Abstract
Tumor necrosis factor (TNF) induces expression of granulocyte-macrophage colony-stimulating factor (GM-CSF) but lymphotoxin β (LTβ) does not. Here we report that priming of cells with agonistic LTβ receptor antibody synergistically enhanced TNF-induced GM-CSF expression. The LTβ priming process was not due to an increase in TNF-mediated nuclear translocation of p65, p65 DNA binding, or NF-κB transactivational activity. The synergistic effect of LTβ priming was not observed with other TNF-responsive genes such as Ccl2 or RelB, which suggested that this effect was not a general increase in TNF signaling. Furthermore, RelB and p65 were both independently recruited to the GM-CSF promoter when cells were primed with LTβ followed by TNF treatment. As a consequence, an increase in both chromatin accessibility and the recruitment of RNA polymerase II were observed to the GM-CSF promoter. Taken together, these findings suggested that LTβ signaling amplified TNF-mediated GM-CSF expression by facilitating chromatin access and the co-recruitment of RNA polymerase II to increase gene transcription. Moreover, the novel priming process described here underscores the complexity of the interactions between the classical and alternative NF-κB signaling pathways.
Keywords: Chromatin, Cytokines Induction, Gene Regulation, NF-κB Transcription Factor, Tumor Necrosis Factor (TNF)
Introduction
The activation of the NF-κB proteins can be through at least two distinct pathways (1, 2). The classical NF-κB pathway involves the activation of the IκB kinase (IKK)2 complex following receptor-ligand interaction. This complex contains three different kinases; IKKα, -β, and -γ, and a major substrate of this kinase complex is IκBα (3). The phosphorylation of IκBα results in its ubiquitinylation and degradation. The p65(RelA)/p50 heterodimer is thereby released to translocate into the nucleus to induce gene transcription. In contrast, the activation of the alternative NF-κB signaling pathway does not involve the IκB kinase complex. A major step in the initiation of the alternative pathway is the activation of the NF-κB-inducing kinase (NIK) (4). The NIK kinase phosphorylates the IKKα kinase, which induces the phosphorylation of p100. The phosphorylation of p100 initiates processing where the ankyrin repeats of p100 are removed to generate p52 and exposes the nuclear localization sequences of RelB. The RelB/p52 complex translocates into the nucleus to bind various promoters and facilitates gene transcription. Several factors distinguish the two pathways. The classical pathway requires IKKγ, the degradation of IκBα, and primarily results in the nuclear translocation of p65/p50. In contrast, the alternative pathway involves the activation of NIK, the processing of p100, and the nuclear translocation of RelB/p52. Although several NF-κB responsive genes are activated by both pathways, numerous genes are uniquely activated by only one pathway (2, 5). Both pathways involve the degradation or processing of inhibitor proteins and the nuclear translocation of the NF-κB transcription factors.
Several studies have shown crosstalk between the two pathways. In some cases, the interaction between the two pathways resulted in a negative response (6). In particular the formation of interchangable dimeric partners has been described with the activation of both pathways. Marienfeld et al. (7) have shown that the formation of a RelA(p65)/RelB heterodimer resulted in a negative response where the RelB protein was sequestering and inhibiting p65 from binding to DNA. Conversely, other studies have shown the formation of the RelA/RelB heterodimer leading to increased gene transcription (8). Recent studies have shown that the priming of cells with tumor necrosis factor (TNF), which activates the classical NF-κB pathway, resulted in the expression of p100 (9). The p100 then acts as a fourth IκB protein; it binds and sequesters the TNF-induced p65/p50 heterodimer by forming a trimolecular complex of p100/RelA/p50 (9). If the alternative pathway is activated through the lymphotoxin β (LTβ) receptor in the TNF-primed cells, the processing of p100 results in the release of the p65/p50 to induce gene transcription. The gene expression profile of the TNF-primed/LTβ-activated cells resembles the gene expression profile of TNF-treated cells. Thus, the two NF-κB pathways may interact with each other to influence either positively or negatively the transcription of NF-κB-responsive genes.
We have previously shown that the phosphorylation of p65 at serine 536 resulted in an increase in GM-CSF gene (Csf2) expression (10). The phosphorylation was responsible for the co-recruitment of p300 and RNA polymerase II to the proximal site of the Csf2 gene promoter. Jiang et al. (11) have shown that the phosphorylation of p65 at serine 536 by IKKα was induced by LTβR in mouse fibroblast cells. Based on these findings, we wondered whether signaling through the LTβR could induce the transcription of Csf2 gene as seen with TNF. Here we have shown that the treatment of 3T3 fibroblast cells with agonistic LTβR mAb resulted in the phosphorylation of p65 on serine 536; however, this was not sufficient to induce the expression of the Csf2 gene. Priming the cells with LTβR mAb resulted in a synergistic increase of TNF-mediated Csf2 expression. The synergistic enhancement required the activation of NIK and signaling through the alternative NF-κB pathway. Furthermore, nuclear translocation and recruitment of both p65 and RelB to the Csf2 promoter were observed during the LTβR priming of TNF-mediated Csf2 gene expression. These findings suggested that RelB binding to the Csf2 promoter synergistically enhances subsequent phospho-p65-driven gene expression.
EXPERIMENTAL PROCEDURES
Cell Culture Condition (Priming Protocol)
NIH 3T3 cells were maintained in DMEM medium supplemented with 10% FBS, l-glutamine, and antibiotics as described elsewhere. For the priming experiments, cells were treated with the priming agent for the indicated time interval. The cells were then washed with PBS to remove the priming agent and then treated with either TNF (25 ng/ml, Peprotech) or agonistic mouse LTβR mAb (0.5 μg/ml, Axxora) for the indicated time interval. In some cases, the cells were pretreated with cycloheximide (CHX) (1 μg/ml, Sigma) for 2 h before priming or treatment. The suppression of Map3k14 and Relb gene expression involved the transfection of NIH 3T3 cells with the Map3k14 and RelB shRNAmir plasmids (OpenBiosystems) with the transfection reagent FuGENE 6 (LaRoche). Stable transfectants were obtained through puromycin selection and confirmed by semi-quantitative real-time qPCR. The MAP3K14 expression plasmid was purchased from OpenBiosystems.
