Abstract
The efficacy of the diphenol curcumin as a cancer chemopreventive agent is limited by its chemical and metabolic instability. Non-enzymatic degradation has been described to yield vanillin, ferulic acid, and feruloylmethane through cleavage of the heptadienone chain connecting the phenolic rings. Here we provide evidence for an alternative mechanism, resulting in autoxidative cyclization of the heptadienone moiety as a major pathway of degradation. Autoxidative transformation of curcumin was pH-dependent with the highest rate at pH 8 (2.2 μm/min) and associated with stoichiometric uptake of O2. Oxidation was also catalyzed by recombinant cyclooxygenase-2 (COX-2) (50 nm; 7.5 μm/min), and the rate was increased ≈10-fold by the addition of 300 μm H2O2. The COX-2 catalyzed transformation was inhibited by acetaminophen but not indomethacin, suggesting catalysis occurred by the peroxidase activity. We propose a mechanism of enzymatic or autoxidative hydrogen abstraction from a phenolic hydroxyl to give a quinone methide and a delocalized radical in the heptadienone chain that undergoes 5-exo cyclization and oxygenation. Hydration of the quinone methide (measured by the incorporation of O-18 from H218O) and rearrangement under loss of water gives the final dioxygenated bicyclopentadione product. When curcumin was added to RAW264.7 cells, the bicyclopentadione was increased 1.8-fold in cells activated by LPS; vanillin and other putative cleavage products were negligible. Oxidation to a reactive quinone methide is the mechanistic basis of many phenolic anti-cancer drugs. It is possible, therefore, that oxidative transformation of curcumin, a prominent but previously unrecognized reaction, contributes to its cancer chemopreventive activity.
Keywords: Cyclooxygenase (COX) Pathway, Lipoxygenase Pathway, Oxidation-Reduction, Peroxidase, Quinones, Autoxidation, Curcumin, Phenoxyl Radical, Quinone Methide
Introduction
The yellow plant phenolic pigment curcumin shows a remarkable ability to affect a wide variety of signaling pathways that are dysregulated during tumorigenesis, including proliferation, invasion, apoptosis, and cell cycle checkpoints (1). Altogether more than one hundred molecular targets of curcumin have been identified using in vitro cell culture-based assays (2). The diversity of biological effects of curcumin has been attributed to its ability to act as an antioxidant, anti-inflammatory, and anti-viral agent (3). As a consequence of promising in vitro results, several clinical trials have been initiated to investigate the effect of dietary curcumin in the prevention of inflammatory bowel disease, colon, and pancreatic cancer, and Alzheimer Disease, among others (3).
Prior to the more recent interest in its chemopreventive properties, curcumin was being considered as a food coloring agent but its chemical and photochemical instability prevented widespread application. Light-induced degradation of curcumin in organic solvents results in cleavage of the heptadienone chain, and the most abundant products have been identified as vanillin, ferulic aldehyde, ferulic acid, and feruloylmethane (4, 5). The same products have been observed as degradation products of curcumin in aqueous buffer solutions (6, 7). The identification of the products, however, was partly based on HPLC retention times and monitoring using fixed wavelength UV detection. Besides the chain cleavage products there is evidence of additional, prominent products in the literature but these were left uncharacterized (8, 9). Degradation of curcumin is also a prominent reaction upon addition to cultured cells (7), and therefore, defining the products and kinetics of degradation of curcumin is relevant to the question whether metabolites could be mediating some of the biological activities, an issue that has been raised frequently in recent years, e.g. Refs. 3, 10–12.
One of the suggested chemopreventive mechanisms of curcumin is the suppression of prostaglandin formation by the cyclooxygenase-2 (COX-2)3 enzyme (13, 14). Induction of COX-2 and the formation of PGE2 are hallmarks in the development of many cancers, best studied in the case of colon carcinogenesis. Suppression of COX-2 activity by selective or non-selective inhibitors of the cyclooxygenase enzymes leads to a significant reduction of colon cancer in animal models and in human populations with a genetically elevated cancer risk (15). Adverse cardiovascular side effects due to inhibition of COX-2 in the vascular endothelium substantially limit the usefulness of these drugs for cancer treatment. Dietary curcumin, in contrast, with its low systemic bioavailability but high concentration in the small intestine and colon (16), may provide suitable pharmacokinetic properties to target COX-2 specifically in the gastrointestinal tract while sparing other organs (17).
