Abstract
Sall1 is a multi-zinc finger transcription factor that regulates kidney organogenesis. It is considered to be a transcriptional repressor, preferentially localized on heterochromatin. Mutations or deletions of the human SALL1 gene are associated with the Townes-Brocks syndrome. Despite its high expression, no function was yet assigned for Sall1 in embryonic stem (ES) cells. In the present study, we show that Sall1 is expressed in a differentiation-dependent manner and physically interacts with Nanog and Sox2, two components of the core pluripotency network. Genome-wide mapping of Sall1-binding loci has identified 591 genes, 80% of which are also targeted by Nanog. A large proportion of these genes are related to self-renewal and differentiation. Sall1 positively regulates and synergizes with Nanog for gene transcriptional regulation. In addition, our data show that Sall1 suppresses the ectodermal and mesodermal differentiation. Specifically, the induction of the gastrulation markers T brachyury, Goosecoid, and Dkk1 and the neuroectodermal markers Otx2 and Hand1 was inhibited by Sall1 overexpression during embryoid body differentiation. These data demonstrate a novel role for Sall1 as a member of the transcriptional network that regulates stem cell pluripotency.
Keywords: Chromatin Immunoprecipitation (ChIP), Differentiation, Embryonic Stem Cell, Gene Expression, Transcription Factors, Nanog, Sall1, Self-renewal, Sox2, Stem Cell Pluripotency
Introduction
Pluripotency of embryonic stem (ES) cells is achieved through the orchestrated function of multiple pathways that activate a large set of transcription factors for regulation of gene expression (1). These factors comprise a transcriptional network with Nanog, Oct4, and Sox2 acting as the master regulators (2, 3), whereas other factors such as Nr0b1, Sall4, c-Myc, Klf4, Zic3, Esrrb, Tcf3, Suz12, Zfp206, and Zfp281 also have important roles in the maintenance of stem cell identity (4, 5). Members of this network have been found to co-exist in large complexes (up to 13 factors) (4) for the regulation of common target gene expression as well as their own. Target genes belong in two major categories, pluripotency-related genes that are activated and differentiation-specific genes that are repressed.
After a transcriptomic analysis of embryonic stem cells treated with a histone deacetylase inhibitor, we observed that the spalt homology 1 (sall1) gene was highly expressed in undifferentiated cells and declined with the onset of differentiation (6), suggesting that Sall1 has a role in the biology of ES cells. The spalt (sal) genes were first isolated in Drosophila. Mice and humans have four known Sal-related genes named sall1–sall4 and SALL1–SALL4, respectively. Spalt genes are homeotic genes that regulate development of the wing, trachea, and sensory organs in Drosophila (7–9). They are important for the development of the limbs, the nervous system, and several organs including the kidney and heart (10, 11). Sall proteins contain zinc finger domains that are arranged in a highly conserved way in all family members (11). Sall1 and Sall4 were intensely studied because they have been associated with human genetic syndromes. Sall2 has been reported as a tumor suppressor factor, whereas Sall4 behaves as an oncogene when up-regulated (11). The Sall2 gene is dispensable for mouse development, but Sall3-deficient homozygous mice die shortly after birth because of the inability to feed properly (11).
Sall1 is involved in mouse kidney organogenesis with kidney agenesis or severe dysgenesis observed in Sall1-deficient animals (12). In humans, SALL1 mutations leading to a truncated molecule cause an autosomal dominant disorder characterized by limb, ear, anal, heart, and limb defects, known as the Townes-Brocks syndrome (11, 13). A truncated Sall1 protein that retains only the N-terminal part can reproduce a phenotype similar to the Townes-Brocks syndrome when expressed in mice, suggesting that it acts in a dominant negative manner (14). Sall4, another spalt factor that shares structural and functional similarities with Sall1, has been shown to contribute in the maintenance of pluripotency in both the inner cell mass (15) and the embryonic stem cells (16). Sall1 and Sall4 have been shown to genetically interact in kidney, heart, and anal development, as observed in mouse Sall1 and Sall4 compound heterozygotes (17). They co-localize in many adult tissues (brain, heart, and anus) as well as in ES cells where both show a heterochromatic localization. Many of the symptoms of Townes-Brocks syndrome overlap with those of the Okihiro syndrome caused by mutations in SALL4.
Sall1 encodes a transcription factor containing 10 zinc finger motifs, most of which are clustered in duplets or triplets (10, 11). It has been reported that Sall1 acts as a transcriptional repressor by localizing in the heterochromatin and interacting with components of the nucleosome remodeling and deacetylase complex (NuRD) (18, 19). Conversely, Sall1 was found to cooperatively activate the Wnt pathway with β-catenin (20) to activate kidney mesenchymal markers (12) and induce angiogenesis by activating VEGF-A (21). The molecular mechanism whereby Sall1 directly targets genes for repression or activation remains unknown.
To identify the role of Sall1 in mouse embryonic stem cell (mESC)2 pluripotency, we have analyzed its interactions with the core pluripotency factors and identified the gene loci where it binds. We found that Sall1 regulates Nanog expression because silencing of Sall1 resulted in Nanog down-regulation. A genome-wide promoter ChIP-on-chip analysis has shown that Sall1 and Nanog bind together to a large number of common target genes that are related to self-renewal and differentiation of mESC. Overexpression of Sall1 during differentiation prevented certain differentiation markers from expressing, especially determinants of mesodermal and ectodermal fate. In complementary fashion, a subset of these genes was up-regulated when Sall1 was silenced in the undifferentiated state. Our findings demonstrate that Sall1 has novel functions in mESC, namely to regulate gene activation and repression in association with Nanog.
EXPERIMENTAL PROCEDURES
Cell Cultures, Antibodies, and siRNAs
CGR8 ES cells were cultivated in GMEM (10% fetal bovine serum, 1000 units of LIF (ESGRO-Chemicon, Temecula, CA)). COS and 293T cells were cultivated in DMEM (10% fetal serum). Antibodies used were α-His (Santa Cruz Biotechnology) and α-Nanog (Chemicon) α-Sall1 (R&D Systems). Polyclonal antibodies employed in ChIP-on-chip experiments were produced in rabbits immunized with a His-Nanog and a His-Sall1(1–702), respectively. siRNAs were: control (scrambled), 5′-CAGUCGCGUUUGCGACUGGUU-3′ (Curevac); Nanog, 5′-AGAAGGAAGGAACCUGGCUUU-3′ (Curevac); and Sall1, 5′-GGGUAAUUUGAAGCAGCACAU-3′ (Metabion). A second siRNA for Sall1, ON-TARGETplus SMARTpool L-062536-01-0010, mouse SALL1, NM_021390 was from Thermo Scientific Dharmacon. CGR8 cells were transfected with siRNAs (final concentration 50 nm) for 2 days using Lipofectamine 2000 (Invitrogen).