Reporter Plasmid Construction and Transfections
The NF-κB responsive region of the Csf2 promoter (12) was synthesized by ligating various oligonucleotides as described by Rouillard et al. (13) (see supplemental Table S1 for primer sequences). The synthesized gene was then directionally subcloned into the pSEAP2-basic reporter plasmid (Clontech). Cells were transfected with either the pSEAP2-NFκB or pSEAP2-Csf2 reporter constructs with FuGENE6 transfection reagent (LaRoche). After an overnight incubation, the cells were treated with the indicated cytokines. On the following day, the culture supernatant was assessed for SEAP activity (source), and the cells were lysed and β-galactosidase activity was measured (source). The transfection efficiency was assessed with the addition of pCMV-βgal plasmid during the transfection process.
p65 DNA Binding Assay
The p65 DNA binding assay was performed according to the protocol provided by the manufacturer (Active Motif). Two micrograms of nuclear extracts from cells treated with the indicated cytokines were analyzed with the kit.
Immunoblot and Immunoprecipitation
Immunoblot and immunoprecipitation analyses were performed as previously described (14). 10 or 5 μg of whole cell lysate or subcellular (cytoplasmic/nuclear) extracts were separated by PAGE and transferred to PVDF membranes. Membranes were blocked and blotted with various antibodies either individually or in combination (see supplemental Table S1 for antibody information). The immune complexes were detected with enhanced chemifluorescence and visualized with the Typhoon scanner (GE Health Science).
For immunoprecipitation experiments with NIK antibody, 1 mg of total protein for each sample was precleared with 40 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) for 1 h at 4 °C. The precleared samples were incubated with 1 μg of NIK antibody (Santa Cruz Biotechnology) overnight at 4 °C. The immunocomplexes were precipitated with protein A/G PLUS-agarose beads and eluted with 1× NuPAGE sample buffer (Invitrogen). Immunoblot analyses with the indicated antibody were conducted to detect the presence of specific proteins in the immunoprecipitated samples.
Semi-quantitative RT-qPCR
Total cellular RNA was isolated using the RNeasy kit (Qiagen), and cDNA was generated using VILO cDNA synthesis kit (Invitrogen). Semi-quantitative real-time PCR was performed as previously described (14). The primer sequences are listed in the supplemental Table S1 (15).
ChIP Assay
Chromatin immunoprecipitation reaction was performed according to the following protocol (16) with a few modifications. Chromatin samples were sonicated four times for 10 s at 40 V on ice. Sheared chromatin samples were precleared with 40 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology). 200 μg of sonicated chromatin was incubated with 2 μg of the indicated antibodies (see supplemental Table S1) overnight at 4 °C. The analysis of fold enrichment was measured by real-time PCR using a previously described method (17). The immunodepletion ChIP assay was conducted according to the following protocol (14).
Chromatin Accessibility by Real-time PCR (CHART) Assay
The CHART assay was performed as previously described (18) with the following modifications. Five million nuclei from treated 3T3 cells were isolated and digested with 100 units of micrococcal nuclease (MNase) (New England Biolab) for 5 min at room temperature. Genomic DNA was isolated with the DNeasy kit (Qiagen), and 36 ng of DNA was used for SYBR green real-time PCR (Bio-Rad). The primer sequences are listed in supplemental Table S2. The endogenous endonuclease activity was assessed with a control that was not treated with micrococcal nuclease for each treatment. The percent MNase cutting was determined as a percentage of digested versus undigested genomic DNA of each sample by converting the threshold cycle values of the amplification plot to a standard curve generated from with the cloned genomic DNA. The percent accessibility was expressed as follows: percent accessibility = 100 − percent MNase cutting.
RESULTS
p65 Phosphorylation with LTβR mAb Treatment
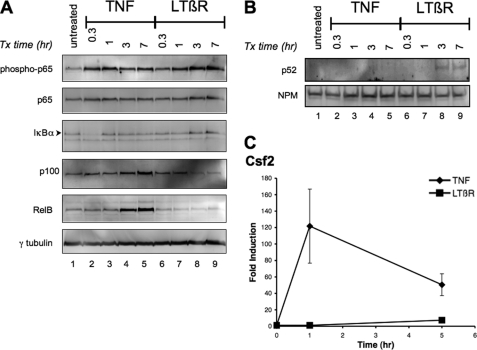
Previously we have shown that the phosphorylation of p65 at serine 536 was required for the induction of TNF-mediated GM-CSF expression (10). Furthermore, Jiang et al. (11) have demonstrated that the phosphorylation of p65 at serine 536 by LTβ receptor (LTβR) signaling involved the NIK-IKKα cascade. To investigate the induction of GM-CSF by LTβR signaling, mouse 3T3 fibroblast cells were treated with either TNF or agonistic LTβR mAb for different time intervals. The rapid phosphorylation of p65 was observed within an hour of TNF treatment (Fig. 1A, lanes 1–5). Likewise, the degradation and resynthesis of IκBα occurred during the same time interval. TNF treatment for 3 h also resulted in an increase in p100 and RelB levels. This was in agreement with previous studies that described the degradation and resynthesis of IκBα and the induction of p100 and RelB by the classical NF-κB signaling pathway following TNF treatment (19). In contrast, a reduction of p100 was observed 3 h after the treatment with agonistic LTβR mAb (compare lanes 1 and 6–9) with a corresponding increase in nuclear p52 (Fig. 1B, lanes 8 and 9). The treatment with LTβR mAb did not affect the amount of IκBα or RelB. These findings were consistent with the activation of the alternative NF-κB signaling pathway where p100 is processed to generate p52, which translocates into the nucleus (20). In contrast to TNF stimulation, a slower gradual increase in phosphorylated p65 was detected following LTβR activation. The results demonstrated that the activation of either the classical (TNF) or the alternative (LTβR) pathway induced the phosphorylation of p65. In contrast to the phosphorylation of p65, TNF, but not LTβR mAb, treatment increased the amount of Csf2 transcripts (Fig. 1C). The kinetics of TNF-mediated Csf2 expression matched that of the phosphorylation of p65. Even with a longer incubation time, the amount of Csf2 induction by LTβR activation did not increase to the level seen with TNF treatment. Although the treatment with the agonistic LTβR mAb resulted in an increase in the phosphorylation of p65, a lack of Csf2 induction was observed. These findings suggested that the phosphorylation of p65 was not sufficient for the induction of Csf2 transcription by LTβR signaling.
FIGURE 1.