The interactions of curcumin with arachidonic acid metabolism and prostaglandin biosynthesis are remarkably complex: curcumin has been shown to down-regulate the expression of COX-2 mRNA and protein (18, 19) and also to directly inhibit the enzymatic activities of COX-2 (20) and of mPGES-1, one of the terminal prostaglandin synthases that converts the COX-derived unstable PGH2 to PGE2 (21). In addition, curcumin has been shown to inhibit the enzymatic activity of 5-lipoxygenase, the enzyme that initiates biosynthesis of leukotrienes from arachidonic acid (22–24), and, unexpectedly, to be a substrate for the lipoxygenase from soybean seeds (25). Here we analyzed the chemical mechanism underlying a previously unrecognized oxidative transformation of curcumin that is in stark contrast to the published literature describing cleavage into smaller fragments as the major non-enzymatic transformation of curcumin.
EXPERIMENTAL PROCEDURES
Materials
Curcumin was synthesized according to the method of Pabon (26) and purified by crystallization from ethyl acetate/MeOH at −20 °C (90% pure by HPLC-DAD analysis; HRMS (ES+) calcd for C21H20O6H 369.1333, found 369.1324). A 5 mm stock solution in ethanol was prepared fresh daily. Hexahydrocurcumin was prepared by reduction of curcumin with H2 in the presence of Pd/Al in ethanol. Vanillin, ferulic acid, indomethacin, and acetaminophen were obtained from Sigma. Feruloylmethane was from ICI. Oxygen-18 labeled water (97 atom-% 18O) was obtained from Isotec. Recombinant His6-tagged human COX-2 was expressed in the baculovirus/Sf9 insect cells system and purified by Ni-NTA affinity chromatography as described (27). The concentration of the purified COX-2 stock solution was 5 μm. Ovine COX-1 was obtained from Cayman Chemical. RAW264.7 were obtained from ATCC.
Synthesis of 4′,4″-Dimethoxycurcumin
The procedure followed the method of Pabon for the synthesis of curcumin (26). Boron oxide (B2O3), 0.5 g (7 mmol), was mixed with 1 g (10 mmol) of acetylacetone and stirred for 3 h to give a thick white suspension. The compound was added to a mixture of 3 g (20 mmol) of 3,4-dimethoxybenzaldehyde and 9.2 g (40 mmol) tributylborate dissolved in 10 ml of ethyl acetate. The entire mixture was stirred for 5 min, and then 4 aliquots of 50 μl of butylamine were added over the course of 30 min (total of 200 μl; 2 mmol), followed by stirring for 4 h and standing overnight. The next day, 15 ml of 0.4 m HCl (heated to 60 °C) were added, and the mixture was stirred for 1 h. The organic layer was collected, and the aqueous phase was extracted twice with ethyl acetate. The combined organic phases were washed with water and dried over sodium sulfate. The organic phase was reduced to 10 ml under a stream of nitrogen, and 10 ml of MeOH were added to induce crystallization of 4′,4″-dimethoxycurcumin. The product was filtered from solvent and allowed to air dry (93% pure by HPLC-DAD analysis; m.p. 128 °C) HRMS (ES+) calcd for C23H24O6H 397.1646, found 397.1648. 1H NMR, 600 MHz, CD3CN (δ 1.93): δ 7.58 (d, J = 15.8 Hz, H1, 2H), δ 7.24 (d, J = 1.7 Hz, ar-H2, 2H), δ 7.20 (dd, J = 8.3/1.8 Hz, ar-H6, 2H), δ 6.96 (d, J = 8.3 Hz, ar-H5, 2H), δ 6.70 (d, J = 15.8 Hz, H2, 2H), δ 5.93 (s, H4, 1H), δ 3.85 (s, -OCH3, 3H), δ 3.84 (s, -OCH3, 3H).