Protein-Protein Interaction Assays
For co-immunoprecipitation assays, protein extracts were incubated overnight with antibodies at 4 °C, and the next day protein G-agarose beads were added (3 h, 4 °C). For GST pulldown assays, GST-enriched bacterial protein extracts were incubated with glutathione-Sepharose beads (1 h, 4 °C), and then His-tagged proteins were added (3 h, 4 °C). His-tagged proteins were purified with the Protino Ni-TED 150 system. GST- and His-fused protein expression was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside in DH5α and BL21 bacterial cells, respectively.
Transfections and Plasmids
Transfection of ES cells was performed by Lipofectamine 2000 (Invitrogen), whereas COS and 293T cells were transfected with the Ca3(PO4)2 method. Nanog promoter plasmids are described in Ref. 6. Nanog-expressing plasmid was provided by P. Savatier. Full-length Nanog cDNA was obtained by PCR (5′-GCAGCTGAGCCATGAGTGTGGGTCTTCCT-3′ and 5′-CCAGCAGAGAGCTCTCATATTTCACCTGGT-3′) and was first cloned in pBSK+ (EcoRV), and from pBSK+, it was further subcloned in GFPC1 (BamHI/SalI). Full-length Nanog, Nanog 1–321 (aa 1–107), and Nanog 1–492 (aa 1–194) cDNA fragments were excised from pBSK+ Nanog using SmaI/SalI, SalI/PvuII, and SalI/XmnI restriction enzymes, respectively, and were cloned in pRSETA (PvuII/XhoI). Nanog cDNA fragments 321–927 (aa 107–309) and 492–927 (aa 194–309) were excised from GFPC1-Nanog using PvuII/SmaI and XmnI/SmaI restriction enzymes, respectively, and were cloned in pRSETC (PvuII). Nanog was expressed in eukaryotic cells using the pCMV-IRES-EGFP vector.
Sall1 cDNA was produced by RT-PCR from CGR8 cells. Four cDNA fragments covering the full-length molecule were constructed using the following primers: fragment 1, 5′-GCATGTCGCGGAAGC-3′ and 5′-GGGGAAGCTGCTTCA-CAC-3′; fragment 2, 5′-GCAGCTGGATTAGCACAG-3 and 5′-TCTCTGAGGCCTGGGCAG-3′; fragment 3, 5′-CTGCCCAGGCCTCAGAGA-3′ and 5′-ATGACTAGTGGGGGC-GTC-3′; and fragment 4, 5′-GACGCCCCCACTAGTCAT-3′ and 5′-TGGCAGCTTTAGCTTGTG-3′. They were cloned successively in pBSK+ vector to reconstitute Sall1 full-length cDNA using the internal restriction sites NheI, StuI, and SpeI. For transfection and ES stable clones, Sall1 cDNA was excised from pBSK+ with SalI/SacII restriction enzymes and was cloned in pCMV-IRES-EGFP in the same sites.
For GST-Sall1, the full-length molecule was excised from pBSK+ using Sal1I/NotI restriction enzymes and was cloned in pGEX4T3 in the same sites. GST-Sall1 aa 1–183 and aa 1–435 were obtained by PCR (forward, 5′-ATCGGGATCCATGTCGCGGAGG-3′, and reverse, 5′-CGATGGATCCCGTCAGGTCCCC-3′; forward, 5′-ATCGGGATCCATGTCGCGGAGG-3′, and reverse, 5′-CGATGGATCCAGTGACATTTGG-3′, respectively) and were cloned in pGEX4T1 (BamHI). GST-Sall1 aa 435–702 was produced by PCR using the forward primer, 5′-GGATCCGCCTTTGAAGCG-3′, and the BamHI restriction site at aa 702. GST-Sall1 aa 702–856 was produced by PCR using the following primers: forward, 5′-GGATCCATCATCTGCCACCGGGTTC-3′, and reverse, 5′-GGATCCGGGCAGAGGCGA-3′. The PCR product was cloned in pGEX3T1 (BamHI).
For GST-Sall1 aa 856–1323, pGEX4T3-Sall1 was digested with SalI/XhoI and was religated. For GST-Sall1 aa 1100–1324, Sall1 cDNA fragment was excised from pIRES-EGFP with SpeI (Klenow-filled)/SmaI restriction enzymes and was inserted in pGEX4T3 (SmaI). Sox2 cDNA was kindly provided by the Dr. E. Reboutsika laboratory as an SV40-Sox2 construct. From this construct, it was excised with BamHI/BglII restriction enzymes and was inserted in pRSETC (BamHI) for His-Sox2 (full length). For His-Sox2 aa 1–54, aa 1–183, aa 1–207, and aa 1–240, His-Sox2 full length was digested with SmaI, PvuII, PstI, and NcoI, respectively, and was religated. For GFP-Sox2, Sox2 cDNA was excised from SV40-Sox2 with EcoRI/BamHI and was first subcloned in pIRES-EGFP in the same sites. It was then re-excised with XhoI/BamHI restriction enzymes and cloned in GFPC1 in the same sites.
Oct4 cDNA was produced by RT-PCR from CGR8 mRNA and cloned in GFPC1 vector. For His-tagged Oct4 (full length), Oct4 cDNA fragment was excised from GFP-Oct4 with BglII/KpnI restriction enzymes and was inserted into pRSETB in the same sites.
Chromatin Immunoprecipitation, Microarray Hybridization, and Data Analysis
Chromatin immunoprecipitation was performed according to Ref. 6. For the ChIP-on-chip analysis, the Affymetrix GeneChip® mouse promoter 1.0R array was used that covers a region 6 kb upstream and 2.5 kb downstream of the gene transcription start site of each mouse gene. Experiments were done in duplicate, and analysis was performed using the Affymetrix web tools and the model-based analysis of tiling-array (MAT) program.