Kinetics of p65 phosphorylation and the induction of Csf2 by either TNF or LTβR mAb. Whole cell lysates (A) and nuclear extracts (B) of 3T3 cells that were treated with either TNF (25 ng/ml) or LTβR-mAb (LTβR) (0.5 μg/ml) for the indicated time period were probed with the indicated antibodies (right). C, induction of Csf2 by TNF but not LTβR-mAb (LTβR). The relative fold induction of Csf2 was determined by RT-qPCR from cDNA of cells treated with TNF or LTβR for the indicated time period. The data points and error bars in the various graphs represent the average and standard error of the means of at least three independent experiments.
LTβR mAb Priming Enhanced TNF-mediated Csf2 Expression
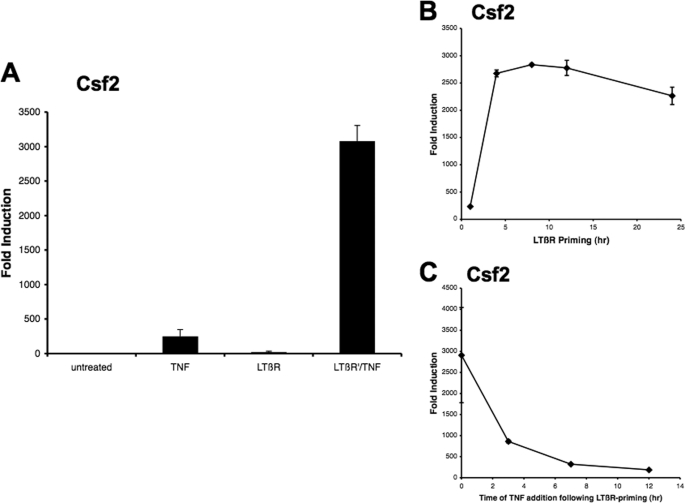
Several studies have shown that the priming of cells with TNF facilitated the induction of classical NF-κB responsive genes by LTβR signaling (21). It has been shown that, during the TNF priming process, p100 expression was induced and functioned as a fourth IκB protein that complexed with p65/p50 heterodimers. The subsequent activation of the LTβR signal resulted in the processing of p100 and the release and nuclear translocation of p65/p50 dimers, which induced the transcription of classical NF-κB responsive genes. In agreement, Csf2 expression was induced by LTβR mAb treatment when the cells were primed with TNF (supplemental Fig. S1). Interestingly, when the sequence of exposure to TNF and LTβR signaling was reversed, a synergistic effect was observed on the induction of Csf2 gene expression. The cells were incubated with the agonistic LTβR mAb for 4 h (priming), washed, and then treated with TNF for an hour. This priming process resulted in a 40-fold increase in the amount of Csf2 transcripts as compared with TNF treatment alone. The synergistic effect was substantially greater than the observed effect when the cells were primed with TNF and then followed with LTβR mAb treatment (compare Fig. 2A with supplemental Fig. S1). This effect seemed to be specific for the Csf2 gene since a synergistic effect was not seen with Ccl2 and RelB expression (supplemental Fig. S2, A and B). Furthermore, a minimum of 4 h of LTβR mAb priming resulted in a synergistic increase in Csf2 transcripts, which corresponds to the kinetics of p100 processing (Fig. 2B). In contrast, when the cells were allowed to rest for various time intervals after LTβR mAb priming before TNF treatment, the synergistic effect was rapidly reduced indicating the transient nature of the priming effect (Fig. 2C). Taken together, these findings indicated that the synergistic effect seen with TNF and agonistic LTβR mAb treatment was crucially influenced by the timing and order of exposure.
FIGURE 2.
LTβR priming increased TNF-mediated Csf2 expression. A, 3T3 cells were treated with either TNF or LTβR mAb individually or primed with LTβR mAb for 4 h and then treated with TNF for 1 h (LTβR′/TNF) as described under “Experimental Procedures.” The amount of Csf2 transcripts was analyzed by RT-qPCR. B, Csf2 transcripts were measured from cDNA sample cells that were primed with LTβR for the indicated time periods, washed, and treated with TNF for 1 h. C, cells were primed for 4 h with LTβR mAb, washed, incubated in medium for the indicated time period, and treated with TNF for 1 h. Complementary DNA from isolated RNA from treated cells were analyzed for Csf2 transcript levels. The data points and error bars in the various graphs represent the average and standard error of the means of at least three independent experiments.
Activation of NIK Is Required for LTβR mAb Priming
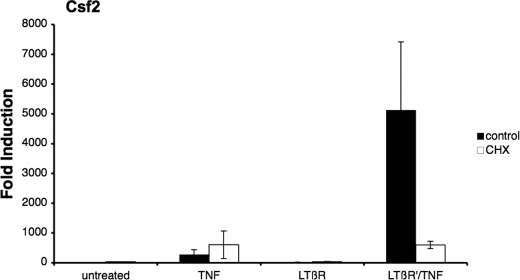
The requirement for de novo protein synthesis is a major difference between the classical and alternative NF-κB signaling pathway (1). The protein synthesis inhibitor CHX enhances the TNF-induced classical NF-κB signal (22), whereas the LTβR signal-mediated alternative NF-κB response is blocked (23). To investigate if the LTβR mAb priming effect was the result of activating the alternative NF-κB pathway, cells were treated with CHX during the LTβR mAb priming process. As seen in Fig. 3, the addition of CHX resulted in an increase in TNF-mediated Csf2 expression (∼3-fold), whereas the LTβR mAb priming effect was completely abrogated by the addition of CHX. These findings indicated that the synergistic response observed with LTβR mAb priming required de novo protein synthesis.
FIGURE 3.
LTβR synergistic effect was sensitive to cycloheximide treatment. Cells were pretreated with either medium or CHX for 2 h before treatment. Complementary DNA from total RNA isolated from TNF, LTβR, or LTβR′/TNF-treated cells were analyzed for Csf2 transcripts.
