google Scholar]
- 28.Noguchi M, Ropars V, Roumestand C, et al. Proto-oncogene TCL1: More than just a coactivator for Akt. FASEB J. 2007;21:2273–2284. doi: 10.1096/fj.06-7684com. [DOI] [PubMed] [Google Scholar]
- 29.Cecil RL, Archer BH. Arthritis of the menopause. JAMA. 1925;84:75–79. [Google Scholar]
- 30.Chlebowski RT, Johnson KC, Kooperberg C, et al. The Women's Health Initiative randomized trial of calcium plus vitamin D: Effects on breast cancer and arthralgias. J Clin Oncol. 2006;24(suppl; abstr LBA):2s. [Google Scholar]