RESULTS
Sall1 Regulates the Expression of Nanog
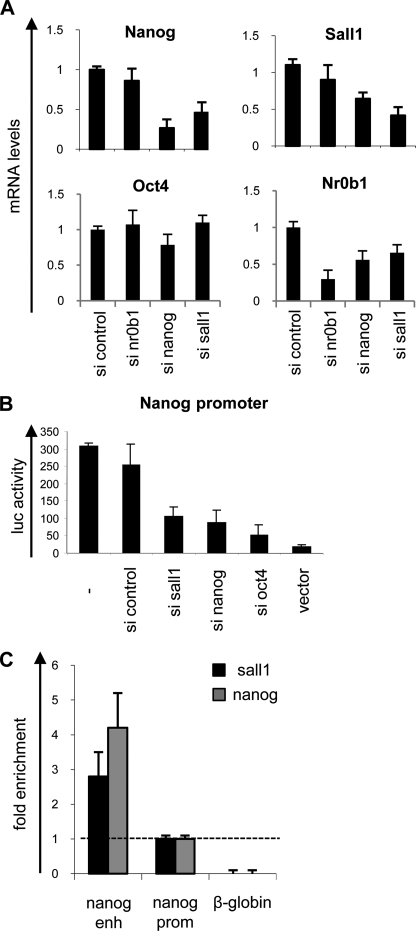
Sall1 is highly expressed in mESC and is rapidly down-regulated during differentiation (6). To study its role in the regulation of pluripotency, we knocked down Sall1 using siRNA technology. The effects of silencing Sall1 were studied along with Nanog and Nr0b1 as controls. Knocking down Sall1 resulted in the reduction of Nanog and Nr0b1 mRNA levels to 45 and 50%, respectively, when compared with the controls, whereas the levels of Sall1 itself were reduced to 35% (Fig. 1A). The expression of Oct4 was not affected by knocking down Sall1, whereas it was reduced by knocking down Nanog. Silencing of Nanog also had an impact on Sall1, with its expression levels being reduced to ∼50% of the initial (Fig. 1A) in accordance with a previous report (3). Moreover, when we used a plasmid carrying ∼1 kb of the Nanog promoter (−966/+50 relative to the transcription start site) cloned upstream of the luciferase reporter gene, we observed that its activity in ES cells was strongly diminished after knocking down Sall1, reaching 35% of its initial levels. Interestingly, the knockdown of either Nanog itself or Oct4, both of which have been shown to affect Nanog expression, resulted in down-regulation of the Nanog promoter activity at comparable levels (Fig. 1B), implying that Sall1 plays an important role in regulating Nanog expression. To exclude the possibility of off-target effects, the above experiments were repeated using a second siRNA for Sall1 (ON-TARGETplus SMARTpool), which generated similar results (not shown).
FIGURE 1.
Sall1 regulates the expression of Nanog. A, Nanog, Sall1, Oct4, and Nr0b1 mRNA levels are shown after silencing of Nanog, Sall1 and Nr0b1. si, siRNA. B, Nanog promoter luciferase (luc) activity is decreased after silencing of Sall1, Nanog, or Oct4. Control in A and B is a scrambled siRNA. C, ChIP using anti-Sall1 (black) or anti-Nanog (gray) antibodies detected Nanog and Sall1 binding on nanog enhancer region (enh) but not on nanog promoter region (prom). β-Globin gene was used as negative control. Error bars in all panels indicate S.D.
To study whether Sall1 regulates nanog via direct binding to the nanog locus, we performed a chromatin immunoprecipitation assay. Because it has been found that a complex that includes Nanog, Sox2, Oct4, and other critical factors binds to the nanog enhancer (3–5, 22), we analyzed this region for Sall1 binding and used the proximal promoter for comparison. As shown in Fig. 1C, Sall1 and Nanog bind to the enhancer region of Nanog, whereas minimal binding is detected on the promoter. These results suggest that Sall1 directly regulates Nanog expression, implicating Sall1 in the regulatory network of pluripotency genes.
Sall1 Interacts with Nanog and Sox2
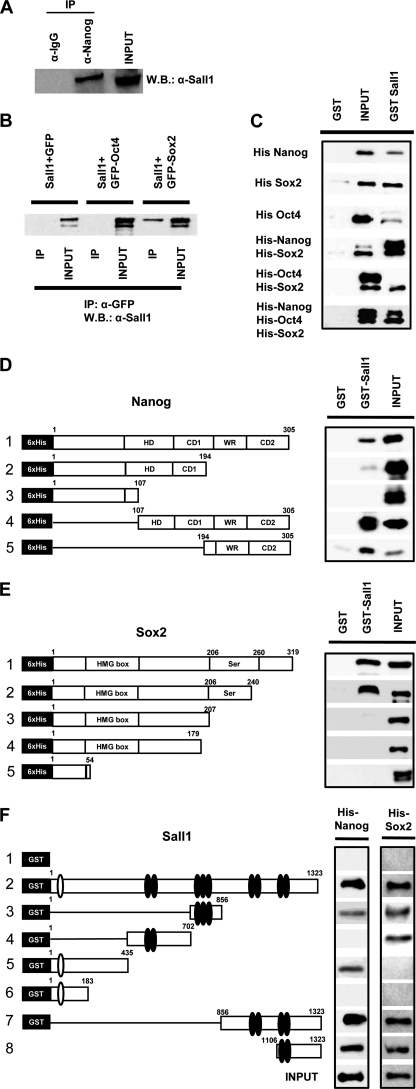
To study the ability of Sall1 to interact with the protein members of the core pluripotency complex such as Nanog, Oct4, and Sox2, we performed co-immunoprecipitation assays. To this end, we used mESC nuclear extracts to check the Sal11-Nanog physical interaction of the endogenous proteins. As depicted in Fig. 2A, the two factors interact in vivo in accordance with a previous study that detected Sall1 in the protein complex that was co-purified with biotinylated Nanog (23). To test the interaction of Sall1 with either Oct4 or Sox2, we used extracts from COS cells transfected with a vector expressing mSall1 and a vector expressing either GFP-mSox2 or GFP-mOct4. We found that Sall1 interacts with Sox2 but not Oct4 (Fig. 2B). To test whether these interactions were direct, we have performed in vitro GST pulldown assays by using a GST-fused Sall1 and His-tagged Nanog, Oct4, and Sox2 proteins. We found that Sall1 interacts with Nanog and Sox2 but only weakly with Oct4, in accordance with the in vivo findings (Fig. 2C). Given the fact that Oct4 cooperates with Sox2 for the regulation of common target genes, we added both protein His-Sox2 and protein His-Oct4 to GST-Sall1 to test whether Sox2 would serve as an intermediate/stabilizer for the weak Oct4-Sall1 interaction. However, no interaction was observed between Oct4 and Sall1 when His-Sox2 was supplied in the reaction mix (Fig. 2C, fifth row) or when Nanog was further added. His-Sox2 and His-Nanog interacted with GST-Sall1 when added together (Fig. 2C, fourth row) or with further addition of His-Oct4 (Fig. 2C, sixth row). Overall these results suggest that the protein complexes of transcription factors required to maintain the pluripotency of ES cells may also include Sall1.
FIGURE 2.
Sall1 interacts with Nanog and Sox2. A, endogenous Sall1 and Nanog proteins were co-immunoprecipitated in ES nuclear extracts. IP, immunoprecipitation; W.B., Western blot. B, co-immunoprecipitation assays after COS cell transfection with vectors expressing Sall1 and GFP-Sox2 or GFP-Oct4. Sall1 co-immunoprecipitated with GFP-Sox2 but not GFP-Oct4. C, GST pulldown experiments using GST-Sall1 and His-tagged Nanog (D) or Sox2 (E) deletions. F, GST pulldown experiments using GST-Sall1 deletions and His-tagged-Nanog or Sox2. HD, Homeodomain; CD1, CD2, C-terminal activation domain 1,2; WR, tryptophan-rich.