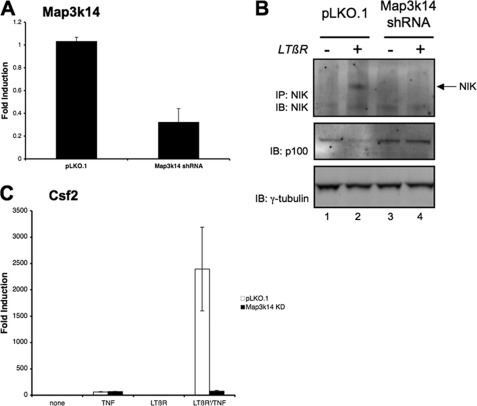
Qing et al. (24) have shown that an increase in NF-κB inducing kinase (NIK) stability following LTβR activation is a key positive regulator in the alternative NF-κB pathway. Furthermore the inhibition of the alternative NF-κB pathway by CHX treatment was due to an increase in NIK turnover (25). To investigate the role of NIK in the LTβR mAb priming process, NIK expression was decreased in cells with the Map3k14 shRNAmir. As seen in Fig. 4A, the level of NIK transcripts was significantly reduced in the cell line stably transfected with Map3K14 shRNA. Furthermore, the induction of NIK protein by LTβR-mAb treatment was abrogated in the cells transfected with the Map3k14 shRNA (upper panel of Fig. 4B). Moreover, LTβR-induced p100 processing was inhibited in the NIK knockdown cells (middle panel of Fig. 4B and quantitated in supplemental Fig. S3A). As seen in Fig. 4C, the amount of TNF-mediated Csf2 expression between the control and NIK knockdown cells were equivalent. These findings suggested that the suppression of NIK expression affected the alternative (LTβR mAb) but not the classical (TNF) NF-κB signaling pathway. Accordingly, the synergistic effect on Csf2 expression seen with LTβR mAb priming followed by TNF stimulation was blocked in the NIK knockdown cells (Fig. 4C). These results indicated that NIK was required for the synergistic effect of LTβR mAb on TNF-mediated Csf2 expression. In addition, the exogenous expression of the human NIK protein (MAP3K14) restored the LTβR mAb priming effect in the NIK knockdown cells (see supplemental Fig. S3, B and C). Thus, the inhibition of the synergistic effects of LTβR priming by CHX and by NIK knockdown indicated that the alternative NF-κB pathway plays a pivotal role in the synergistic effect of LTβR priming on TNF-induced Csf2 expression.
FIGURE 4.
Suppression of NIK (Map3k14) inhibited LTβR-mediated priming. A, complementary DNA from total RNA isolated from cells transfected with the control (pLKO.1) or Map3k14 shRNA plasmid was analyzed for Map3k14 transcripts. B, transfected cells with either the control (pLKO.1) or NIK shRNAi (Map3k14 shRNA) plasmid were treated with LTβR mAb (0.5 μg/ml) for 2 h. Lysates were immunoprecipitated and immunoblotted with the NIK antibody. Whole cell lysate from the same samples were immunoblotted for p100 levels, as seen in the middle panel. The intensity of p100 bands following LTβR-mAb treatment was measured, and the results from three separate experiments were summarized in supplemental Fig. S3A. C, total RNA isolated from transfected cells with the control (pLKO.1) or Map3k14 shRNA plasmid (Map3k14 kd) treated with the indicated agents was analyzed for Csf2 transcripts. The amount of Csf2 expression following either TNF or LTβR-mAb treatment is shown in supplemental Fig. S3B.
Nuclear Translocation of RelB, but Not p65, Occurred during LTβR mAb Priming
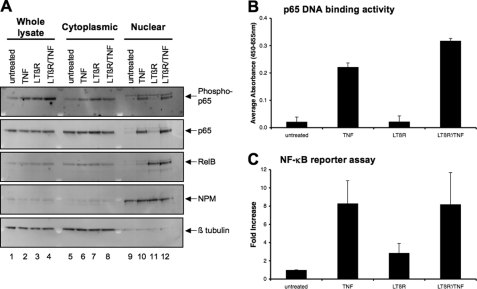
The nuclear translocation of the various NF-κB proteins plays a pivotal role in the activation of both the classical and alternative pathway. It was conceivable that the priming effect by LTβR mAb was mediated by an increase in the amount of nuclear translocation of p65/p50 dimers to induce Csf2 expression. To investigate this possibility, cytoplasmic and nuclear extracts from treated cells were analyzed for the nuclear translocation of NF-κB proteins. The treatment with either LTβR mAb or TNF resulted in an increase in the phosphorylation of p65 in the whole cell lysates and cytoplasmic fractions (lanes 1–8 of the top panel of Fig. 5A). However, an increase in nuclear phospho-p65 was detected only after TNF but not after LTβR mAb treatment (lanes 9 and 10). The absence of either phospho- or total p65 in the nucleus following LTβR mAb treatment could explain the failure of LTβR signaling alone to augment Csf2 expression as seen in Fig. 1. In contrast, the activation of LTβR signaling resulted in an increase in nuclear RelB (middle panel). These findings were consistent with the activation of the alternative NF-κB pathway through LTβR signaling. With TNF treatment, an increase in nuclear p65, but not p52 (see supplemental Fig. S4) or RelB, was observed. These findings were consistent with the activation of the classical NF-κB pathway by TNF signaling. The amount of nuclear NF-κB proteins was not significantly different between the LTβR-primed nuclear samples and those stimulated with TNF alone. In particular, although a slight increase was observed in phospho-p65 in the whole cell lysate (lane 4 of the top panel), the amount of nuclear phospho-p65 was similar between the LTβR-primed and unprimed TNF-treated samples (compare lanes 10 and 12 of the top panel). Likewise, no difference in the amount of nuclear RelB (compare lanes 11 and 12) was observed between the primed and LTβR mAb alone treated samples. Therefore, subsequent TNF stimulation did not augment nuclear translocation of the p52 and RelB that had been induced by LTβR signaling. It is important to note that the priming with LTβR mAb did not affect the total amount of p65 and RelB (see lanes 1 through 4). Furthermore, immunoprecipitation experiments of nuclear lysates from primed cells demonstrated that RelB and p65 were not associated with each other (supplemental Fig. S6A). The major difference between the primed cells and those treated with TNF alone was the nuclear accumulation of both phospho-p65 and RelB in the primed cell nucleus. These findings suggested that qualitative, but not quantitative, differences in the nuclear translocation of both p65 and RelB might be involved in synergistic effect observed after LTβR priming followed by TNF treatment.
FIGURE 5.