Synthesis of 4′-Methoxycurcumin
The same procedure as for synthesis of 4′,4″-dimethoxycurcumin was used. Instead of 3 g of 3,4-dimethoxybenzaldehyde, 1.5 g (10 mmol) each of vanillin and 3,4-dimethoxybenzaldehyde were used. After extraction of the reaction mixture with ethyl acetate the crude products were purified using preparative thin layer chromatography developed with a solvent of chloroform/EtOH (25:1, by vol). Regions of the plate containing 4′-methoxycurcumin were scraped off and extracted with ethyl acetate. A 5 mm stock solution was prepared in MeOH and stored at −20 °C (90% pure by HPLC-DAD analysis; HRMS (ES+) calculated for C22H22O6H 383.1489, found 383.1482). ESI-MS: m/z 381.25 ([M-H]−). 1H NMR, 600 MHz, CD3COCD3 (δ 2.04): δ 7.59 (d, J = 15.8 Hz, H1&H7, 2H), δ 7.32 (d, J = 1.6 Hz, H2″, 1H), δ 7.31 (d, J = 1.6 Hz, H2′, 1H), δ 7.22 (dd, J = 8.3/1.7 Hz, H6′, 1H), δ 7.17 (dd, J = 8.2/6.1 Hz, H6″, 1H) δ 7.00 (d, J = 8.3 Hz, H5′, 1H), δ 6.88 (d, J = 8.1 Hz, H5, 1H), δ 6.73 (d, J = 15.8, H2, 1H), δ 6.69, (d, J = 15.8, H6, 1H), δ 5.98 (s, H4, 1H), δ 3.91 (s, 3″-OCH3, 3H), δ 3.87 (s, 3′-OCH3, 3H), δ 3.86 (s, 4′-OCH3, 3H).
Autoxidative Transformation of Curcumin
Curcumin was diluted to a concentration of 70 μm in 500 μl of 100 mm Tris-HCl buffer pH 8 in a cuvette. The sample was scanned repetitively from 700 to 220 nm in 2-min intervals using a Perkin Elmer PE35 UV/Vis spectrophotometer. When >70% of the absorbance at 430 nm had disappeared the sample was acidified to pH 4 using 1 n HCl and loaded on a preconditioned 30 mg of Waters HLB cartridge. After washing with water the cartridge was eluted with 2 × 500 μl of methanol. The solvent was evaporated under a stream of nitrogen, and the residue was dissolved in 50 μl of MeOH for RP-HPLC analysis. For product identification, 20 separate 2-ml reactions were conducted in parallel using 100 μm curcumin in the same buffer and a reaction time of 30 min. Extraction was performed as described above.
COX-2 Catalyzed Transformation of Curcumin
COX-2, 5 μl (50 nm final), was diluted in a cuvette using 500 μl of 100 mm K-phosphate buffer pH 9 containing 1 μm hematin for reconstitution of the holoenzyme. Curcumin (100 μm) was added, and the reaction was followed at 430 nm using the spectrophotometer in the time drive mode. In some of the reactions, 5 nm COX-2 and 300 μm of H2O2 were added. For reactions in the presence of the inhibitors (indomethacin and acetaminophen) the enzyme, hematin, and inhibitors were preincubated for 1 min before addition of curcumin to start the reaction. All enzymatic transformations were conducted in the absence of arachidonic acid and phenol.
Oxygen Electrode Assay
A Clark-type oxygen electrode (Hansatech Instruments, obtained through PP Systems, Amesbury, MA) was maintained at 30 °C using a circulating water bath. The calibration was performed using air-saturated distilled water and sodium dithionite assuming 235 μm for the maximum concentration of oxygen. 1 ml of Na-phosphate/citrate buffer pH 8.0 was added to the chamber and allowed to equilibrate for 5 min before addition of 12 μl of curcumin from a 5 mm stock in ethanol or the same amount of ethanol.
Reaction in H218O
Autoxidative transformation of curcumin was conducted using 100 μl of buffer. 5 μl of a 1 m stock solution of Tris-HCl buffer pH 8 in water were diluted with 95 μl of H218O (97 atom-% 18O). Curcumin, 3 μl from a 5 mm stock solution in ethanol, was added, and the reaction was allowed to proceed for 30 min followed by acidification to pH 4 using 1 n HCl and extraction using a HLB cartridge as described above. A reaction using unlabeled H2O was conducted in parallel. In a control reaction curcumin was autoxidized in 100 μl of buffered H218O, extracted, and then incubated in unlabeled water for 60 min followed by acidification and extraction. Only a minimal exchange of 18O content of the main oxidized product was observed.
Reaction of 4′-Methoxycurcumin in H218O
The transformation of 4′-methoxycurcumin was conducted in the presence of COX-2. 5 μl of a 1 m Tris-HCl pH 8 stock solution in water were diluted with 95 μl of H218O (97 atom-% 18O) and 5 μl of COX-2 and 1 μm heme (final) were added. After 2 min, 100 μm 4′-methoxycurcumin and 300 μm H2O2 were added. After 10 min of reaction time, a second 5 μl aliquot of COX-2 was added, and the reaction was allowed to proceed for additional 60 min before acidification and extraction as described above. After elution from the HLB cartridge and evaporation of MeOH, the sample was dissolved in 50 μl of MeOH, and analyzed by LC-ESI-MS using a Thermo Finnigan LTQ ion trap instrument.
