Abstract
Radixin is a member of the ezrin/radixin/moesin (ERM) family of proteins, which play a role in the formation of the membrane-associated cytoskeleton by linking actin filaments and adhesion proteins. This cross-linking activity is regulated by phosphoinositides such as phosphatidylinositol 4,5-bisphosphate (PIP2) in the downstream of the small G protein Rho. The X-ray crystal structures of the radixin FERM domain, which is responsible for membrane binding, and its complex with inositol-(1,4,5)-trisphosphate (IP3) have been determined. The domain consists of three subdomains featuring a ubiquitin-like fold, a four-helix bundle and a phosphotyrosine-binding-like domain, respectively. These subdomains are organized by intimate interdomain interactions to form characteristic grooves and clefts. One such groove is negatively charged and so is thought to interact with basic juxta-membrane regions of adhesion proteins. IP3 binds a basic cleft that is distinct from those of pleckstrin homology domains and is located on a positively charged flat molecular surface, suggesting an electrostatic mechanism of plasma membrane targeting. Based on the structural changes associated with IP3 binding, a possible unmasking mechanism of ERM proteins by PIP2 is proposed.
Keywords: CD44/cell adhesion/cytoskeleton/ICAM/PIP2
Introduction
Radixin is a member of the ezrin/radixin/moesin (ERM) family of proteins, which function as cross-linkers between plasma membranes and actin filaments (Arpin et al., 1994; Bretscher, 1999; Mangeat et al., 1999; Tsukita and Yonemura, 1999). These proteins have been found in eukaryotic cells in organisms ranging from Caenorhabditis elegans to humans. Immunofluorescence studies of cultured cells have revealed the localization of ERM proteins in cell-surface structures such as microvilli, ruffling membranes and cell-adhesion sites where actin filaments associate with plasma membranes (Sato et al., 1991, 1992; Amieva and Furthmayr, 1995; Franck et al., 1993; Serrador et al., 1997).
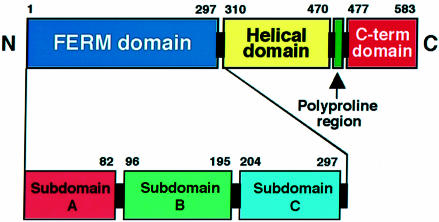
Radixin, which was originally isolated from rat liver as a component of cell–cell adherens junctions (Tsukita et al., 1989), is a 583 amino acid protein consisting of three domains, an N-terminal FERM (4.1 and ERM) domain (residues 1–297), a central helical domain (residues 310–470) and a C-terminal domain (477–583) (Figure 1). The radixin FERM domain displays high (>84%) sequence homology to the corresponding domains of ezrin and moesin. In all three ERM proteins, the FERM domain has been shown to mediate binding to the cytosolic parts of integral membrane proteins that play key roles in cell adhesion and cell–cell communication. To date, three membrane proteins, CD44, CD43 and ICAMs 1, 2 and 3, have been characterized as possible membrane partners of ERM proteins. CD44 is a cell-surface hyaluronate receptor protein that precisely colocalizes with ERM proteins in cultured fibroblasts (Tsukita et al., 1994). In some cell types, a cell-surface glycoprotein of the sialomutin family, CD43 (Yonemura et al., 1993), and a membrane protein of the immunoglobulin superfamily, ICAM-2 (Helander et al., 1996), are also colocalized with ERM proteins. Recently, possible ERM-binding regions of these adhesion proteins have been identified in their cytoplasmic domains or tails (Yonemura et al., 1998). The C-terminal halves of ERM proteins bind F-actin through their conserved actin-binding sites, which consist of 34 residues (550–583 for radixin) (Turunen et al., 1994; Pestonjamasp et al., 1995). This domain organization resembles that of band 4.1 protein, which links membrane proteins such as glyco phorin C/D to actin cytoskeleton (Gould et al., 1989; Algrain et al., 1993). In particular, their FERM domains exhibit significant homology (∼30%) (Chishti et al., 1998).
Fig. 1. Diagram of radixin domains. The N-terminal FERM domain (blue), helical domain (yellow), polyproline region (green) and the C-terminal autoinhibitory domain (red) are indicated. Three subdomains in the FERM domain found by this crystallographic analysis are also indicated.
Several other proteins exhibit sequence similarity with the FERM domain. These proteins contain merlin/schwannomin, the neurofibromatosis type 2 (NF2) tumor-suppressor gene product (Rouleau et al., 1993; Trofatter et al., 1993), a subset of protein tyrosine phosphatases such as PTP-D1 (Moller et al., 1994) and membrane-associated unconventional myosins such as myosins VIIa and X. Moreover, recent sequence analyses suggest that FERM-like domains may exist in non-receptor tyrosine kinases (JAK1, JAK2, JAK3 and TRK2) of the Janus kinase family (Girault et al., 1999) as well as focal adhesion kinase (FAK) and the related PYK2/CAKβ/RAFTK (Schultz et al., 1998). Therefore, the FERM domains have been thought to form a newly discovered protein module that mediates protein–protein or protein–membrane (or both) interactions.
Full-length ERM proteins show a low level of binding activity to both membrane and actin. These inactive states are believed to be expressed by a masking mechanism in which the FERM domain binds the C-terminal half to suppress the actin filament and membrane-binding activities (Andreoli et al., 1994; Gary and Bretcher, 1995; Magendantz et al., 1995; Hirao et al., 1996). Recent biochemical studies have shown that phospholipids such as phosphatidylinositol 4,5-bisphosphate (PIP2) bind the FERM domains and stimulate the binding of ERM proteins to CD44 in the downstream Rho (Niggli et al., 1995; Hirao et al., 1996). In fact, the Rho-induced phosphorylation of radixin Thr564, which is located in the C-terminal region, maintains radixin in the active state by suppressing the intermolecular masking (Matsui et al., 1998, 1999). Interestingly, the unmasked FERM domains bind Rho-specific GDP-dissociation inhibitor (RhoGDI) and accelerate its release of Rho to activate Rho-dependent processes (Hirao et al., 1996; Takahashi et al., 1997). ERM proteins also interact with the Rho-specific guanine nucleotide exchange factor Dbl, although this interaction does not affect the exchange reaction (Takahashi et al., 1998). These findings suggest that the Rho signaling pathway regulates ERM proteins both upstream and downstream of Rho. Moreover, several experiments have indicated that ERM proteins respond to extracellular signals. For example, epidermal growth factor (EGF) stimulation of parietal cells by gastric acid secretion, or activation of platelets by thrombin rapidly recruits ERM proteins to the cortical actin layer with concomitant phosphorylation (Bretscher, 1989; Hanzel et al., 1989; Krieg and Hunter, 1992; Takeda et al., 1995; Crepaldi et al., 1997). Thus, it is conceivable that extracellular and intracellular signaling pathways dynamically regulate ERM proteins. Another ERM-binding partner is the ERM-binding 50 kDa phosphoprotein (EBP50) that binds the FERM domain (Reczek et al., 1997). EBP50 is identical to the Na+/H+ exchange regulator factor (NHE-RF), a cofactor necessary for protein kinase A regulation of the renal Na+/H+ exchanger, a membrane ion channel protein (Weinman et al., 1993). NHE-RF and its isoform, E3KARP (Yun et al., 1997), bind the Na+/H+ exchanger through their PDZ domains.
Here, we report the crystal structures of the radixin FERM domain and its complex with inositol-(1,4,5)-trisphosphate (IP3), which is the headgroup of PIP2. The structures reveal that the FERM domain consists of three subdomains that form characteristic grooves and clefts. Features of the grooves and clefts suggest possible mechanisms for the interaction of the FERM domain with its binding partners. The structural changes induced by IP3 binding suggest a possible unmasking mechanism of ERM proteins by PIP2.