Nuclear translocation of p65 and RelB during LTβR priming. A, whole cell lysates (Whole lysate), cytoplasmic, and nuclear extracts from treated cells were fractionated as described under “Experimental Procedures.” 10 μg of total protein of each fraction were loaded into each lane. The extracts were probed with the indicated antibodies. NPM and β-tubulin served as the nuclear and cytoplasmic controls, respectively. This experiment has been repeated at least twice. B, p65 DNA binding activity was analyzed from nuclear extracts from cells treated with either TNF (25 ng/ml for 1 h), LTβR (0.5 μg for 4 h) or LTβR′/TNF (primed with LTβR mAb for 4 h, washed, and then treated with TNF for 1 h). C, cells were transfected with reporter plasmids containing the Csf2 promoter, and treated with the indicated agents for 24 h. SEAP and β-gal activity were assessed, and the data were represented as a fold induction as compared with the untreated sample.
It was also conceivable that either the DNA binding activity or the transactivation potential of the NF-κB transcription factors was increased during the LTβR mAb priming process. To investigate these possibilities, the DNA binding capacity of p65 was measured in nuclear extracts from cells that were primed with LTβR mAb. As seen in Fig. 5B, an increase in p65 DNA binding activity was observed in nuclear extracts from cells that were treated with TNF but not LTβR mAb. Priming with LTβR mAb followed by TNF treatment resulted in an increase in p65 DNA binding activity to a level similar to TNF treatment alone. The p65 DNA binding activity was blocked with an oligonucleotide containing the wild type, but not a mutated, NF-κB consensus sequence (supplemental Fig. S5). Furthermore, a reporter plasmid containing the cis acting NF-κB response element from the Csf2 promoter (12) was transfected into cells and the amount of reporter expression was assessed after treating the cells with TNF or LTβR mAb alone or primed with LTβR mAb before TNF treatment. As seen in Fig. 5C, an increase in NF-κB transcriptional activity was seen with TNF, but not LTβR, treatment. The amount of transcriptional activity did not significantly increase over TNF treatment alone when the cells were primed with LTβR mAb and then treated with TNF. Taken together, these findings indicated that the LTβR-priming effect was not due to an increase in either the p65 DNA binding activity or NF-κB transactivational activity.
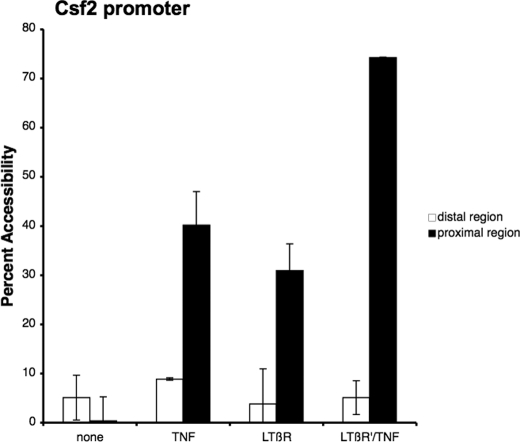
Although the amount of nuclear translocation and the transactivational potential of the NF-κB transcriptional factors were not further increased, it was possible that an increase in chromatin accessibility could explain the synergistic effect observed during LTβR mAb priming of TNF-mediated Csf2 transcription. Several studies (18, 26, 27) have shown that the activation of T cells resulted in an increase in chromatin accessibility at the Csf2 promoter during transcription. To study this possibility, chromatin remodeling of the Csf2 promoter was monitored with the chromatin accessibility assay from cells that were primed with LTβR mAb and then treated with TNF. For the chromatin accessibility assay, nuclei from treated cells were incubated with MNase, quantitatively assessed by real-time PCR, and expressed as a percentage of the amount of undigested genomic DNA samples (26, 28). As seen in the Fig. 6, TNF or LTβR mAb treatment resulted in an increase in chromatin accessibility to the proximal, but not the distal, region of the promoter. The proximal region has been shown to contain a cis acting NF-κB responsive element, whereas the distal region does not. Moveover, accessibility to the proximal region was dramatically increased when the cells were primed with LTβR mAb and then treated with TNF. In contrast, accessibility of the distal region was not affected which indicated that the LTβR priming process was specific to the proximal region of the Csf2 promoter. These findings suggested that a significant increase in accessibility to the Csf2 promoter during the LTβR mAb priming process could account for the synergistic induction seen with TNF-mediated Csf2 expression.
FIGURE 6.
An increase in chromatin accessibility to the Csf2 promoter during LTβR priming. Nuclei from either untreated or treated cells were isolated and digested with micrococcal nuclease as indicated under “Experimental Procedures.” The percent MNase cutting was determined by the amount of remaining DNA after MNase incubation as compared with undigested DNA and the percent accessibility is the difference between 100% minus the percent MNase cutting. The average and standard error of means of three experiments were graphed.
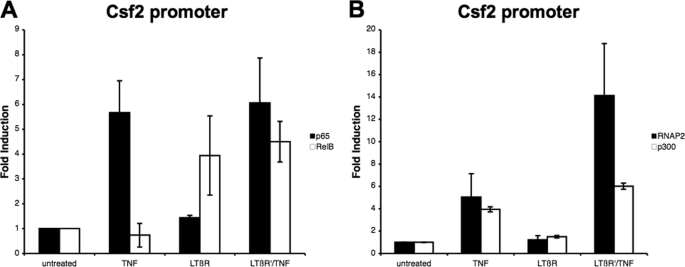
Co-recruitment of RelB and p65 to the Csf2 Promoter during LTβR Priming and TNF Treatment
Several studies have shown that the recruitment of p65 to the Csf2 promoter results in an increase in gene transcription (27). To investigate if p65 and RelB were recruited to the Csf2 promoter, a chromatin immunoprecipitation (ChIP) assay was performed on cells that were LTβR mAb primed followed by TNF treatment or treated with TNF or LTβR mAb alone. As seen in Fig. 7A, p65 but not RelB was recruited to the Csf2 promoter following TNF treatment. In contrast, LTβR mAb treatment induced the recruitment of RelB, but not p65, to the Csf2 promoter. When the cells were primed with LTβR mAb and then treated with TNF, the recruitment of both RelB and p65 to the Csf2 promoter was observed. The amount of p65 and RelB recruited to the Csf2 promoter did not change with LTβR priming. In addition, it was possible that the RelB and p65 were recruited the Csf2 promoter on different alleles. To address this possibility, chromatin from LTβR mAb primed and TNF-treated samples were immunodepleted with antibodies against RelB and the remaining chromatin was precipitated with antibodies against p65. As seen in the supplemental Fig. S7, the amount of DNA precipitated with anti-p65 was reduced when the samples were immunodepleted with anti-RelB as compared with the control sample (immunodepletion with normal immunoglobulin). A similar finding was observed when the samples were immunodepleted with anti-p65 and precipitated with anti-RelB. These findings indicated that RelB and p65 were recruited to the Csf2 promoter on the same allele. Taken together, these findings demonstrated that there were qualitative (the recruitment of both RelB and p65) and not quantitative differences between the priming followed by TNF treatment and the individual treatments. Furthermore, we have previously described the co-recruitment of p300 and RNA polymerase II to the Csf2 promoter following TNF treatment, which resulted in an increase in Csf2 transcription (10). As expected, the treatment with TNF alone resulted in the recruitment of p300 and RNA polymerase II to the Csf2 promoter (Fig. 7B). In contrast, p300 or RNA polymerase II was not recruited to the Csf2 promoter after LTβR mAb treatment. The levels of RNA polymerase II recruited to the Csf2 promoter were higher in the LTβR mAb-primed sample treated with TNF than in cells stimulated with TNF alone. These differences in the levels of RNA polymerase II could be responsible for the synergistic effect observed with LTβR mAb priming of TNF-mediated Csf2 expression.