To better delineate the interactions between Sall1 and Nanog or Sox2, we used His-tagged deletions of Nanog or Sox2 and GST-tagged deletions of Sall1 in GST pulldown experiments. Fig. 2D shows that the C-terminal region of Nanog covering amino acids 194–306 (lane 5) and containing the transcriptional activation domain (24) was sufficient to interact with the full-length Sall1 protein, whereas no interaction was detected when this region was absent. Sox2, on the other hand (Fig. 2E), could interact with Sall1 when its C-terminal domain (aa 240–320, lane 2) was absent, but this ability was lost when a domain covering amino acids 207–240 (lane 3) was further deleted, indicating that this region, which contains the transcriptional activation domain of Sox2 (25), is critical for the interaction with Sall1. Furthermore, we used GST-tagged Sall1 in binding assays with the full-length His-Nanog or Sox2 proteins. We observed that both Nanog and Sox2 interact with the extreme C-terminal domain (aa 1106–1323, lane 8), which includes the last double Zn+ finger motif. In addition, Nanog interacts with the regions covering aa 1–435 and 702–856, whereas Sox2 interacts with the central region 435–856. Therefore, multiple domains of Sall1 protein are involved in interaction with Nanog and Sox2.
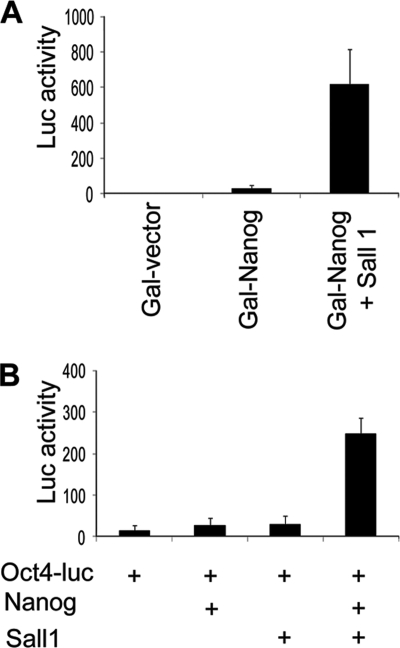
To examine the functional impact of Sall1 interaction with Nanog, we performed transient transfection assays to quantify the transcriptional activatory function of Nanog. Sall1 was able to enhance the transactivation potential of a Gal4-Nanog fusion protein on a Gal4-luciferase reporter (Fig. 3A). Moreover, the addition of Sall1 along with Nanog strongly activated an Oct-4 promoter-luciferase construct in 293 cells (Fig. 3B). Therefore, although Sall1 has a reported repressor function (19, 26), our results show that it can also act as a transcriptional co-activator when interacting with Nanog.
FIGURE 3.
Sall1 activates transcription in cooperation with Nanog. A, luciferase (Luc) activity in extracts from 293 cells transfected with a 5× Gal4-Luc reporter and a Gal4-Nanog-expressing construct in the absence or presence of Sall1-expressing plasmid. B, luciferase activity in extracts from 293 cells transfected with an Oct4-Luc reporter containing a promoter region covering 3 kb upstream from the transcription start site and plasmids expressing Nanog or Sall1. Error bars in both panels indicate S.D.
Sall1 and Nanog Bind to Common Target Genes
Our finding that Sall1 interacts physically and cooperates functionally with members of the core pluripotency complex suggests that these factors are linked in a regulatory network in mES cells. Nanog and Sox2 are known transcription factors that exert their function through binding to their target genes. To elucidate the role of Sall1 in mES pluripotency, we performed a genome-wide promoter ChIP-on-chip analysis to identify its putative target genes. Moreover, because we have already found that Sall1 binds to Nanog enhancer, which is a target of Nanog and Sox2, we expanded this analysis to Nanog-regulated genes to investigate whether the two factors share common target genes. For the ChIP-on-chip analysis, we used the Affymetrix GeneChip® mouse promoter 1.0R array, which covers a region 6 kb upstream and 2.5 kb downstream of the gene transcription start site of each gene. Experiments were done in duplicate, and analysis was performed using the Affymetrix web tools and the MAT program. Antibodies used were specific for the proteins (supplemental Fig. S1). As a negative control, we used an antibody against Fras, a cytoplasmic protein that is not expressed in mESC.
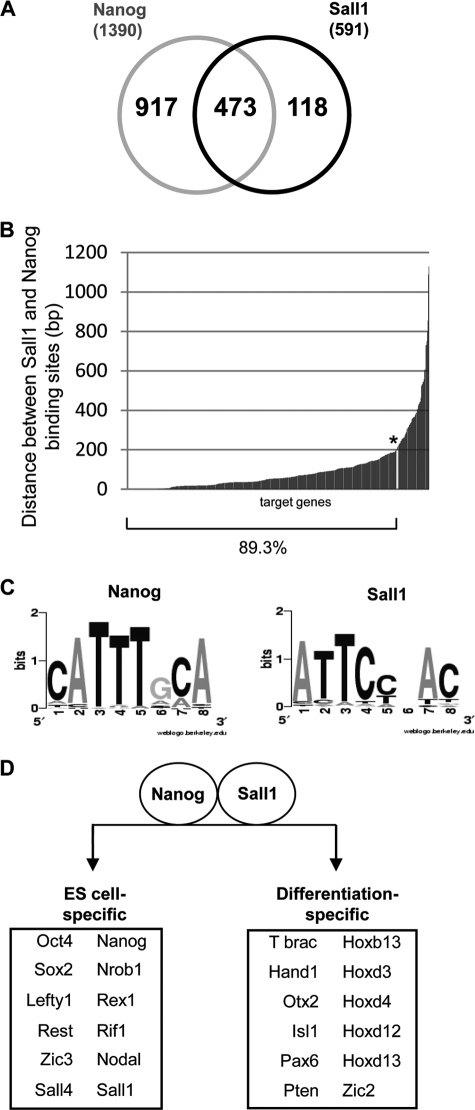
The microarray analysis revealed 591 putative target genes for Sall1 and 1390 target genes for Nanog, 473 of which were common for both factors (Fig. 4A and supplemental Table 1). The microarray data for 13 pluripotency and differentiation genes (supplemental Fig. S2) were validated by chromatin immunoprecipitation and real-time PCR experiments (supplemental Fig. S3). We tested the expression status of Nanog and Sall1 target genes based on our previous publications (6, 27) and found that they included both transcriptionally active and transcriptionally inactive genes. Human Nanog, as well as Oct4 and Sox2, have also been found to bind on promoters of transcriptionally active and inactive genes (2, 22). It seems that Sall1 follows the same binding pattern with these factors. To test whether the corresponding Sall1- and Nanog-binding sites of the common 473 target genes are located in close proximity, we analyzed the distribution of distances between the two binding sites. As shown in Fig. 4B, in 89% of the common target genes, Nanog-and Sall1-binding sites are located within 200 bp. This strongly suggests that the two factors co-exist on most gene promoters, possibly within the same protein complex. In support of this idea, we have found that both Sall1 and Nanog bind to Nanog enhancer (Fig. 1C), and this observation is in accordance with the microarray data where we also detected the binding of both factors in the same region. For the latter experiment, the distance between Sall1- and Nanog-binding sites is 17 bp, a finding that implies that Sall1 is part of the complex found on Nanog enhancer that also contains Nanog itself.