Results and discussion
Structure determination and overall structure
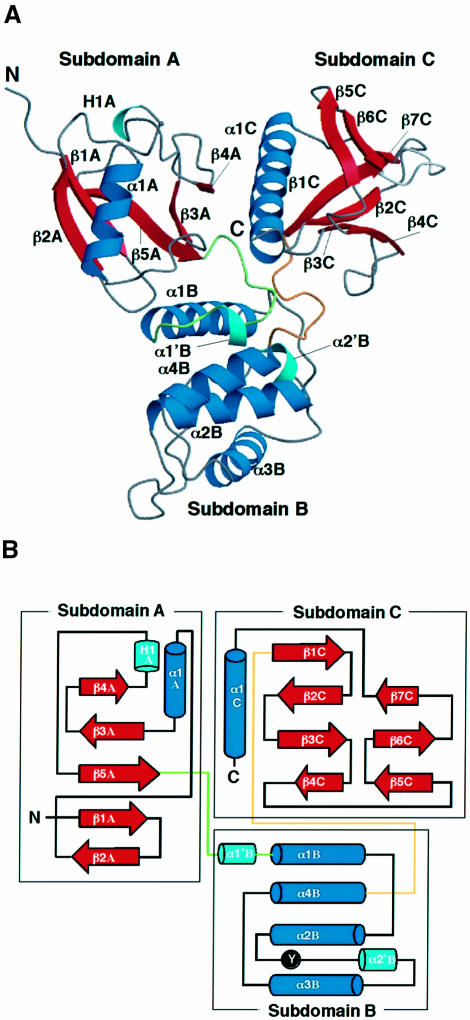
The FERM domain of mouse radixin containing residues 1–310 is sufficient for binding to phospholipids and cell-adhesion proteins as described above. Crystals of the radixin FERM domain (1–310) containing one molecule per asymmetrical unit were obtained. The structure was determined by the multiple isomorphous replacement method and refined to 2.8 Å resolution. The current structure includes 297 residues of the FERM domain. The FERM domain contains three subdomains, A, B and C, which interact with each other to fold into a compact globular shape (Figure 2A). The radixin FERM domain has a somewhat trigonal shape with overall dimensions of ∼70 Å by 70 Å, with a thickness of 40 Å. For convenience, the secondary structure elements in each subdomain are numbered with a subdomain identifier, A, B or C. The complex crystals between the radixin FERM domain and IP3 were obtained both by soaking and by co-crystallization. All these crystals are isomorphous to the native crystals of the free radixin FERM domain. The structures of the complexes were determined by molecular replacement using the unliganded radixin FERM domain. The complex structure with IP3 was refined at 2.9 Å resolution.
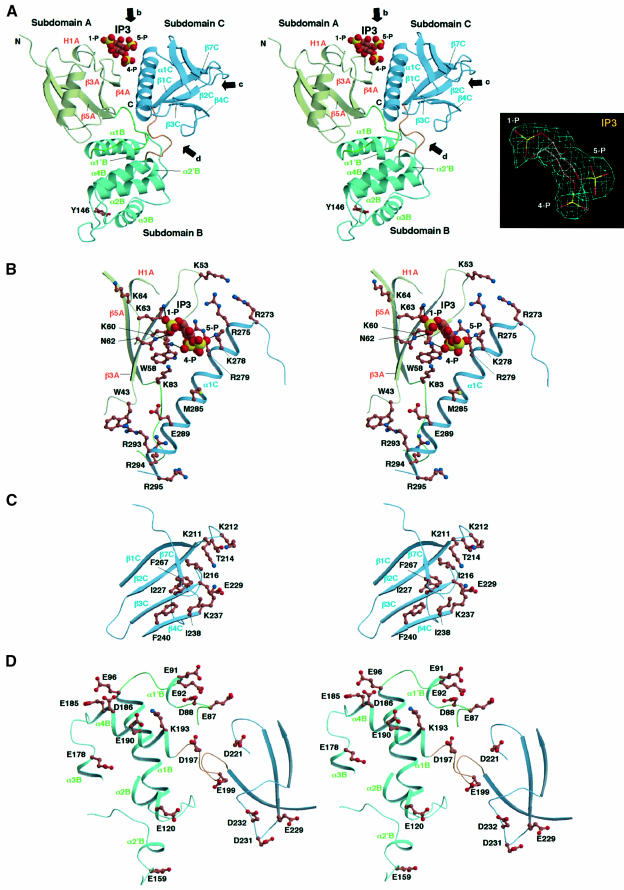
Fig. 2. Overall structure of the radixin FERM domain. (A) View of the radixin FERM domain by ribbon representation with secondary structure elements: α-helices (blue) and β-strands (red). Two short α-helices and one 310-helix are colored in light blue. The linkers A–B and B–C are colored in green and brown, respectively. (B) Topology diagram of the radixin FERM domain. Helices are denoted by cylinders and strands by arrows. Tyr146, which is phosphorylated by the protein tyrosine kinase v-Src, is marked.
Subdomain structures
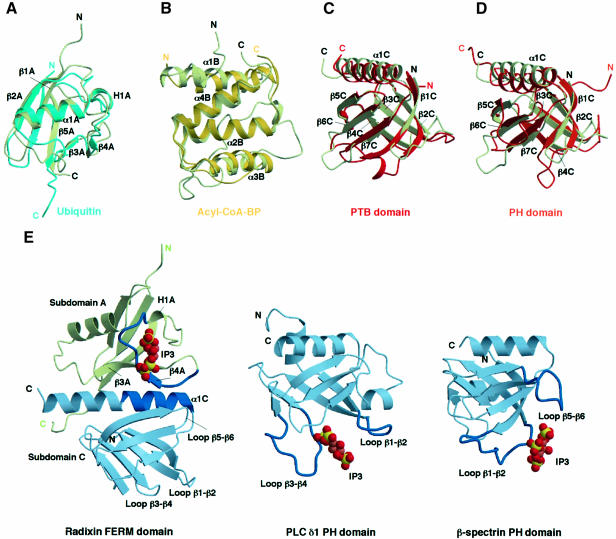
Subdomain A, consisting of the 82 N-terminal residues, folds into an α+β structure with one long α-helix (helix α1A) and a five-stranded mixed β-sheet (strands β1A–β5A) (Figures 2 and 3). A structural comparison with the DALI database (Holm and Sander, 1993) shows that subdomain A has a typical ubiquitin fold, which is commonly found in functionally unrelated proteins. This fold superfamily contains the Ras-binding domains of Raf1 (Nassar et al., 1995) and RalGDS (Huang et al., 1998), whereas no sequence similarity is detected among these domains. Superimposition of ubiquitin (Ramage et al., 1994) and subdomain A gives an average root mean square (r.m.s.) deviation of 1.9 Å over the superimposed Cα atoms (Figure 4A). Compared with the ubiquitin structure, subdomain A lacks a short helix located in the ubiquitin loop α1–β3.
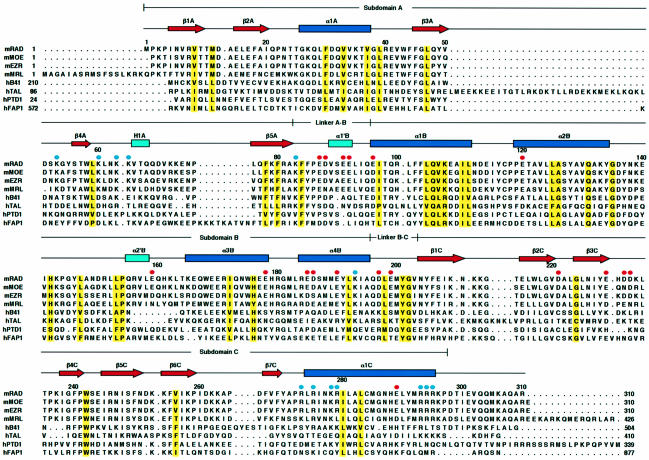
Fig. 3. Secondary structure elements and sequence alignment of the FERM domains. The FERM domains of mouse radixin (mRAD) and the related proteins are aligned with the secondary structure elements of the radixin FERM domain at the top: α-helices (blue rectangles) and β-strands (red arrows). Conserved residues are highlighted in yellow. The aligned FERM domains are mouse moesin (mMOE), ezrin (mEZR), merlin (mMRL) and human band 4.1 (hB41), talin (hTAL), protein tyrosine phosphatase D1 (hPTD1) and FAP-1 (hFAP1). The mouse FERM domains exhibit 100% identities with the human FERM domains and >99.7% identities with other mammalian homologs. Acidic residues of the acidic groove between subdomains B and C are indicated by red circles. Basic residues of the basic cleft between subdomains A and C are indicated by blue circles.