FIGURE 7.
Recruitment of p65 and RelB to Csf2 promoter during LTβR priming. Chromatin from cells treated with the indicated agents were immunoprecipitated with antibodies against p65 and RelB (A) or p300 and RNA polymerase II (RNAP2) (B). Immunoprecipitated chromatin was probed with primers against the Csf2 promoter and measured with real-time PCR.
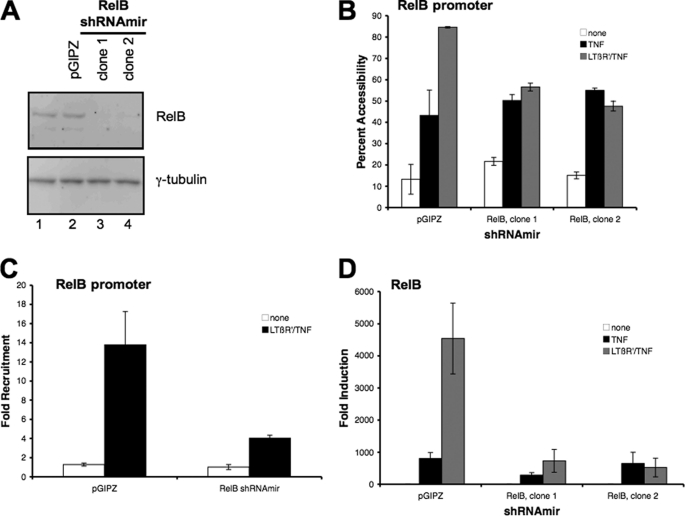
Based on our findings, the recruitment of RelB to the Csf2 promoter during the priming process could account for the increased robustness in TNF-mediated Csf2 expression. To investigate the role of RelB in the enhancement of TNF-mediated Csf2 transcription, the expression of endogenous RelB was suppressed with the transfection of Relb shRNAmir into 3T3 fibroblast cells. As seen in Fig. 8, the amount of Relb protein was reduced in the two clones transfected with the Relb shRNAmir as compared with the cells transfected with the control plasmid (pGIPZ) (Fig. 8A). Chromatin accessibility to the proximal region of the Csf2 promoter was reduced in the cells lacking Relb protein when primed with LTβR and treated with TNF, although the TNF-mediated chromatin accessibility was not affected (Fig. 8B). Similarly, the recruitment of RNA polymerase II to the Csf2 promoter was suppressed in the cells lacking RelB when primed with LTβR and treated with TNF (Fig. 8C, data not shown for second clone). Lastly the synergistic enhancement of Csf2 expression by LTβ signaling of TNF-mediated Csf2 expression was abrogated with the suppression of RelB protein expression (Fig. 8D). These findings demonstrated that RelB was required for the increase in chromatin accessibility and recruitment of RNA polymerase II to the Csf2 promoter during the LTβ-priming/TNF treatment. Hence the RelB may be playing a role in increasing chromatin accessibility to the Csf2 promoter, which then increases the recruitment of RNA polymerase II for the transcription of the Csf2 gene.
FIGURE 8.
Knocking down RelB expression resulted in the mitigation of LTβR mAb priming of TNF-mediated Csf2 expression. A, lysates from cells stably transfected with either the control (pGIPZ) or two clones of RelB (RelB shRNAmir clones 1 and 2) shRNAmir plasmids were probed with the RelB antibody. B, nuclei from cells transfected with the control (pGIPZ) or RelB shRNAmir (clones 1 and 2), that were treated with TNF or primed with LTβR mAb and then treated with TNF (LTβR′/TNF) were isolated and the accessibility to the proximal region of the Csf2 promoter was assessed as described earlier. C, chromatin immunoprecipitation analyses for the recruitment of RNA polymerase II to the proximal region of the Csf2 promoter were conducted on sheared chromatin from cells transfected with either the control (pGIPZ) or RelB shRNAmir (clone 1) that were either untreated (none) or primed with LTβR mAb and then treated with TNF (LTβR′/TNF). Similar results were obtained from chromatin immunoprecipitation analyses performed with RelB shRNAmir clone 2 (data not shown). D, inhibition of LTβR mAb priming synergy of TNF-mediated Csf2 expression with the suppression of RelB expression. Semi-quantitative RT-qPCR analyses for Csf2 transcripts were performed on complementary DNA isolated from cells transfected with either the control or RelB shRNAmir plasmids that were either treated with TNF alone (TNF) or primed with LTβR mAb and then treated with TNF (LTβR′/TNF).
DISCUSSION
Many characteristics distinguish the two NF-κB signaling pathways. However, as seen in many biological signaling systems, there exist numerous intersections that allow the pathways to influence each other. Here we described a synergistic relationship between the classical and alternative NF-κB signaling pathway that regulates the expression of GM-CSF. The transcription of the Csf2 gene has been shown to respond to stimuli that activate the classical NF-κB signaling pathway such as signaling through the TNF receptor. We have previously shown that this process involves the phosphorylation of p65 on serine 536 (10). The activation of the alternative NF-κB signaling pathway through the LTβR resulted in the phosphorylation of serine 536 on p65 but did not induce Csf2 gene transcription. However if the cells were primed with agonistic LTβR antibody and then treated with TNF, a synergistic induction of Csf2, but not Ccl2, another TNF responsive gene, expression was observed. The synergistic effect was dependent on de novo protein synthesis and involved the nuclear translocation and recruitment of RelB and p65 proteins to the Csf2 promoter. In addition, suppression of NIK expression resulted in the loss of the synergistic effect induced by agonistic LTβR mAb, which indicated that the alternative NF-κB signaling pathway was required. Taken together these findings suggested that RelB facilitated the induction of p65-mediated Csf2 expression.