FIGURE 4.
Gene targets of Sall1 and Nanog detected by ChIP-on-chip analysis. A, diagram of Sall1 and Nanog target genes identified by Chip-on-chip analysis. B, distribution of Nanog and Sall1 binding distances on common target genes. The various distances between Nanog- and Sall1-binding sites on the 473 common target genes were calculated and plotted. The y axis represents the distance between the binding sites in base pairs. C, consensus binding motifs for Nanog and Sall1 identified using the Weeder algorithm. D, selected Sall1 and Nanog common targets expressed in ES cells or during differentiation.
We next analyzed the sequences of the top 50 Nanog and Sall1 target genes by using the Weeder algorithm (28) to identify a potential consensus binding site. In accordance with the motif previously predicted by Loh et al. (3), the program identified a CATT-containing motif (Fig. 4C and supplemental Fig. S4) for Nanog binding. When we analyzed the top 50 Sall1 target genes, we obtained a consensus containing the motif ATTCC shown in Fig. 4C (see also supplemental Fig. S5) that is different from the AT-rich sequence previously reported for Sall1 binding to heterochromatin major satellite DNA (29).
Functional categorization of the target genes was done by analyzing the Gene Ontology (GO) terms obtained after using the DAVID web tool (david.abcc.ncifcrf.gov). Within the 473 common target genes, which belong in both expressed and non-expressed classes, the most important biological process categories were related to metabolism, transcription, embryonic development, differentiation, and stem cell maintenance (Table 1). Interestingly, many pluripotency regulators such as Nanog, Oct4, Sox2 Sall4, Nr0b1, and Sall1 itself and many differentiation-related genes such as Hox genes, T brachyury, Isl1, Hand1, and Otx2 (Fig. 4D) are included in the aforementioned categories, suggesting that Sall1 is part of an elaborate network of cross-regulated factors. Moreover, Sall1 and Nanog target genes are also related to organ (gland, lung, heart, brain, ear, liver, and kidney) and tissue (nervous, respiratory, endocrine, exocrine, urogenital, and skeletal) development and differentiation along the trophectodermal, mesodermal, and endodermal lineages (Table 1).
TABLE 1.
Functional categorization (biological process) of Sall1and Nanog common target genes
The number of genes and the statistical values are shown for each category.
| Biological Process | Genes | p value |
|---|---|---|
| System development | 87 | 2.53 × 10−8 |
| Nervous | 47 | 9.04 × 10−8 |
| Respiratory | 14 | 3.09 × 10−6 |
| Endocrine | 10 | 4.54 × 10−5 |
| Exocrine | 5 | 4.25 × 10−3 |
| Urogenital | 8 | 2.73 × 10−2 |
| Skeletal | 6 | 4.49 × 10−2 |
| Organ development | 72 | 4.67 × 10−7 |
| Gland | 17 | 3.58 × 10−7 |
| Lung | 12 | 3.73 × 10−5 |
| Heart | 16 | 2.69 × 10−4 |
| Brain | 16 | 2.71 × 10−3 |
| Inner ear | 8 | 4.27 × 10−3 |
| Liver | 5 | 1.71 × 10−2 |
| Metanephros | 5 | 3.64 × 10−2 |
| Tissue development | 37 | 1.45 × 10−6 |
| Epithelium | 18 | 2.71 × 10−6 |
| Transcription | 102 | 8.30 × 10−11 |
| Embryonic development | 44 | 2.72 × 10−10 |
| Gene expression | 121 | 9.53 × 10−9 |
| Chromatin modification | 13 | 9.04 × 10−3 |
| Stem cell maintenance | 5 | 1.66 × 10−3 |
| Stem cell development | 5 | 2.08 × 10−3 |
| Cell differentiation | 69 | 9.39 × 10−7 |
| Cell fate commitment | 16 | 1.38 × 10−6 |
| Cell development | 42 | 2.57 × 10−6 |
| Cell proliferation | 32 | 8.17 × 10−4 |
| Cell death | 36 | 1.29 × 10−3 |
| Cell cycle | 26 | 4.54 × 10−2 |
| Gastrulation | 11 | 1.08 × 10−5 |
| Blastocyst formation | 6 | 1.16 × 10−4 |
| Trophectodermal cell differentiation | 6 | 3.53 × 10−5 |
| Mesodermal differentiation | 4 | 4.03 × 10−4 |
| Endodermal differentiation | 2 | 6.94 × 10−2 |
Sall1 Suppresses Differentiation
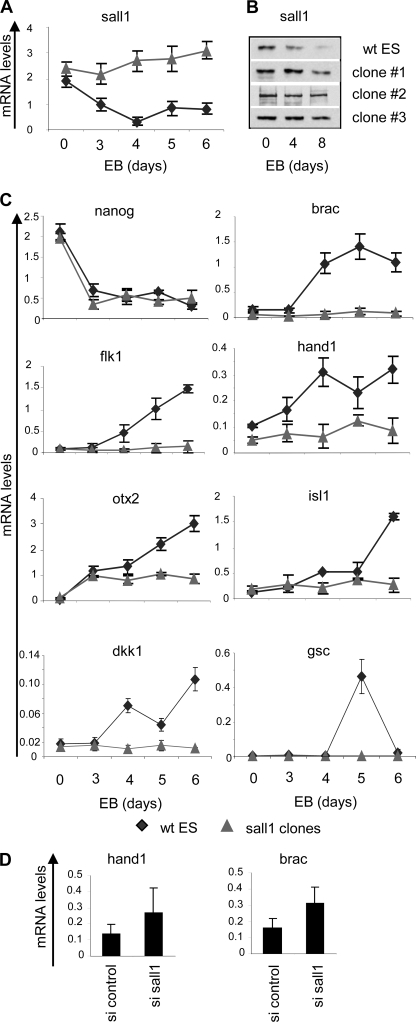
Because Sall1 target genes are both transcriptionally active and transcriptionally inactive genes, we examined whether it functions as an activator or repressor of gene expression in ES cells. For this purpose, we have generated ES clones stably expressing Sall1 protein and induced differentiation via embryoid body (EB) formation. Sall1 mRNA and protein levels remained stable during this process (Fig. 5, A and B). We then analyzed mRNA levels of various genes at days 0, 3, 4, 5, and 6 of differentiation to study the potential effect of Sall1 on their expression. Because a large number of Sall1 target genes encode for pluripotency factors, we first analyzed mRNA levels of Nanog, Oct4, and Nr0b1. All three genes were down-regulated in a way similar to the control ES cells (Fig. 5C and supplemental Fig. S5), showing that gain of Sall1 function cannot maintain these genes in the active state during ES cell differentiation.