Fig. 4. Subdomain structures of the radixin FERM domain. The color codes used are light green for radixin subdomains, light blue for ubiquitin, yellow for acyl-coenzyme A binding protein, red for the PTB domain and orange for the PH domain. (A) Superimposition of radixin subdomain A on ubiquitin (PDB code 1UBI, blue). (B) Superimposition of radixin subdomain B on E.coli acyl-coenzyme A binding protein (yellow, 1ACA). (C) Superimposition of radixin subdomain C on the IRS-1 PTB domain (1QQG, red). (D) Superimposition of radixin subdomain C on the PH domain (1PLS, orange). (E) Comparison of the IP3-binding sites found in the radixin FERM domain (left), the phospholipase C δ1 PH domain (middle) and the β-spectrin PH domain (right). Two loops forming the binding site of each PH domain are colored in blue. The binding site of the radixin FERM domain is located at the basic cleft between subdomains A (a ubiquitin-like fold in light green) and C (a PTB-like fold in light blue) (see text). The N-terminal half of helix α1C of subdomain C and the protruding loop between strands β3A and β5A of subdomain A form the IP3-binding site of the radixin FERM domain and are colored in blue.
Subdomain B (residues 96–195) is an all-α structure with four longer α-helices (α1B–α4B) and one short helix (α2′B) and is tethered to subdomain A by a linker (linker A–B) of 13 residues that contains a short helix (α1′B). Helices α2B and α4B interact intimately to form an antiparallel bundle with an inter-helix angle of 17° and also play a role as a subdomain core sandwiched by two other helices, α1B and α3B. This architecture, which is composed of four helices, is classified as an acyl-coenzyme A binding protein-like fold in the SCOP protein structure database (Murzin et al., 1995). The Escherichia coli acyl-coenzyme A binding protein (Kragelund et al., 1993) can be superimposed on subdomain B with an r.m.s. deviation of 2.0 Å, but the total identity is poor (14% identity) (Figure 4B).
The protein tyrosine kinase v-Src is known to phosphorylate ERM proteins (Takeda et al., 1995). This phosphorylation is also induced by EGF (Krieg and Hunter, 1992) or hepatocyte growth factor stimulation (Crepaldi et al., 1997). One of the phosphorylation sites, Tyr146, is conserved in the FERM domain. The amino acids neighboring this tyrosine residue are also conserved in ERM proteins and in two protein tyrosine phosphatases having FERM-like domains, PTP-H1 and PTP-MEG, but not in merlin, band 4.1 protein or talin (Figure 3). In our structure, Tyr146 is located in the C-terminal flanking loop of helix α2B, with its side-chain phenol ring intruding inside the loop (Figure 2B; also see Figure 6A). Therefore, a local conformational change of the loop is necessary for tyrosine phosphorylation.
Fig. 6. Close-up stereo views of the radixin FERM domain. (A) Main chains of subdomains A (light green), B (blue green) and C (blue) are represented by ribbons. Arrows b, c and d indicate views of the basic cleft, the small basic pocket and the acidic groove shown in (B), (C) and (D), respectively. The IP3 molecule is shown in a space-filled model. In subdomain B, the side chain of Tyr146, which is phosphorylated by v-Src kinase, is shown in a stick model. The inset shows the IP3 model in a 2Fo – Fc electron density map contoured at the 1σ level. (B) The basic cleft between subdomains A and C. The protein side chains are shown in ball-and-stick models with color codes of brown for carbon, blue for nitrogen, red for oxygen and yellow for phosphorous atoms. The IP3 molecule is shown in a space-filled model. (C) The hydrophobic hole and the small basic pocket of subdomain C. (D) The acidic groove between subdomains B and C.
Subdomain C (residues 204–297) is tethered to subdomain B by a linker (linker B–C) consisting of eight conserved residues, most of which are buried inside the interdomain region. This subdomain exhibits a typical phosphotyrosine-binding (PTB) or pleckstrin homology (PH) domain with a standard seven-stranded β-sandwich core (strands β1C–β7C) with one α-helix (α1C). A DALI search indicates high structural alignment scores for the PTB domains from several sources, with r.m.s. deviations ranging from 1.7 to 2.1 Å, as well as for the PH domains with r.m.s. deviations of ∼2.5 Å. The highest scores have been obtained for PTB domains from a human insulin receptor substrate 1 (IRS-1) (Dhe-Paganon et al., 1999) (Figure 4C) and a neuron-specific X11 protein (Zhang et al., 1997), as well as for the PH domain of human pleckstrin (Yoon et al., 1994) (Figure 4D). No significant sequence homology has been detected with these PTB and PH domains.
The overall structures of radixin subdomain C and the IRS-1 PTB domain are quite similar, with closely corresponding helices and strands (Figure 4C). In contrast, subdomain C and the human PH domain display several significant differences in their structures (Figure 4D). In particular, the position of the radixin helix α1C is shifted away from the β-sandwich fold. It has been well established by several structural investigations that the PTB domains bind peptides containing a phosphotyrosine in the C-terminal region. In radixin subdomain C there is a groove between strand β5C and helix α1C that corresponds to the phosphotyrosine-containing peptide-binding site found in structures in which the IRS-1 PTB domain bound to such peptides from the juxta-membrane region of the insulin receptor (Eck et al., 1996) and from interleukin-4 (Zhou et al., 1996). Two basic residues interacting with phosphotyrosine in the IRS-1 PTB–peptide complexes (Arg246 and Lys262) are also conserved in subdomain C. The site corresponding to the phosphotyrosine-binding pocket of IRS-1 PTB, however, is occupied by a bulky hydrophobic residue (Ile260). At present, no experimental evidence has been obtained to demonstrate whether the FERM domain binds to a phosphotyrosine-containing peptide. This possibility will be investigated in further experiments.
Subdomain interfaces
Subdomain A forms intimate interdomain interactions with two other subdomains. The major interactions at both interdomain interfaces occur primarily at the side-chain level and are a mixture of hydrophobic and polar contacts, including hydrogen bonds and salt bridges. The interdomain interfaces between subdomains A and B include helix α1B and loop β1A–β2A, the C-terminus of helix α1A and its flanking loop. In addition, the linker A–B, which interacts with strand β3A and its N-terminal flanking loop region, folds on to helices α1B and α4B to make several contacts.
The interfaces between subdomains A and C are created primarily by contacts between strand β4A and helix α1C. As in the interfaces between subdomains A and B, the linker A–B contributes to form the B–C interfaces by interacting with strand β1C and helix α1C. Notably, this loop has phenylalanine (Phe85) at the center of the hydrophobic core, surrounded by three subdomains. In contrast to the wealth of interactions between sub domains A and B and between A and C, the direct interactions between subdomains B and C are few. Again, the linker A–B contacts the linker B–C to stabilize its conformation. In conclusion, subdomain A and the linker A–B play a key role in building up the overall structure of the FERM domain studied here.
Molecular surface properties
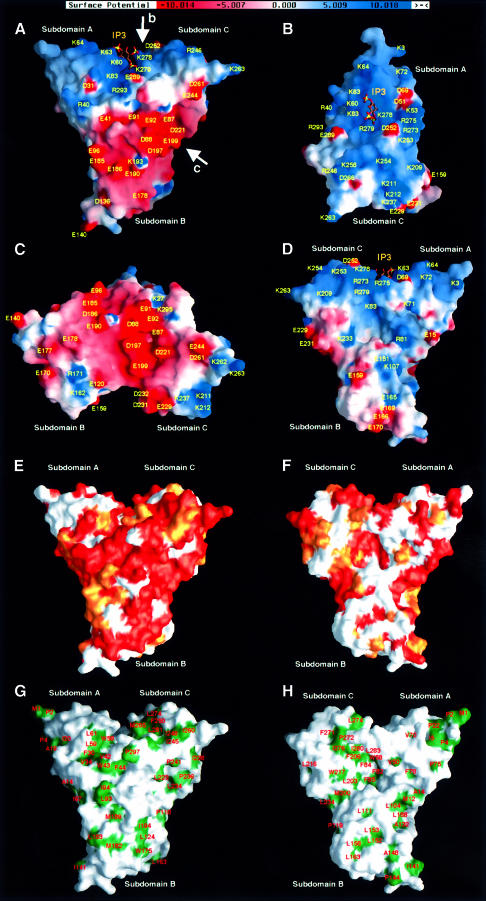
The radixin FERM domain possesses pronounced grooves formed by each pairing of the three subdomains (Figure 5A). The first is a deep cleft created by the interdomain region between subdomains A and C and is positively charged by clustered basic residues (Figure 5B). This deep basic cleft has a width of ∼7 Å. Other basic residues are scattered around this basic cleft to form a positively charged, rather flat molecular surface. The second groove is negatively charged and is found between subdomains B and C (Figure 5C). This acidic groove has hydrophobic residues at its base and is surrounded by remote acidic residues; it is thus located in the midst of a negatively charged molecular surface. These two positively and negatively charged surfaces produce marked polarized electrostatic potentials around the FERM domain. Interestingly, surface residues located at the basic cleft, the acidic groove and their vicinities are largely conserved in ERM proteins (Figure 5E), implying that these residues may have possible molecular functions such as interactions with binding partners as described below.