Reaction of 4′,4″-dimethoxycurcumin
4′,4″-dimethoxycurcumin (50 μm) was dissolved in 500 μl of 100 mm potassium phosphate buffer, pH 8, in a cuvette and the absorbance at 430 nm was monitored using the time-drive mode of the spectrophotometer. COX-2 (50 nm final) and/or H2O2 (300 μm final) were added as indicated.
Determination of Superoxide
The amount of superoxide formed during transformation of curcumin was measured as the reduction of cytochrome c (from bovine heart). Cytochrome c, 12 μm, was dissolved in 500 μl of 100 mm potassium phosphate buffer pH 8 in a cuvette and 50 μm curcumin were added. The solution was scanned form 700–200 nm in the spectrophotometer in 2 min intervals. Reduction of cytochrome c was measured as the increase in absorbance at 550 nm using ϵ 29.5 mm−1 cm−1 (28). In some of the reactions, superoxide dismutase (450 units) or catalase (50 units) was included.
Cell Culture
RAW264.7 cells were cultured in DMEM and grown in 6-well plates at 37 °C in an atmosphere of 5% CO2. Cells of passages 5 to 8 were used. Some of the cells were stimulated by treatment with 100 ng/ml LPS for 5 h in DMEM adjusted to pH 7.6 using 20 mm sodium phosphate buffer and in the absence of FBS. Curcumin (20 μm) was added from a stock solution in DMSO. After 2 h of incubation the culture medium was removed, acidified to pH 4, and extracted using ethyl acetate/isopropanol (90/10, by vol). Products were analyzed by RP-HPLC with diode array detection as described below.
HPLC and LC-MS Analyses
The transformation reactions were analyzed using a Waters Symmetry C18 5-μm column (4.6 × 250 mm) eluted with a gradient of 20% acetonitrile to 80% acetonitrile in 0.01% aqueous acetic acid over 20 min followed by 10 min of isocratic elution at a flow rate of 1 ml/min. Elution of the products was monitored using an Agilent 1200 diode array detector.
For LC-MS analyses a Finnigan TSQ Quantum triple quadrupole MS instrument equipped with an electrospray interface was used. The instrument was operated in the negative ion mode, and mass spectra were acquired at a rate of 2 s/scan. The heated capillary was set at 300 °C, and the spray voltage was set at 4.4 kV. For LC-MS analysis of oxygen incorporation from 18O-buffer (MSn analyses), a Thermo Finnigan LTQ ion trap instrument was used. The settings for the heated capillary (300 °C), spray voltage (4.0 kV), spray current (0.22 μA), auxiliary (37 mTorr) and sheath gas (16 mTorr) were optimized using direct infusion of a solution of curcumin (20 ng/μl) in acetonitrile/water 95/5, by vol., containing 10 mm NH4OAc. Samples were introduced into both instruments using a Waters Symmetry Shield C18 3.5-μm column (2.1 × 100 mm) eluted with a gradient of acetonitrile/water (5/95, by vol., containing 10 mm NH4OAc) to acetonitrile/water (95/5, by vol., containing 10 mm NH4OAc) over 10 min followed by 3 min of isocratic elution and re-equilibration in the starting solvent.
For HRMS a Waters Synapt hybrid quadrupole orthogonal access time-of-flight mass spectrometer was used. Samples were analyzed in the positive mode using a dual ESI/CI ion source with lock spray channel.
NMR
Samples were dissolved in 150 μl of acetone-d6 or acetonitrile-d3 in a 3 mm tube, and the NMR spectra were recorded using a Bruker DRX 600 MHz spectrometer equipped with a cryoprobe. Chemical shifts are reported in ppm relative to residual non-deuterated solvent (δ 2.04 for acetone-d6 and δ 1.93 for acetonitrile-d3). The pulse frequencies for the H,H-COSY, HSQC, and HMBC experiments were taken form the Bruker library. The spectra were acquired at 285 K.