FIGURE 5.
Sall1 suppresses differentiation. Sall1 mRNA (A) and protein levels (B) are decreased in WT ES, whereas they remain unaltered in clones stably expressing Sall1 (clones 1, 2, and 3) during EB formation. C, mRNA levels of selected genes that are up-regulated during EB formation from WT ES (⧫) but not from clones stably expressing Sall1 (▴). Data were obtained from three independent clones. D, Hand1 and T brachyury mRNA levels are increased after silencing of Sall1. si, siRNA. Error bars in panels A, C, and D indicate S.D.
Next, we studied the expression of transcriptionally inactive genes of all three germ layers that are normally induced during EB differentiation. Interestingly, we found that the activation of the mesodermal markers T-brac, Gsc, Flk-1, and Dkk was totally impaired in EBs expressing Sall1 (Fig. 5C). T-brac and Gsc are early mesodermal (gastrulation) markers, whereas Flk1 marks mesodermal and then hematopoietic and endothelial lineages. The fact that Flk-1 was not part of the Sall1 target list implies that it is possibly affected indirectly through a direct Sall1 target. In addition, the trophectodermal factor Otx2 and two genes involved in both cardiac and neuronal development, Hand1 and Isl-1, were not induced in the Sall1-expressing EBs. Isl1 and Hand1 are cardiogenic genes and appear later during cardiac mesoderm patterning. On the contrary, endodermal differentiation was not affected, as exemplified by the induction of Sox17 and Gata 6 (supplemental Fig. S5). The above data were obtained from three independent clones (Fig. 5B) and collectively suggest that maintenance of Sall1 expression during EB differentiation is not sufficient to sustain expression of pluripotency factors but can prevent certain differentiation markers from being expressed, especially those of mesodermal and ectodermal lineage. A genome-wide expression analysis under these experimental conditions would clarify the differentiation pathways that are blocked by Sall1.
Because overexpression of Sall1 during differentiation prevents the expression of the above genes, we tested whether its silencing in the undifferentiated state would induce their expression. After knocking down Sall1, we observed a 2-fold up-regulation of T-brac and Hand1 (Fig. 5D), whereas the other examined genes did not show any changes in their expression levels (not shown). It is possible that loss of Sall1 alone is not sufficient for the derepression of these genes or that the presence of Sall4 could compensate for the loss of Sall1.
DISCUSSION
The pluripotent state of embryonic stem cells is maintained by a complex transcriptional regulatory network that sustains the undifferentiated cell functions while silencing the differentiation-specific genes. This network contains a growing list of transcription factors, chromatin modifiers, and microRNAs (30). The multiplicity of interaction possibilities among different factors, the cross- and co-regulation mechanisms, and the resulting functional redundancies endow the system with stability and sensitivity (31).
In the present study, we examined the role of Sall1, a member of a small multi-zinc finger transcription factor family, in ES cells. The rapid down-regulation of Sall1 during ES cell differentiation (6) led to the hypothesis that it may play a role in the maintenance of the undifferentiated state. In agreement with this hypothesis, we have shown that silencing of Sall1 leads to down-regulation of Nanog promoter activity and mRNA levels, whereas Oct4 and Sox2 expression levels were not affected. Using chromatin immunoprecipitation experiments, we have detected Sall1 binding to the Nanog enhancer. Therefore, Sall1 is a member of the group of Nanog-positive regulators along with Sall4 (32), Stat3 (33), Zfp143 (34), Zfp281 (35), Zic3 (36), and Klf4 (37). The redundant function of these proteins is counteracted by Nanog-negative regulators Tcf3 (38) and p53 (39).
Protein-protein interactions are critical for stabilization of transcription factor complexes on target gene chromatin. Oct4, Sox2, and Nanog reside at the core of an intricate protein interaction network that operates in embryonic stem cells and also includes Sall1 (23). In this report, we have shown that Sall1 physically interacts with both Nanog and Sox2 and have dissected in vitro the domains involved. Sall1 interacts with the carboxyl-terminal domain of Nanog that is involved in transcriptional activation, homodimerization, and the promotion of stem cell pluripotency (40). Similarly, the Sall1-interacting region of Sox2 was found to be the carboxyl-terminal domain that is required for transcriptional activation (25). Multiple domains of Sall1 residing at the amino-, central, and carboxyl-terminal part are involved in interactions with Nanog and Sox2, suggesting that Sall1 might bind simultaneously to both factors.
One approach to answer the question how Sall1 functions in ES cells was to map the gene loci where Sall1 binds using the ChIP-on-chip methodology. Our analysis has shown that Sall1 binds to DNA loci that harbor the consensus site ATTCCNAC. This motif is different from the AT-rich major satellite sequences where Sall1 has been previously reported to bind (29). Therefore, it is likely that Sall1 is a DNA-binding factor of dual specificity. One form is located in heterochromatin and binds to AT-rich sequences (29, 41), whereas another form is recruited to euchromatic gene promoter areas and binds to the ATTCCNAC consensus. Despite previous reports stating that Sall1 is involved in transcriptional repression (19, 26), its target genes in undifferentiated embryonic stem cells contained approximately equal numbers of silenced and active genes. Most importantly, 80% of Sall1 targets were also bound by Nanog, and in 89% of the regions, the two factors were located less than 200 bp apart. Hence Sall1, in agreement with an earlier report (4), has the property of binding in close proximity to Nanog. Transcriptionally active target genes included known pluripotency transcription factors (Nanog, Oct4, Nr0b1, and Sall4) and Sall1 itself, whereas inactive genes were mostly associated with differentiation and development. To gather insight into the functional role of Sall1 in ESC, we performed Gene Ontology (GO) analysis on the 473 common Sall1 and Nanog targets. In addition to transcriptional regulators, we detected lineage-specific genes that participate in the development of the endocrine, circulatory, muscle, nervous, and skeletal systems and of organs such as gland, heart, and brain. Overall our results show that Sall1 acts in a context-dependent manner either as a gene activator or as a suppressor. These properties may thus account for its previously reported functions in tissue development and homeostasis in normal or diseased cells (10, 11, 41).