Fig. 5. Molecular surface properties of the radixin FERM domain. (A) Surface electrostatic potentials of the radixin FERM domain viewed from the same direction as in Figure 2A. Positive (blue) and negative (red) potentials are mapped on the van der Waals surfaces. The IP3 molecule found in the complex crystal is shown in a stick model. (B) Surface electrostatic potentials viewed along the arrow b in (A) to show the basic cleft between subdomains A and C. The IP3 molecule found in the complex crystal is shown in a stick model. (C) Surface electrostatic potentials viewed along arrow c in (A) to show the acidic groove between subdomains B and C. (D) A backside view of surface electrostatic potentials seen in (A). The IP3 molecule found in the complex crystal is shown in a stick model. (E) Conserved residues of the radixin FERM domain mapped on the molecular surfaces. A front view of the radixin FERM domain depicted as a colored molecular surface using a gradient; orange indicates conserved identical residues and white non-conserved residues, while lighter shades of orange indicate semi-invariant residues. A view from the same direction as in (A) and Figure 2A. (F) Back view of conserved residues of the radixin FERM domain. (G) Front view of hydrophobic residues of the radixin FERM domain mapped on the molecular surfaces. (H) Back view of hydrophobic residues of the radixin FERM domain.
It is noteworthy that the front molecular surface shown in Figure 5A contains clusters of negatively and positively charged residues from the acidic groove and the basic cleft, respectively. In contrast, no such clusters are seen on the surface at the back of the molecule (Figure 5D). A similar contrast is found in the distribution of conserved surface residues, which are distributed mainly on the front of the molecule rather than on the back (Figure 5E and F). One interesting feature of the surface on the back of the molecule is that a deep hydrophobic pocket exists in the center of the three subdomains (Figure 5H). Subdomains A and B form a large open concavity that has no characteristic charge distribution (Figure 5D).
Binding site for IP3
In all the complex crystals obtained under the various conditions, including both soaking and cocrystallization, IP3 is located at the center of the basic cleft between subdomains A and C (Figure 5A and B). This positively charged cleft is formed by the C-terminal helix (α1C) of subdomain C and the protruding loops flanking strands β3A and β4A of subdomain A [Figure 4E (left) and Figure 6A and B]. The positive charges are due to seven lysines and four arginines in these regions. One lysine protrudes from the C-terminal end of strand β5A.
The IP3 molecule binds a different site from those found in PH domains (Figure 4E), although subdomain C is folded similarly to the PH domain, as described. In general, the IP3- and PIP2-binding sites of PH domains are known to be primarily formed by two loops, β1–β2 and β3–β4, as has been reported for pleckstrin (Harlan et al., 1994), the phospholipase C δ1 PH domain (Ferguson et al., 1995) and the dynamin PH domain (Salim et al., 1996) (Figure 4E, middle). Another binding site has been observed in the β-spectrin PH domain, whose site is formed by two loops, β1–β2 and β5–β6 (Hyvonen et al., 1995) (Figure 4E, right). Compared with these PH domains, the short radixin loops, β1C–β2C and β5C–β6C, are projected towards the different molecular surfaces. Moreover, the radixin loop β3C–β4C is acidic. In addition to PH domains, the IP3-binding site of the radixin FERM domain is also distinct from that of the Vps27p FYVE domain for binding to phosphatidylinositol 3-monophosphate, PI3P (Misra et al., 1999).
In the complex studied here, three lysines (Lys60 and Lys63 from subdomain A and Lys278 from subdomain C) and one asparagine (Asn62 from subdomain A) contact the three phosphate groups of IP3. These contacts suggest stereochemically cooperative interactions that enhance specificity. The hydrophobic base of the cleft contains one tryptophan from strand β4A but does not interact with IP3. In the orientation of the bound IP3 molecule, the 1-phosphate group is exposed to solvent, which enables PIP2 to reach the binding site from the membrane surface (Figure 7A). The proposed orientation of the radixin FERM domain associated with the membrane enables the positively charged molecular surface between subdomains A and C to interact with the negatively charged membrane surface. This relatively flat molecular surface seems to maximize the interactions with the membrane surface.
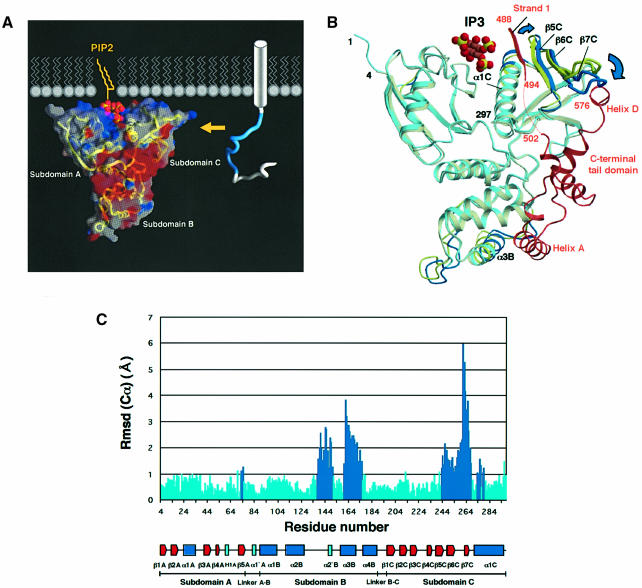
Fig. 7. Schematic representation of the radixin FERM domain bound to PIP2 in a membrane and its possible unmasking mechanism. (A) The radixin FERM domain is shown in terms of surface electrostatic potentials, with a ribbon representation of the main-chain tracing. Positive (blue) and negative (red) potentials are mapped on the van der Waals surfaces. PIP2 is shown bound to the FERM domain as seen in the current crystal structure. Diacylglycerol (yellow) has been attached to the IP3 head and is shown in a highly schematized lipid layer. A cytoplasmic tail of an adhesion protein such as ICAM-2 is shown in a tube representation with its basic juxta-membrane region in blue. Part of the transmembrane α-helix of the adhesion protein is shown as a cylinder. A yellow arrow indicates a possible binding of the basic juxta-membrane region to the acidic groove of the FERM domain. (B) Superimposition of the radixin FERM domain (light blue) on the moesin FERM domain (pale yellow) complexed with the C-terminal tail domain (brown). Segments displaying large displacements (>1 Å) are highlighted in blue (radixin) and yellow (moesin). (C) A histogram showing the distances of the corresponding Cα atoms between the IP3-bound radixin FERM domain and the moesin FERM domain complexed with the C-terminal tail domain. Large displacements are highlighted in blue.
In the IP3 molecule bound to the radixin FERM domain, the 4-phosphate group intrudes into the cleft, whereas the 5-phosphate group resides at the molecular surface. In the IP3–PH domain complexes (Ferguson et al., 1995; Hyvonen et al., 1995; Salim et al., 1996), both the 4- and 5-phosphate groups are anchored at the binding pocket. Moreover, in the phospholipase Cδ1 PH domain complexed with IP3, the 5-phosphate group is completely buried (Ferguson et al., 1995). These differences may have possible biological significance. In the case of radixin, the buried 4-phosphate group seems to be consistent with a relatively strong binding of the radixin FERM domain to phosphatidylinositol 4-monophosphate (PI4P; Hirao et al., 1996).
Subdomain C has a small basic pocket containing two lysines from loop β1C–β2C and one lysine from β4C (Figure 6C). No electron density of IP3, however, was observed in this pocket in the crystals of the complex studied here. This small pocket, which is distinct from the phospholipid binding sites in the PH domains reported thus far, has its base ‘tiled’ with hydrophobic residues from β-strands (β3C, β4C, β6C and β7C) and is adjacent to a hydrophobic hole.