A feature that distinguishes the alternative from the classical NF-κB pathway is the lack of NEMO (IKKγ) activity (29). In contrast, the activity of IKKα has been reported to be involved in both pathways (30, 31). The activation of IKKα is responsible for the phosphorylation of p65 on serine 536 by both TNF (32) and agonistic LTβR mAb (11) treatment. The degradation of IκBα induced by TNF and the processing of p100 induced by LTβR signaling are both mediated by IKKα activity; IκBα degradation is a NEMO-dependent process and p100 processing is a NEMO-independent process. The kinetics of maximum TNF- and LTβR mAb-induced phosphorylation of p65 coincided with the kinetics of degradation of IκBα and processing of p100, which was consistent with the activation of the two IKKα complexes. However, because the alternative NF-κB pathway is not dependent on NEMO activity, the LTβR mAb-induced phospho-p65 was sequestered in the cytoplasm by IκBα. This explains the lack of Csf2 induction following the LTβR mAb treatment despite the phosphorylation of p65. This also confirms the specificity of the agonistic LTβR mAb antibody in stimulating only the alternative NF-κB signaling pathway.
Based on the data shown here, the synergistic effect of LTβR priming on TNF-mediated Csf2 induction involved the recruitment of p65 and RelB independently to the Csf2 promoter, which resulted in an increase in chromatin remodeling and the co-recruitment of RNA polymerase II. In accordance, Brettingham-Moore et al. (18) have shown that the presence of the NF-κB transcription factor c-Rel in the nucleus resulted in the remodeling of the chromatin surrounding the Csf2 promoter and Csf2 transcription in activated T cells. Although we used a different cell-type and stimuli, the co-recruitment of p65, p300, and RNA polymerase II to the Csf2 promoter following TNF treatment was sufficient to induce gene transcription. With LTβR priming, RelB was also recruited to the Csf2 promoter, which resulted in a dramatic increase in Csf2 transcription. Furthermore, the co-recruitment of RelB to the Csf2 promoter increased the amount of RNA polymerase II recruitment, which is consistent with an increase in Csf2 transcription. RelB alone, as seen with LTβR treatment, was not responsible for the co-recruitment of RNA polymerase II and p300 to the Csf2 promoter. Furthermore, the suppression of RelB expression resulted in the abrogation of the synergistic effect on TNF-mediated Csf2 expression by LTβR mAb priming. Hence it is possible that recruitment of RelB (during LTβR priming) to the Csf2 promoter makes the chromatin accessible and facilitates the p65-mediated recruitment of RNA polymerase II (during TNF activation), which results in an increase in gene transcription.
The findings from the sequential immunoprecipitation experiment in supplemental Fig. S5 demonstrated that p65 and RelB exist as independent dimers in lysates from LTβR mAb-primed and TNF-treated cells. In agreement, the amount of p65 recruitment to the Csf2 promoter was not affected in RelB knock-down cells that were primed with LTβR-mAb and then treated with TNF (see supplemental Fig. S6B). Furthermore, the results from the immunodepletion ChIP assay suggested that p65 and RelB were recruited to the Csf2 promoter on the same allele, as seen in supplemental Fig. S6, when cells were primed with LTβR mAb and treated with TNF. In addition, a single cis acting NF-κB response element has been described to the region of the Csf2 promoter that was precipitated in the ChIP assays. It is conceivable that the LTβR mAb priming process induces RelB to act as an enhancer that binds to a distal site which alters the chromatin architecture to promote p65-mediated Csf2 transcription following TNF treatment. Osborne et al. (12) have described a 400 bp element, which shares 67% homology to the human CSF2 enhancer and is 2 kb upstream from the mouse Csf2 gene. However, we could not detect the recruitment of either RelB or p65 to this distal 400 bp element following LTβR priming/treatment or TNF treatment (data not shown). It is possible that there could be other sites within the Csf2 promoter that facilitate p65-mediated Csf2 expression after TNF treatment.
Remodeling of chromatin involves the post-translational modification of histones, which influences gene transcription. The treatment with the histone deacetylase inhibitor trichostatin A has been shown to increase the acetylation of histones and promote the transcription of GM-CSF in activated T cells (26). In contrast, the treatment with trichostatin A followed by TNF stimulation did not result in a synergistic effect as seen with LTβR mAb priming (data not shown). In addition, the histone acetyltransferase p300 was not recruited to the promoter with LTβR mAb treatment, which suggested that the chromatin remodeling seen with LTβR mAb priming process does not involve histone acetylation. It is not known if LTβR mAb priming process induces other histone modifications, such as phosphorylation or methylation, to boost TNF-mediated Csf2 transcription. Defining the role of RelB in promoting chromatin accessibility to the Csf2 promoter will be pivotal in understanding the priming process.
In studies involving multiple cytokines, an autocrine or paracrine relationship could be responsible for the observed synergistic effect. It was conceivable that LTβR mAb priming induced the expression of TNF or TNF receptor, which could explain the synergistic response to TNF-mediated Csf2 expression. Although the amount of TNF or TNFR was not investigated, the response to the LTβR mAb priming process was specific to Csf2 expression. The other TNF-responsive genes such as Ccl2 and RelB were not affected by the LTβR mAb priming process. This would argue against the possibility of LTβR mAb increasing the amount of TNF or TNFR during the priming process. Nevertheless, the findings indicated that neither an autocrine nor paracrine relationship between TNF and LTβR signaling was responsible for the synergistic effect on Csf2 expression. Moreover, these findings underscore the complexity between the classical (TNF) and alternative (LTβR) signaling pathways and their interactions in regulating Csf2 expression.