In line with the above, forced expression of Sall1 during embryoid body formation was able to prevent the up-regulation of mesodermal and ectodermal but not endodermal differentiation markers. This effect reveals the ability of Sall1 to inhibit multiple differentiation pathways similarly to other factors such as Nr0b1(Dax1) (43), Zfp281 (35), and Zfp143 (34). Our analysis provides evidence that Sall1, besides its known function in kidney development (43), also affects the induction of neuronal and cardiogenic differentiation markers in ES. These data are in agreement with the expression of Sall1 in the developing mouse heart (42, 45), the presence of heart defects in Townes-Brocks syndrome (46), and the involvement of Sall proteins in neuronal development (45, 47). Because the vast majority of Sall1 targets are common with Nanog and because these factors bind in close proximity, we suggest that Sall1 functions in ES cells in association with Nanog.
Sall4 was the first spalt gene family member found to be involved in the regulation of embryonic stem cell pluripotency. Sall4 was reported to interact with and regulate the expression of Nanog (32) and preserve ESC pluripotency (16). Following the identification of its target genes in embryonic stem cells (48, 49), Sall4 has been included as a member of the Oct4, Sox2, and Nanog interconnected regulatory circuit. Yuri et al. (50) have examined a double Sall1/Sall4 knock-out ES cell line, and although they reported that Nanog expression was reduced in the double KO but not in the single Sall4 KO, they concluded that Sall1 had a minimal effect. We have compared Sall1 (this report) and Sall4 (48) ChIP-on-chip data and found ∼30% overlap in target genes. Thus despite their heterodimerization (17), Sall1 and Sall4 have non-redundant functions in embryonic stem cells and target both distinct and common gene groups.
The molecular mechanism of transcriptional repression exerted by pluripotency factors is poorly understood because the majority of them are transcriptional activators. Both Sall1 and Sall4 contain at their amino-terminal ends short homologous protein domains that recruit NuRD (19, 51). Thus Sall1 and Sall4 can assist Nanog (52) and other pluripotency factors in the silencing of differentiation genes. Despite the ability of Sall1 to activate gene expression observed in this and previous reports (12, 20, 21), it does not contain a transcription activation domain. Thus it is likely that activation or repression by Sall1 is determined by signal-regulated switches and/or by the presence of distinct promoter-specific factors.
Collectively, this work presents novel functions of Sall1 in mouse embryonic stem cells. Sall1 is a partner of Nanog and Sox2 that is recruited on promoters of both active and silenced genes. The repertoire of Sall1 target genes includes new signaling and development-related genes that are interesting candidates for future investigations (53). Sall1 regulates the expression of Nanog and may cooperate with it in transcriptional regulation. In addition, Sall1 suppresses mesodermal and ectodermal differentiation. These findings suggest that Sall1 is a novel component of stemness. Taking into account that Sall1 mutations cause tissue-specific defects in human patients, it will be important to investigate the role of Sall1 in tissue repair and regeneration after injury.
Supplementary Material
Acknowledgments
We are grateful to G. Chalepakis for helping produce the rabbit α-Nanog and α-Sall1 antibodies. We thank Chrysa Deligianni for the Oct4-GFP construct, Anna Vardi for GST-Sall1 constructs, P. Savatier for Nanog-expressing plasmid, and E. Remboutsika for CMV-Sox2. We thank V. Makatounakis and G. Vretzos for excellent technical assistance. Special thanks to Hedi Peterson and Jaak Vilo for the help with the analysis of ChIP-on-chip data. We thank A. K. Hatzopoulos and C. Spilianakis for critical reading of the manuscript and G. Papagiannakis and G. Garinis for assistance to the Institute of Molecular Biology and Biotechnology (IMBB) Affymetrix facility.
This work was supported by the European Union FP6 I. P. FunGenES Grant LSHG-CT 2003-503494 and IMBB intramural funding (to A. K.).

The on-line version of this article (available at http://www.jbc.org) contains primers, supplemental Table S1, and Figs. S1–S5.
The data sets reported in this paper have been deposited in the GEO database (GSE 25523).
- mESC
- mouse embryonic stem cell
- EB
- embryoid body
- m
- mouse
- aa
- amino acids.
REFERENCES
- 1. Boiani M., Schöler H. R. (2005) Nat. Rev. Mol. Cell Biol. 6, 872–884 [DOI] [PubMed] [Google Scholar]
- 2. Boyer L. A., Mathur D., Jaenisch R. (2006) Curr. Opin. Genet. Dev. 16, 455–462 [DOI] [PubMed] [Google Scholar]
- 3. Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H. (2006) Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- 4. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 5. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karantzali E., Schulz H., Hummel O., Hubner N., Hatzopoulos A., Kretsovali A. (2008) Genome Biol. 9, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kühnlein R. P., Brönner G., Taubert H., Schuh R. (1997) Mech. Dev. 66, 107–118 [DOI] [PubMed] [Google Scholar]
- 8. de Celis J. F., Barrio R., Kafatos F. C. (1996) Nature 381, 421–424 [DOI] [PubMed] [Google Scholar]
- 9. de Celis J. F., Barrio R., Kafatos F. C. (1999) Development 126, 2653–2662 [DOI] [PubMed] [Google Scholar]
- 10. Sweetman D., Münsterberg A. (2006) Dev. Biol. 293, 285–293 [DOI] [PubMed] [Google Scholar]
- 11. de Celis J. F., Barrio R. (2009) Int. J. Dev. Biol. 53, 1385–1398 [DOI] [PubMed] [Google Scholar]
- 12. Nishinakamura R., Matsumoto Y., Nakao K., Nakamura K., Sato A., Copeland N. G., Gilbert D. J., Jenkins N. A., Scully S., Lacey D. L., Katsuki M., Asashima M., Yokota T. (2001) Development 128, 3105–3115 [DOI] [PubMed] [Google Scholar]