Superimposition of the IP3-binding form of the radixin FERM domain with the free form gives a small r.m.s. deviation of 0.28 Å for all Cα atoms. Therefore, the overall structures of the two states are essentially the same. However, relatively small (0.6–1.2 Å) but interesting structural changes have been found in the N-terminal region of helix α1C and strand β5C of subdomain C. The bound IP3 molecule pushes the N-terminal region of helix α1C so as to widen the basic cleft for its accommodation. This movement induces a shift of the C-terminal end of strand β5C and its N-terminal flanking loop. These structural changes are important for the unmasking of the ERM domain, as described below.
Possible unmasking mechanism of ERM proteins by PIP2
PIP2 binds the full-length ERM proteins in the masked state by the C-terminal tail domain and markedly stimulates the binding of ERM proteins to adhesion proteins. For example, in the presence of PIP2, moesin binds CD44 with a dissociation constant value of 9.3 nM at a physiological ionic strength, under which conditions, in the absence of PIP2, moesin does not bind CD44. These results indicate that phospholipid binding induces an unmasking state in full-length ERM proteins. Recently, the crystal structure of the moesin FERM domain masked by the C-terminal tail domain has been determined (Pearson et al., 2000) and the reported structure shows that the negatively charged surface of the putative-binding site to the adhesion proteins is completely masked by the C-terminal tail domain. A structural comparison of the current IP3-bound radixin FERM domain with the moesin FERM domain shows essentially the same folding of the three subdomains and their domain organization. However, local but large structural changes are found in subdomains B and C (Figure 7B). These contain loop α2B–α2′B (residues 138–150) and helix α3B (residues 160–178) in subdomain B and the β-sheet formed by three strands, β5C/β6C/β7C (residues 243–269), and the N-terminal portion (residues 272–280) of helix α1C in subdomain C (Figure 7C). Excluding these segments displaying large displacements, superimposition of the IP3-bound radixin FERM domain with the moesin FERM domain gives a small r.m.s. deviation of 0.5 Å for the selected Cα atoms. Using all Cα atoms, superimposition of these two domains gives an overall r.m.s. deviation of 1.2 Å.
The superimposed structures show that the bound IP3 molecule has no direct contact with the C-terminal tail domain, although the IP3-binding site is close to the binding site of strand 1 (moesin residues 488–493) of the C-terminal tail domain (Figure 7B). The structural changes in response to IP3 binding may be described as follows, in light of the stimulatory effect of PIP2 binding on the unmasking of ERM proteins. PIP2 binding induces a displacement (1.6 Å) of the N-terminal region (residues 272–280) of helix α1C, which directly participates in IP3 binding. This displacement widens the basic cleft forming the IP3-binding site for the accommodation of the IP3 molecule. The N-terminal region of helix α1C moves towards the β-sheet β5C/β6C/β7C and pushes up one edge of this β-sheet so as to cause a tilting of the β-sheet, with the largest displacement of 6.0 Å being located at Asp261. Consequently, this tilting of the β-sheet β5C/β6C/β7C results in a narrowing of the groove between two β-sheets of the β-sandwich of subdomain C to squeeze out the C-terminal helix D of the tail domain from the groove of the β-sandwich.
Since residues located in the C-terminal helix D are essential for the binding of the tail domain to the FERM domain, the release of the C-terminal helix D from the groove of subdomain C could trigger a dissociation of the whole tail domain from the FERM domain. All ERM proteins have hydrophobic residues such as methionine or leucine at the C-terminal ends of their tail domain, i.e. in C-terminal helix D. It has been shown that the removal of two C-terminal residues (Ala and Leu) destroys the masking state of ezrin (Gary and Bretscher, 1995). The displacements of helix α1C and the β-sheet β5C/β6C/β7C also result in a shallowing of the groove formed by helix α1C and strand β5C, where strand 1 of the tail domain is located. This may destabilize the binding of strand 1 of the tail domain to the FERM domain and contribute the unmasking of the FERM domain.
In subdomain B, helix A (moesin residues 516–530) of the tail domain binds to the hydrophobic groove formed by helices α2B, α3B and α4B of the FERM domain. This binding accompanies a movement (∼3 Å) of helices α3B towards the tail domain, probably an induced fit to helix A. This movement propagates a similar movement of loop α2B–α2′B that contacts helix α3B. Since these regions are far from the IP3-binding site, it is unlikely that these structural changes are associated with the IP3 binding.
The principal structural changes in the FERM domains described above are also seen in a comparison of the free radixin FERM domain and the masked moesin FERM domain. However, the structural changes in subdomain C of the free FERM domain are smaller than those found in the IP3-bound FERM domain. For example, the displacement (0.7 Å) of helix α1C in the free radixin FERM domain is less than half that occurring in the IP3-bound radixin FERM domain. These differences in the degree of structural change are due to the changes induced by IP3 binding, as described above. In conclusion, when the C-terminal tail domain binds the free FERM domain, induced fits occur in subdomains B and C to accommodate helices A and D of the tail domain, respectively. The PIP2 binding to the cleft between subdomains A and B of the masked FERM domain induces relatively subtle changes in helix α1C and strand β5C. These changes would stimulate the release of the induced fits in subdomain C so that subdomain C goes back to a state similar to the free state, with extensive displacements of the β-sheet to release the tail domain.
Potential binding site for adhesion proteins and other binding partners
CD44 and CD43 have cytosolic domains consisting of 70 and 140 residues, respectively. In contrast, ICAM-1, -2 and -3 have short peptide tails as their cytoplasmic portions. For example, the ICAM-2 tail consists of 28 residues. Although unrelated in their overall sequences and different in size, all these cytoplasmic portions have basic juxta-membrane regions that have been identified as their ERM-binding sites (Yonemura et al., 1998). Unfortunately, no obvious consensus sequence motif has been found in these binding regions, which are all characterized by repeated basic residues. Interestingly, the binding of the FERM domains to these regions is sensitive to ionic strength, suggesting that the polar interactions between them predominate in binding. Mutation experiments have suggested that a triplet of basic residues, such as that of lysine or arginine, in the cytoplasmic tails of adhesion proteins is important for binding to the FERM domain. Therefore, the acidic groove between subdomains B and C of the radixin FERM domain is the first candidate for binding to the basic juxta-membrane regions of adhesion proteins. This groove contain >15 negatively charged glutamate/aspartate residues primarily from two linkers, A–B and B–C, loop β3C–β4C and helix α4B (Figure 6D). A close look at the basic juxta-membrane regions of the adhesion proteins reveals glycines spread all over these regions, implying that these regions are probably flexible. The acidic groove spans ∼40 Å, which may be sufficient for the accommodation of ∼10 residues of an extended peptide. The acidic groove of the proposed FERM domain associated with membrane (Figure 7A) seems to be accessible to the juxta-membrane basic regions of adhesion proteins. Although unrelated in sequence, the ezrin-binding site of EBP50 also contains several basic residues, implying that it also binds the acidic groove.
Based on deletion experiments, the second actin-binding site has been mapped at residues 288–310 of ezrin, in addition to the conserved C-terminal 33 residues of ERM proteins (Roy et al., 1997). The N-terminal half of this binding region is located on helix α1C, which includes the basic cleft, but the C-terminal half of this binding region is unstructured in the present crystal. Deletion experiments have suggested that the 30 N-terminal residues of the ezrin FERM domain are required for actin binding to this site. These N-terminal residues are essential for the ubiquitin fold of subdomain A, implying that their deletion would destroy the subdomain A structure and thus also destroy the basic cleft between subdomains A and C.