Here we showed that the alternative NF-κB pathway influences the classical NF-κB pathway in a synergistic manner with respect to Csf2 expression. The activation of lymphocytes results in the expression of TNF and LTβ, and their receptors are found on macrophages and stromal fibroblast cells (1, 33). In addition, TNF is soluble and secreted as a trimer whereas LTβ is cell membrane-bound (34). Furthermore, lack of development of the secondary lymph node organs is the phenotype seen in LTβR knock-out mice (35). It is conceivable that the interactions between these cell types are influenced by soluble TNF and cell membrane-associated LTβ in a temporal and spatial manner to maintain the function of the lymph node. Furthermore, because GM-CSF may act as an immune adjuvant, it is possible that the synergistic relationship between the alternative and classical NF-κB signaling pathways could facilitate the generation of a robust immune response when amounts of TNF and LTβ are limiting.
Supplementary Material
This work was supported, in whole or in part, by the Intramural Research Program of the NIA, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Tables S1 and S2.
- IKK
- IκB kinase
- NIK
- NF-κB-inducing kinase
- CHX
- cycloheximide
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- LTβ
- lymphotoxin β
- CHART
- chromatin accessibility by real-time PCR
- RT-qPCR
- real-time qPCR.
REFERENCES
- 1. Vallabhapurapu S., Karin M. (2009) Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 2. Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z. W., Karin M., Ware C. F., Green D. R. (2002) Immunity 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 3. Karin M., Ben-Neriah Y. (2000) Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 4. Bonizzi G., Karin M. (2004) Trends Immunol. 25, 280–288 [DOI] [PubMed] [Google Scholar]
- 5. Lovas A., Radke D., Albrecht D., Yilmaz Z. B., Möller U., Habenicht A. J., Weih F. (2008) BMC Genomics 9, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacque E., Tchenio T., Piton G., Romeo P. H., Baud V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14635–14640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marienfeld R., May M. J., Berberich I., Serfling E., Ghosh S., Neumann M. (2003) J. Biol. Chem. 278, 19852–19860 [DOI] [PubMed] [Google Scholar]
- 8. Saito T., Sasaki C. Y., Rezanka L. J., Ghosh P., Longo D. L. (2010) Biochem. Biophys. Res. Commun. 391, 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Basak S., Kim H., Kearns J. D., Tergaonkar V., O'Dea E., Werner S. L., Benedict C. A., Ware C. F., Ghosh G., Verma I. M., Hoffmann A. (2007) Cell 128, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sasaki C. Y., Slemenda C. F., Ghosh P., Barberi T. J., Longo D. L. (2007) Cancer Res. 67, 11218–11225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang X., Takahashi N., Matsui N., Tetsuka T., Okamoto T. (2003) J. Biol. Chem. 278, 919–926 [DOI] [PubMed] [Google Scholar]
- 12. Osborne C. S., Vadas M. A., Cockerill P. N. (1995) J. Immunol. 155, 226–235 [PubMed] [Google Scholar]
- 13. Rouillard J. M., Lee W., Truan G., Gao X., Zhou X., Gulari E. (2004) Nucleic Acids Res. 32, W176–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasaki C. Y., Barberi T. J., Ghosh P., Longo D. L. (2005) J. Biol. Chem. 280, 34538–34547 [DOI] [PubMed] [Google Scholar]
- 15. Cui W., Taub D. D., Gardner K. (2007) Nucleic Acids Res. 35, D805–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nelson J. D., Denisenko O., Bomsztyk K. (2006) Nat. Protoc. 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 17. Geisberg J. V., Struhl K. (2004) Nucleic Acids Res. 32, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brettingham-Moore K. H., Rao S., Juelich T., Shannon M. F., Holloway A. F. (2005) Nucleic Acids Res. 33, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bren G. D., Solan N. J., Miyoshi H., Pennington K. N., Pobst L. J., Paya C. V. (2001) Oncogene 20, 7722–7733 [DOI] [PubMed] [Google Scholar]
- 20. Xiao G., Harhaj E. W., Sun S. C. (2001) Mol. Cell 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 21. Basak S., Shih V. F., Hoffmann A. (2008) Mol. Cell Biol. 28, 3139–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klement J. F., Rice N. R., Car B. D., Abbondanzo S. J., Powers G. D., Bhatt P. H., Chen C. H., Rosen C. A., Stewart C. L. (1996) Mol. Cell Biol. 16, 2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coope H. J., Atkinson P. G., Huhse B., Belich M., Janzen J., Holman M. J., Klaus G. G., Johnston L. H., Ley S. C. (2002) EMBO J. 21, 5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qing G., Qu Z., Xiao G. (2005) J. Biol. Chem. 280, 40578–40582 [DOI] [PubMed] [Google Scholar]
- 25. Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 26. Brettingham-Moore K. H., Sprod O. R., Chen X., Oakford P., Shannon M. F., Holloway A. F. (2008) Nucleic Acids Res. 36, 2639–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holloway A. F., Rao S., Chen X., Shannon M. F. (2003) J. Exp. Med. 197, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutcliffe E. L., Parish I. A., He Y. Q., Juelich T., Tierney M. L., Rangasamy D., Milburn P. J., Parish C. R., Tremethick D. J., Rao S. (2009) Mol. Cell Biol. 29, 1972–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rudolph D., Yeh W. C., Wakeham A., Rudolph B., Nallainathan D., Potter J., Elia A. J., Mak T. W. (2000) Genes Dev. 14, 854–862 [PMC free article] [PubMed] [Google Scholar]
- 30. Hu Y., Baud V., Delhase M., Zhang P., Deerinck T., Ellisman M., Johnson R., Karin M. (1999) Science 284, 316–320 [DOI] [PubMed] [Google Scholar]
- 31. Senftleben U., Cao Y., Xiao G., Greten F. R., Krähn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S. C., Karin M. (2001) Science 293, 1495–1499 [DOI] [PubMed] [Google Scholar]
- 32. Sakurai H., Chiba H., Miyoshi H., Sugita T., Toriumi W. (1999) J. Biol. Chem. 274, 30353–30356 [DOI] [PubMed] [Google Scholar]
- 33. Vassalli P. (1992) Annu. Rev. Immunol. 10, 411–452 [DOI] [PubMed] [Google Scholar]
- 34. Beutler B., Cerami A. (1989) Annu. Rev. Immunol. 7, 625–655 [DOI] [PubMed] [Google Scholar]
- 35. Fütterer A., Mink K., Luz A., Kosco-Vilbois M. H., Pfeffer K. (1998) Immunity 9, 59–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.