- 13. Kohlhase J., Wischermann A., Reichenbach H., Froster U., Engel W. (1998) Nat. Genet. 18, 81–83 [DOI] [PubMed] [Google Scholar]
- 14. Kiefer S. M., Ohlemiller K. K., Yang J., McDill B. W., Kohlhase J., Rauchman M. (2003) Hum. Mol. Genet. 12, 2221–2227 [DOI] [PubMed] [Google Scholar]
- 15. Elling U., Klasen C., Eisenberger T., Anlag K., Treier M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16319–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 17. Sakaki-Yumoto M., Kobayashi C., Sato A., Fujimura S., Matsumoto Y., Takasato M., Kodama T., Aburatani H., Asashima M., Yoshida N., Nishinakamura R. (2006) Development 133, 3005–3013 [DOI] [PubMed] [Google Scholar]
- 18. Kiefer S. M., McDill B. W., Yang J., Rauchman M. (2002) J. Biol. Chem. 277, 14869–14876 [DOI] [PubMed] [Google Scholar]
- 19. Lauberth S. M., Rauchman M. (2006) J. Biol. Chem. 281, 23922–23931 [DOI] [PubMed] [Google Scholar]
- 20. Sato A., Kishida S., Tanaka T., Kikuchi A., Kodama T., Asashima M., Nishinakamura R. (2004) Biochem. Biophys. Res. Commun. 319, 103–113 [DOI] [PubMed] [Google Scholar]
- 21. Yamamoto C., Fukuda N., Matsumoto T., Higuchi T., Ueno T., Matsumoto K. (2010) Hypertens Res. 33, 143–148 [DOI] [PubMed] [Google Scholar]
- 22. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006) Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 24. Pan G., Pei D. (2005) J. Biol. Chem. 280, 1401–1407 [DOI] [PubMed] [Google Scholar]
- 25. Nowling T. K., Johnson L. R., Wiebe M. S., Rizzino A. (2000) J. Biol. Chem. 275, 3810–3818 [DOI] [PubMed] [Google Scholar]
- 26. Lauberth S. M., Bilyeu A. C., Firulli B. A., Kroll K. L., Rauchman M. (2007) J. Biol. Chem. 282, 34858–34868 [DOI] [PubMed] [Google Scholar]
- 27. Schulz H., Kolde R., Adler P., Aksoy I., Anastassiadis K., Bader M., Billon N., Boeuf H., Bourillot P. Y., Buchholz F., Dani C., Doss M. X., Forrester L., Gitton M., Henrique D., Hescheler J., Himmelbauer H., Hübner N., Karantzali E., Kretsovali A., Lubitz S., Pradier L., Rai M., Reimand J., Rolletschek A., Sachinidis A., Savatier P., Stewart F., Storm M. P., Trouillas M., Vilo J., Welham M. J., Winkler J., Wobus A. M., Hatzopoulos A. K. (2009) PLoS One 4, e6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pavesi G., Mauri G., Pesole G. (2001) Bioinformatics 17, Suppl. 1, S207–S214 [DOI] [PubMed] [Google Scholar]
- 29. Yamashita K., Sato A., Asashima M., Wang P. C., Nishinakamura R. (2007) Genes Cells 12, 171–182 [DOI] [PubMed] [Google Scholar]
- 30. Do J. T., Schöler H. R. (2009) Trends Pharmacol. Sci. 30, 296–302 [DOI] [PubMed] [Google Scholar]
- 31. Ng J. H., Heng J. C., Loh Y. H., Ng H. H. (2008) Mutat. Res. 647, 52–58 [DOI] [PubMed] [Google Scholar]
- 32. Wu Q., Chen X., Zhang J., Loh Y. H., Low T. Y., Zhang W., Zhang W., Sze S. K., Lim B., Ng H. H. (2006) J. Biol. Chem. 281, 24090–24094 [DOI] [PubMed] [Google Scholar]
- 33. Suzuki A., Raya A., Kawakami Y., Morita M., Matsui T., Nakashima K., Gage F. H., Rodríguez-Esteban C., Izpisúa Belmonte J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X., Fang F., Liou Y. C., Ng H. H. (2008) Stem Cells 26, 2759–2767 [DOI] [PubMed] [Google Scholar]
- 35. Wang Z. X., Teh C. H., Chan C. M., Chu C., Rossbach M., Kunarso G., Allapitchay T. B., Wong K. Y., Stanton L. W. (2008) Stem Cells 26, 2791–2799 [DOI] [PubMed] [Google Scholar]
- 36. Lim L. S., Hong F. H., Kunarso G., Stanton L. W. (2010) Stem Cells 28, 1961–1969 [DOI] [PubMed] [Google Scholar]
- 37. Zhang P., Andrianakos R., Yang Y., Liu C., Lu W. (2010) J. Biol. Chem. 285, 9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cole M. F., Johnstone S. E., Newman J. J., Kagey M. H., Young R. A. (2008) Genes Dev. 22, 746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin T., Chao C., Saito S., Mazur S. J., Murphy M. E., Appella E., Xu Y. (2005) Nat. Cell Biol. 7, 165–171 [DOI] [PubMed] [Google Scholar]
- 40. Wang J., Levasseur D. N., Orkin S. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6326–6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Netzer C., Bohlander S. K., Hinzke M., Chen Y., Kohlhase J. (2006) Biochim. Biophys. Acta 1762, 386–391 [DOI] [PubMed] [Google Scholar]
- 42. Buck A., Kispert A., Kohlhase J. (2001) Mech. Dev. 104, 143–146 [DOI] [PubMed] [Google Scholar]
- 43. Khalfallah O., Rouleau M., Barbry P., Bardoni B., Lalli E. (2009) Stem Cells 27, 1529–1537 [DOI] [PubMed] [Google Scholar]
- 44. Nishinakamura R., Osafune K. (2006) J. Physiol. Sci. 56, 131–136 [DOI] [PubMed] [Google Scholar]
- 45. Ott T., Parrish M., Bond K., Schwaeger-Nickolenko A., Monaghan A. P. (2001) Mech. Dev. 101, 203–207 [DOI] [PubMed] [Google Scholar]
- 46. Kohlhase J., Liebers M., Backe J., Baumann-Müller A., Bembea M., Destrée A., Gattas M., Grüssner S., Müller T., Mortier G., Skrypnyk C., Yano S., Wirbelauer J., Michaelis R. C. (2003) J. Med. Genet. 40, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Böhm J., Buck A., Borozdin W., Mannan A. U., Matysiak-Scholze U., Adham I., Schulz-Schaeffer W., Floss T., Wurst W., Kohlhase J., Barrionuevo F. (2008) Am. J. Pathol. 173, 1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lim C. Y., Tam W. L., Zhang J., Ang H. S., Jia H., Lipovich L., Ng H. H., Wei C. L., Sung W. K., Robson P., Yang H., Lim B. (2008) Cell Stem Cell 3, 543–554 [DOI] [PubMed] [Google Scholar]
- 49. Yang J., Chai L., Fowles T. C., Alipio Z., Xu D., Fink L. M., Ward D. C., Ma Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19756–19761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuri S., Fujimura S., Nimura K., Takeda N., Toyooka Y., Fujimura Y., Aburatani H., Ura K., Koseki H., Niwa H., Nishinakamura R. (2009) Stem Cells 27, 796–805 [DOI] [PubMed] [Google Scholar]
- 51. Lu J., Jeong H. W., Kong N., Yang Y., Carroll J., Luo H. R., Silberstein L. E., Yupoma, Chai L. (2009) PLoS One 4, e5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 53. Inoue S., Inoue M., Fujimura S., Nishinakamura R. (2010) Genesis 48, 207–212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.