Relationship with other related proteins
The FERM domain of band 4.1 protein exhibits ∼30% sequence identity to those of ERM proteins and binds the basic regions of the band 3 cytosolic domain, which contains a triplet of arginines (Jons and Drenckhahn, 1992). An arginine/histidine/lysine triplet is also found in the juxta-membrane region of the glycophorin cytosolic tail. Interestingly, in vitro experiments have shown that band 4.1 binds the CD44 basic regions characterized by two triplets of arginines/lysines (Nunomura et al., 1997). The FERM domains of protein tyrosine phosphatases such as PTP-D1 and FAP-1 also exhibit ∼30% sequence identity to those of the ERM proteins. Recently, a kinesin-like protein, KIF1C, has been identified as a binding partner for the PTP-D1 FERM domain, and its binding site has been analyzed (Dorner et al., 1998). This FERM-binding site also contains basic regions characterized by triplets of arginines/lysines. In contrast, the talin FERM domain, which exhibits ∼24% sequence identity to those of ERM proteins, binds the integrin-β cytoplasmic tail, which has no basic residue cluster (Calderwood et al., 1999). Recently, this talin domain has been found to bind the integrin-β juxta-membrane region containing one acidic cluster with some basic residues (Patil et al., 1999). FAK also binds the integrin-β tail (Lewis and Schwartz, 1995; Schaller et al., 1995). Moreover, the FERM-like domain of the JAK kinase Tyk2 binds the juxta-membrane region of an α-interferon receptor 1 subunit, which is characterized by a number of hydrophobic and acidic residues (Yan et al., 1996; Richter et al., 1998). These findings suggest that well-conserved FERM domains bind peptide regions containing positively charged residues, whereas FERM-like domains exhibiting poor sequence homology would bind more diverse sequences. In the FERM-like domains of FAK and JAK, the acidic residues corresponding to those forming the radixin acidic groove are poorly conserved. In addition, several insertion and deletion sites are found, in contrast with ERM proteins.
Conclusion
The present work reveals a novel structure of the FERM domain as a protein–protein interaction module, although its subdomains are folded into common domain architectures. Subdomains A and C form a positively charged molecular surface containing the basic cleft for binding to PIP2. Subdomains B and C form a negatively charged molecular surface containing the acidic groove for possible binding to the positively charged cytoplasmic tails of adhesion proteins. IP3 binding to the basic cleft between domains A and C of the free FERM domain induces relatively subtle changes in helix α1C and strand β5C. Compared with the masked FERM domain, these changes associated with the IP3 binding are linked with extensive displacements of the β-sheet β5C/β6C/β7C, resulting in a release of the C-terminal tail domain from the FERM domain. The structure reported here provides a framework for further experiments designed to probe the molecular basis of ligand recognition by ERM proteins and other proteins containing the FERM domains.
Materials and methods
Expression, crystallization and data collection
The FERM domain (1–310 residues) of mouse radixin was expressed as a fusion protein with glutathione S-transferase (GST) (Matsui et al., 1998). Details of the purification and crystallization screening of the protein have been described previously (Hamada et al., 2000). In brief, the protein was purified using a GST affinity column with glutathione–Sepharose 4B (Pharmacia Biotech) and ion-exchange columns. To prepare the selenomethionyl protein, host cells were exchanged with E.coli B834(DE3) pLysS (Met auxotroph; parent of BL21; Novagen) and grown with selenomethionine in LeMaster broth. Selenomethionyl proteins were purified using the same method as that used for the native protein. The purified radixin FERM domain was verified with matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (JMS Elite, PerSeptive Inc.) and N-terminal analysis (M492, Applied Biosystems). The crystals of the domain were obtained from a solution containing 10 mg/ml protein, 50 mM MES–Na pH 6.0, 2% polyethylene glycol 6000, 150 mM NaCl and 0.5 mM dithiothreitol (DTT) using hanging drop vapor-diffusion at 4°C. The crystals belong to space group P41212, which was determined by the following structure analysis with unit-cell parameters a = b = 96.36 Å and c =133.16 Å. X-ray diffraction data of the crystals were collected by a Rigaku imaging-plate area-detector (R-AXIS IV) using Cu–Kα radiation for native (native-2) and heavy atom-derivative crystals at 15°C (Table I). After determining the structure, a native data (native-1) set was collected using a charge-coupled area detector (ADSC Quantum 4R) installed on the beam line BL6A at the Photon Factory, Tsukuba, Japan. The wavelength was set to 1.00 Å with a crystal–detector distance of 200 mm. The data were collected at 15°C from three crystals with angular ranges of 30–60° with step sizes of 1–2° for an exposure time of 20–30 s. Intensity data were processed using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997) or DPS-Mosfilm (Rossmann and van Beek, 1999).
Table I. Crystallographic analysis.
| Phasing | Native-1(PF) | IP3 complex | Native-2 | K2Pt(CN)4 | K2PtCl6 | SeMeta |
|---|---|---|---|---|---|---|
| Resolution (Å) | 2.80 | 2.90 | 3.00 | 3.50 | 3.60 | 3.50 |
| Reflections | ||||||
| measured | 161 048 | 88 728 | 52 320 | 33 602 | 22 284 | 37 209 |
| unique | 23 751 | 14 511 | 12 482 | 7885 | 6582 | 7840 |
| Completeness (%)b | 99.8/99.9 | 99.5/98.8 | 95.3/91.7 | 94.3/96.4 | 85.2/80.4 | 93.4/88.3 |
| Rsymc (%)b | 4.6/39.3 | 8.1/48.5 | 6.5/30.8 | 9.9/25.5 | 9.3/24.2 | 9.2/20.1 |
| Mean I/σb | 11.8/2.0 | 17.9/1.9 | 16.8/2.5 | 11.6/3.4 | 8.7/2.1 | 12.8/3.6 |
| RDeri (%)b,c | – | – | 22.9 | 19.3 | 13.4 | |
| Phasing powerc (centric/acentric) | – | – | 0.64/0.93 | 0.57/0.80 | 0.56/0.80 | |
| RCullis (%)d (centric/acentric) | – | – | 0.85/0.90 | 0.85/0.92 | 0.84/0.91 | |
| Mean figure of merit |
|
– |
0.39 |
|
|
|
| Refinement | Native-1 | IP3 complex | ||||
| Resolution range | 30.0–2.80 | 30.0–2.90 | ||||
| Number of residues | 279 | 279 | ||||
| Atoms included | 2482 for protein | 2482 for protein, 24 for IP3 | ||||
| Rcryst/Rfree (%)e | 21.5/25.5 | 22.9/28.3 | ||||
| Mean B-factor (Å2) | 64.2 | 63.6 | ||||
| R.m.s. deviations | ||||||
| bond length (Å) | 0.007 | 0.011 | ||||
| bond angle (°) | 1.442 | 1.478 | ||||
| improper (°) | 0.863 | 1.205 | ||||
aSelenomethionyl protein.
bEach pair of values is for overall/outer shell. The resolution ranges of their outer shells are 2.90–2.80 Å for native-1, 3.00–2.90 Å for IP3 complex, 3.11–3.00 Å for native-2, 3.62–3.50 Å for K2Pt(CN)4 and SeMet, and 3.73–3.60 Å for K2PtCl6.
cRsym = Σ |I – < I >|/ΣI, calculated for all data; RDeri = Σ||FPH| – |FP||/Σ|FP|. Phasing power = r.m.s. heavy atom structure factor/residual lack of closure.
dRCullis = Σ||FPH – FP| – |FH (calc)||/Σ|FPH – FP|.
eRcryst and Rfree = Σ||Fo| – |Fc||/Σ|Fo|, where the free reflections (5% of the total used) were held aside for Rfree throughout refinement.
As described in the Results and discussion, the crystals have huge solvent channels with a large calculated fraction of solvent region (71%). The radixin FERM domain has two possible binding sites for PIP2, a basic cleft between subdomains A and C, and a loop region of domain C. The molecular surfaces containing these sites are open to the solvent channels. We therefore tried to soak IP3 into the crystals as well as to co-crystallize the radixin FERM domain with IP3. Soaking of inositol phosphates into the crystals was performed with 10–20 mM IP3 for 10–48 h. A crystal soaked with IP3 had unit cell parameters (a = b = 96.84 Å and c = 132.75 Å) isomorphous to the native crystals of the free radixin FERM domain. Cocrystals of the radixin FERM domain and IP3 were also obtained under similar conditions to those used for the native crystals and were also found to be isomorphous to the native crystals. X-ray diffraction data of these crystals were collected by R-AXIS IV using Cu–Kα radiation at a resolution range of 3.2–2.9 Å. The crystal structures were solved by the molecular replacement method using the program AMoRe (Navaza, 1994).