RESULTS
Autoxidative Transformation of Curcumin
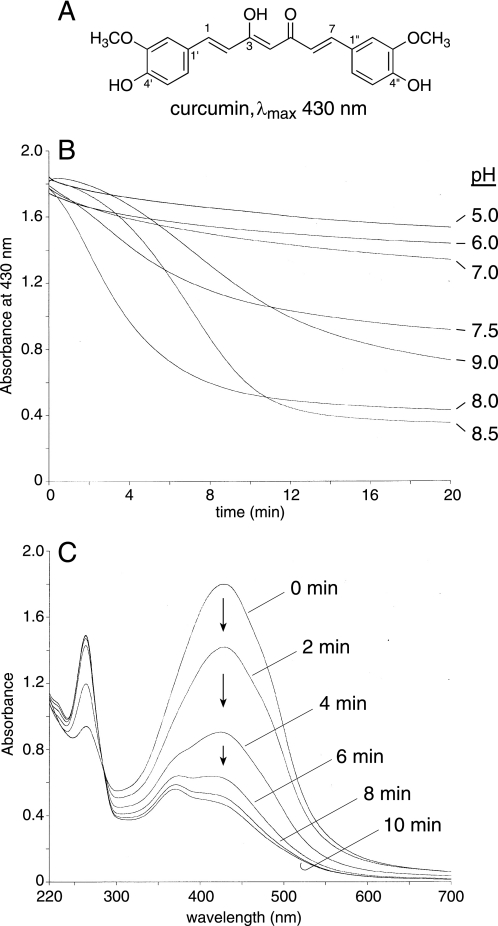
The degradation of curcumin (Fig. 1A) in aqueous buffers of pH values ranging from 5 to 9 was monitored by the loss of the absorbance at the λmax of 430 nm using a UV/Vis spectrophotometer. Curcumin was unstable at basic pH (7.5–9) but its stability was increased at acidic pH (Fig. 1B), similar to findings reported by (7). Curcumin was most unstable at pH 8, and, therefore, most of the following assays were performed at pH 8. Repetitive scanning from 700–220 nm confirmed that the chromophore at 430 nm was lost during the transformation indicating that the conjugated double bond system between the phenolic rings of curcumin was disrupted (Fig. 1C).
FIGURE 1.
Effect of pH on the transformation of curcumin. A, extended conjugation of the seven-carbon heptadienone chain connecting the two phenolic rings results in the yellow-orange color of curcumin. In 4′-methoxy- and 4′,4′-dimethoxycurcumin the phenolic hydroxyl group is replaced by a methoxy group. B, curcumin was added to buffer of the pH indicated and the decrease in absorbance at 430 nm at room temperature was monitored for 20 min using a UV/Vis spectrophotometer. For all reactions 70 μm curcumin was added except for pH 9.0 where 50 μm curcumin was added. The buffers pH 5 to 8 were generated using mixtures of 200 mm Na2HPO4 and 100 mm citric acid, pH 8.5 and 9.0 were 100 mm K2HPO4 adjusted with NaOH. C, repetitive scans in 2-min intervals of a solution of 70 μm curcumin in 100 mm Tris-HCl buffer, pH 8.
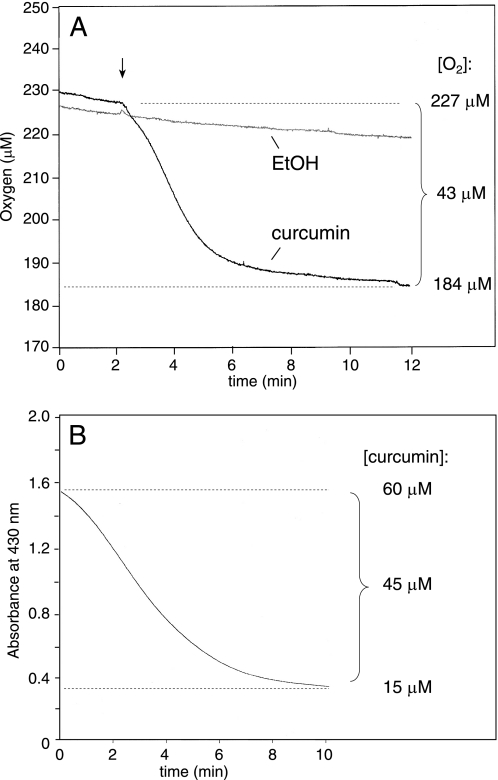
The reaction observed in the spectrophotometer was compared with the amount and rate of oxygen consumed during the transformation of curcumin. Addition of 60 μm curcumin (dissolved in 5 μl of ethanol) to 1 ml of Na-phosphate/citrate buffer pH 8.0 in a Clark-type oxygen electrode resulted in immediate and rapid uptake of oxygen (Fig. 2A). The total reduction of the oxygen concentration in the buffer was calculated as 43 μm. Addition of the ethanol vehicle resulted in a drift of about 5 μm oxygen over the same time. When the transformation of 60 μm curcumin was monitored in the spectrophotometer, the rate of disappearance of the signal at 430 nm was similar to the rate of oxygen consumption measured in the oxygen electrode (Fig. 2B). The amount of curcumin degraded in the spectrophotometric assay (about 75% or 45 μm) was equivalent to the amount of oxygen consumed (≈38 μm) implicating that curcumin reacted with an equimolar amount of oxygen during degradation.