Structure determination and refinement
Conventional multiple isomorphous replacement was used for phase determination. Phases to 3.5 Å were calculated using MLPHARE (Collaborative Computational Project, 1994) and SHARP (de la Fortelle and Bricogne, 1997). At this stage, the total mean figure of merit was 0.39. The phases were improved by solvent-flattening and histogram-matching using Solomon (Abrahams and Leslie, 1996) and extended to 3.0 Å. An initial model was built into the electron density map using the graphic program O (Jones et al., 1991). This model was refined by the method of simulated annealing using CNS (Brunger et al., 1998) to an R factor of 29%. After several cycles of rebuilding and refinement, the model was refined to an R value of 21.5% (free R value of 25.5%) for intensity data, including 297 of 310 residues at 2.8 Å resolution. The side chains of Lys296 and Pro297 were poorly defined in the current structure and replaced with alanines. No model was built for the 13 C-terminal residues, which were poorly defined in the current map. At present, no water molecules were included. As defined in PROCHECK (Laskowski et al., 1993), there were no residues in the disallowed main-chain torsion angle regions. Ribbon representations of the main-chain folding of the molecule were drawn using the program Molscript (Kraulis, 1991), while molecular surface representations were drawn using the program GRASP (Nicholls et al., 1991). During the review process for this paper, the crystal structure of the moesin FERM domain masked by the C-terminal tail domain was published (Pearson et al., 2000). Structural comparison between the radixin and moesin FERM domains was accomplished using the coordinates (PDB code 1EF1) of the moesin FERM–tail complex.
Acknowledgments
Acknowledgements
We would like to thank Drs N.Igarashi, M.Suzuki and K.Okada for their technical assistance in data collection at the Photon Factory, Tsukuba. This work was supported by Grants-in-Aid for Scientific Research from the MESSC of Japan to T.H. (10359003, 10179103-4). K.H. was supported by a research fellowship for Young Scientists from JSPS. T.H. is a member of the TARA project of Tsukuba University. Coordinates for the radixin FERM domain in the IP3-bound and free forms have been deposited in the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics under accession Nos 1ge6 and 1ge7, respectively.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Algrain M., Turunen,O., Vaheri,A., Louvard,D. and Arpin,M. (1993) Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J. Cell Biol., 120, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva M.R. and Furthmayr,H. (1995) Subcellular localization of moesin in dynamic filopodia, retraction fibers, and other structures involved in substrate exploration, attachment, and cell–cell contacts. Exp. Cell Res., 219, 180–196. [DOI] [PubMed] [Google Scholar]
- Andreoli C., Martin,M., Le Borgne,R., Reggio,H. and Mangeat,P. (1994) Ezrin has properties to self-associate at the plasma membrane. J. Cell Sci., 107, 2509–2521. [DOI] [PubMed] [Google Scholar]
- Arpin M., Algrain,M. and Louvard,D. (1994) Membrane–actin microfilament connections: an increasing diversity of players related to band 4.1. Curr. Opin. Cell Biol., 6, 136–141. [DOI] [PubMed] [Google Scholar]
- Bretscher A. (1989) Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J. Cell Biol., 108, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. (1999) Regulation of cortical structure by the ezrin–radixin–moesin protein family. Curr. Opin. Cell Biol., 11, 109–116. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Zent,R., Grant,R., Rees,D.J., Hynes,R.O. and Ginsberg,M.H. (1999) The talin head domain binds to integrin β-subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem., 274, 28071–28074. [DOI] [PubMed] [Google Scholar]
- Chishti A.H. et al. (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci., 23, 281–282. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Conti E. and Kuriyan,J. (2000) Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure, 8, 329–338. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy,M., Leighton,L., Blobel,G. and Kuriyan,J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Crepaldi T., Gautreau,A., Comoglio,P.M., Louvard,D. and Arpin,M. (1997) Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J. Cell Biol., 138, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S., Ottinger,E.A., Nolte,R.T., Eck,M.J. and Shoelson,S.E. (1999) Crystal structure of the pleckstrin homology–phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc. Natl Acad. Sci. USA, 96, 8378–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner C., Ciossek,T., Muller,S., Moller,N.P.H., Ullrich,A. and Lammers,R. (1998) Characterization of KIF1C, a new kinesin-like protein involved in vesicle transport from the Golgi apparatus to the endoplasmic reticulum. J. Biol. Chem., 273, 20267–20275. [DOI] [PubMed] [Google Scholar]
- Eck M.J., Dhe-Paganon,S., Trub,T., Nolte,R.T. and Shoelson,S.E. (1996) Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell, 85, 695–705. [DOI] [PubMed] [Google Scholar]
- Ferguson K.M., Lemmon,M.A., Schlessinger,J. and Sigler,P.B. (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell, 83, 1037–1046. [DOI] [PubMed] [Google Scholar]
- Franck Z., Gary,R. and Bretscher,A. (1993) Moesin, like ezrin, colocalizes with actin in the cortical cytoskeleton in cultured cells, but its expression is more variable. J. Cell Sci., 105, 219–231. [DOI] [PubMed] [Google Scholar]
- Gary R. and Bretscher,A. (1995) Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell, 6, 1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault J.A., Labesse,G., Mornon,J.P. and Callebaut,I. (1999) The N-termini of FAK and JAKs contain divergent band 4.1 domains. Trends Biochem. Sci., 24, 54–57. [DOI] [PubMed] [Google Scholar]
- Gould K.L., Bretscher,A., Esch,F.S. and Hunter,T. (1989) cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1. EMBO J., 8, 4133–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K., Matsui,T., Tsukita,S., Tsukita,S. and Hakoshima,T. (2000) Crystallographic characterization of the membrane-binding domain of radixin. Acta Crystallogr. D, 56, 922–923. [DOI] [PubMed] [Google Scholar]
- Hanzel D.K., Urushidani,T., Usinger,W.R., Smolka,A. and Forte,J.G. (1989) Immunological localization of an 80-kDa phosphoprotein to the apical membrane of gastric parietal cells. Am. J. Physiol., 256, G1082–G1089. [DOI] [PubMed] [Google Scholar]
- Harlan J.E., Hajduk,P.J., Yoon,H.S. and Fesik,S.W. (1994) Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature, 371, 168–170. [DOI] [PubMed] [Google Scholar]
- Helander T.S., Carpen,O., Turunen,O., Kovanen,P.E., Vaheri,A. and Timonen,T. (1996) ICAM-2 redistributed by ezrin as a target for killer cells. Nature, 382, 265–268. [DOI] [PubMed] [Google Scholar]
- Hirao M., Sato,N., Kondo,T., Yonemura,S., Monden,M., Sasaki,T., Takai,Y., Tsukita,S. and Tsukita,S. (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol., 135, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Huang L., Hofer,F., Martin,G.S. and Kim,S.-H. (1998) Structural basis for the interaction of Ras with RalGDS. Nature Struct. Biol., 5, 422–426. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Nelson,W.J. and Weis,W.I. (1997) Three-dimensional structure of the armadillo repeat region of β-catenin. Cell, 90, 871–882. [DOI] [PubMed] [Google Scholar]
- Hyvonen M., Macias,M.J., Nilges,M., Oschkinat,H., Saraste,M. and Wilmanns,M. (1995) Structure of the binding site for inositol phosphates in a PH domain. EMBO J., 14, 4676–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jons T. and Drenckhahn,D. (1992) Identification of the binding interface involved in linkage of cytoskeletal protein 4.1 to the erythrocyte anion exchanger. EMBO J., 11, 2863–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nature Struct. Biol. 6, 388–397. [DOI] [PubMed] [Google Scholar]
- Kragelund B.B., Andersen,K.V., Madsen,J.C., Knudsen,J. and Poulsen, F.M. (1993) Three-dimensional structure of the complex between acyl-coenzyme A binding protein and palmitoyl-coenzyme A. J. Mol. Biol., 230, 1260–1277. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Krieg J. and Hunter,T. (1992) Identification of the two major epidermal growth factor-induced tyrosine phosphorylation sites in the microvillar core protein ezrin. J. Biol. Chem., 267, 19258–19265. [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lewis J.M. and Schwartz,M.A. (1995) Mapping in vivo associations of cytoplasmic proteins with integrin β1 cytoplasmic domain mutants. Mol. Biol. Cell, 16, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magendantz M., Henry,M.D., Lander,A. and Solomon,F. (1995) Interdomain interactions of radixin in vitro. J. Biol. Chem., 270, 25324–25327. [DOI] [PubMed] [Google Scholar]
- Mangeat P., Roy,C. and Martin,M. (1999) ERM proteins in cell adhesion and membrane dynamics: authors’ correction. Trends Cell Biol., 9, 289. [DOI] [PubMed] [Google Scholar]
- Matsui T., Maeda,M., Doi,Y., Yonemura,S., Amano,M., Kaibuchi,K., Tsukita,S. and Tsukita,S. (1998) Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol., 140, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Yonemura,S., Tsukita,S. and Tsukita,S. (1999) Activation of ERM proteins in vivo by Rho involves phosphatidyl-inositol 4-phosphate 5-kinase and not ROCK kinases. Curr. Biol., 9, 1259–1262. [DOI] [PubMed] [Google Scholar]
- Misra S. and Hurley,J.H. (1999) Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell, 97, 657–666. [DOI] [PubMed] [Google Scholar]
- Moller N.P., Moller,K.B., Lammers,R., Kharitonenkov,A., Sures,I. and Ullrich,A. (1994) Src kinase associates with a member of a distinct subfamily of protein-tyrosine phosphatases containing an ezrin-like domain. Proc. Natl Acad. Sci. USA, 91, 7477–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) ‘scop’: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Nassar N., Horn,G., Herrmann,C., Scherer,A., McCormick,F. and Wittinghofer,A. (1995) The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature, 375, 554–560. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Niggli V., Andreoli,C., Roy,C. and Mangeat,P. (1995) Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett., 376, 172–176. [DOI] [PubMed] [Google Scholar]
- Nunomura W., Takakuwa,Y., Tokimitsu,R., Krauss,S.W., Kawashima,M. and Mohandas,N. (1997) Regulation of CD44–protein 4.1 interaction by Ca2+ and calmodulin. Implications for modulation of CD44–ankyrin interaction. J. Biol. Chem., 272, 30322–30328. [DOI] [PubMed] [Google Scholar]
- Orsulic S. and Peifer,M. (1996) An in vivo structure–function study of armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J. Cell Biol., 134, 1283–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pai L.M., Kirkpatrick,C., Blanton,J., Oda,H., Takeichi,M. and Peifer,M. (1996) Drosophila α-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. J. Biol. Chem., 271, 32411–32420. [DOI] [PubMed] [Google Scholar]
- Patil S., Jedsadayanmata,A., Wencel-Drake,J.D., Wang,W., Knezevic,I. and Lam,S.C. (1999) Identification of a talin-binding site in the integrin β3 subunit distinct from the NPLY regulatory motif of post-ligand binding functions. The talin N-terminal head domain interacts with the membrane-proximal region of the β3 cytoplasmic tail. J. Biol. Chem., 274, 28575–28583. [DOI] [PubMed] [Google Scholar]
- Pearson M.A., Reczek,D., Bretscher,A. and Karplus,P.A. (2000) Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell, 101, 259–270. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp K., Amieva,M.R., Strassel,C.P., Nauseef,W.M., Furthmayr,H. and Luna,E.J. (1995) Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol. Biol. Cell, 6, 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage R., Green,J., Muir,T.W., Ogunjobi,O.M., Love,S. and Shaw,K. (1994) Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem. J., 299, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D., Berryman,M. and Bretscher,A. (1997) Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin–radixin–moesin family. J. Cell Biol., 139, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M.F., Dumenil,G., Uze,G., Fellous,M. and Pellegrini,S. (1998) Specific contribution of Tyk2 JH regions to the binding and the expression of the interferon α/β receptor component IFNAR1. J. Biol. Chem., 273, 24723–24729. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G. and van Beek,C.G. (1999) Data processing. Acta Crystallogr. D, 55, 1631–1653. [DOI] [PubMed] [Google Scholar]
- Rouleau G.A. et al. (1993) Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature, 363, 515–521. [DOI] [PubMed] [Google Scholar]
- Roy C., Martin,M. and Mangeat,P. (1997) A dual involvement of the amino-terminal domain of ezrin in F- and G-actin binding. J. Biol. Chem. 272, 20088–20095. [DOI] [PubMed] [Google Scholar]
- Salim K. et al. (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J., 15, 6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Sato N., Yonemura,S., Obinata,T., Tsukita,S. and Tsukita,S. (1991) Radixin, a barbed end-capping actin-modulating protein, is concentrated at the cleavage furrow during cytokinesis. J. Cell Biol., 113, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Funayama,N., Nagafuchi,A., Yonemura,S., Tsukita,S. and Tsukita,S. (1992) A gene family consisting of ezrin, radixin and moesin. Its specific localization at actin filament/plasma membrane association sites. J. Cell Sci., 103, 131–143. [DOI] [PubMed] [Google Scholar]
- Schaller M.D., Otey,C.A., Hildebrand,J.D. and Parsons,J.T. (1995) Focal adhesion kinase and paxillin bind to peptides mimicking β integrin cytoplasmic domains. J. Cell Biol., 130, 1181–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador J.M., Alonso-Lebrero,J.L., del Pozo,M.A., Furthmayr,H., Schwartz-Albiez,R., Calvo,J., Lozano,F. and Sanchez-Madrid,F. (1997) Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol., 138, 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert J. and Kemler,R. (1994) A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Adhes. Commun., 2, 319–327. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Sasaki,T., Mammoto,A., Takaishi,K., Kameyama,T., Tsukita,S., Tsukita,S. and Takai,Y. (1997) Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J. Biol. Chem., 272, 23371–23375. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Sasaki,T., Mammoto,A., Hotta,I., Takaishi,K., Imamura,H., Nakano,K., Kodama,A. and Takai,Y. (1998) Interaction of radixin with Rho small G protein GDP/GTP exchange protein Dbl. Oncogene, 16, 3279–3284. [DOI] [PubMed] [Google Scholar]
- Takeda H., Nagafuchi,A., Yonemura,S., Tsukita,S., Behrens,J., Birchmeier,W. and Tsukita,S. (1995) V-src kinase shifts the cadherin-based cell adhesion from the strong to the weak state and β catenin is not required for the shift. J. Cell Biol., 131, 1839–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofatter J.A. et al. (1993) A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell, 72, 791–800. [DOI] [PubMed] [Google Scholar]
- Tsukita S. and Yonemura,S. (1999) Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J. Biol. Chem., 274, 34507–34510. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Hieda,Y. and Tsukita,S. (1989) A new 82-kD barbed end-capping protein (radixin) localized in the cell-to-cell adherens junction: purification and characterization. J. Cell Biol., 108, 2369–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S., Oishi,K., Sato,N., Sagara,J., Kawai,A. and Tsukita,S. (1994) ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol., 126, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen O., Wahlstreom,T. and Vaheri,A. (1994) Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol., 126, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman E.J., Steplock,D., Corry,D. and Shenolikar,S. (1993) Identification of the human NHE-1 form of Na+-H+ exchanger in rabbit renal brush border membranes. J. Clin. Invest., 91, 2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Krishnan,K., Lim,J.T., Contillo,L.G. and Krolewski,J.J. (1996) Molecular characterization of an α interferon receptor 1 subunit (IFNαR1) domain required for TYK2 binding and signal transduction. Mol. Cell. Biol., 16, 2074–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Nagafuchi,A., Sato,N. and Tsukita,S. (1993) Concentration of an integral membrane protein, CD43 (leukosialin, sialophorin), in the cleavage furrow through the interaction of its cytoplasmic domain with actin-based cytoskeletons. J. Cell Biol., 120, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Hirao,M., Doi,Y., Takahashi,N., Kondo,T., Tsukita,S. and Tsukita,S. (1998) Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43 and ICAM-2. J. Cell Biol., 140, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.S., Hajduk,P.J., Petros,A.M., Olejniczak,E.T., Meadows,R.P. and Fesik,S.W. (1994) Solution structure of a pleckstrin homology domain. Nature, 369, 672–675. [DOI] [PubMed] [Google Scholar]
- Yun C.H., Oh,S., Zizak,M., Steplock,D., Tsao,S., Tse,C.M., Weinman,E.J. and Donowitz,M. (1997) cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl Acad. Sci. USA, 94, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lee,C.H., Mandiyan,V., Borg,J.P., Margolis,B., Schlessinger,J. and Kuriyan,J. (1997) Sequence-specific recognition of the internalization motif of the Alzheimer’s amyloid precursor protein by the X11 PTB domain. EMBO J., 16, 6141–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.M., Huang,B., Olejniczak,E.T., Meadows,R.P., Shuker,S.B., Miyazaki,M., Trub,T., Shoelson,S.E. and Fesik,S.W. (1996) Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain. Nature Struct. Biol., 3, 388–393. [DOI] [PubMed] [Google Scholar]