FIGURE 2.
Oxygen consumption during the transformation of curcumin. A, 1 ml of Na-phosphate/citrate buffer pH 8.0 was placed in a Clark-type oxygen electrode and allowed to adjust to 30 °C. Curcumin (60 μm) or vehicle (ethanol) was added after 2 min (arrow), and the oxygen concentration was monitored for 10 min. B, curcumin (60 μm) was added to 1 ml of Na-phosphate/citrate buffer pH 8.0 in a spectrophotometer cuvette, and the absorbance at 430 nm was recorded for 10 min.
HPLC Analysis of the Reaction Products
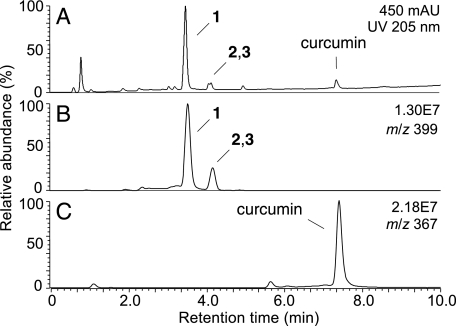
LC-ESI-MS analysis with online diode array detection and recording of the absorbance at 205 nm showed a major transformation product (1) of curcumin eluting at 3.5 min and two less abundant products 2 and 3 eluting as a double peak at 4.2 min (Fig. 3A). Concomitant ESI-MS analysis with detection in the negative ion mode gave a molecular ion of m/z 399 for the three products 1, 2, and 3, equivalent to a molecular weight of 400 (Fig. 3B). This is an increase of 32 amu over curcumin (7.4 min retention time; m/z 367; MW 368) suggesting that the products had been formed by reaction with molecular oxygen (Fig. 3C).
FIGURE 3.
RP-HPLC analysis of the transformation products of curcumin. The transformation reaction of curcumin as shown in Fig. 1B was extracted and analyzed by RP-HPLC with detection by (A) UV 205 nm and (B and C) LC-ESI-MS. Three products 1, 2, and 3 were detected in the UV chromatogram and gave a signal in the ion trace m/z 399 (panel B), equivalent to a molecular weight of 400 for all three products. C, unreacted curcumin was detected as the [M-H]− molecular ion in the ion trace m/z 367.
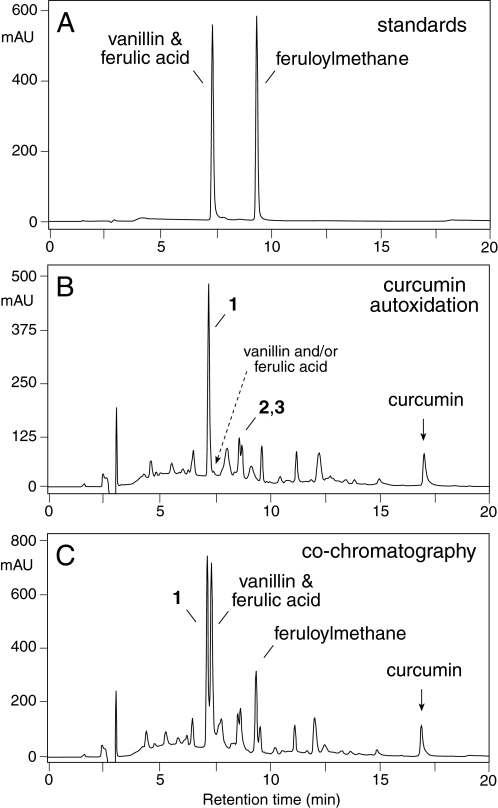
Products 1-3 were compared with authentic standards of three of the four reported major degradation products of curcumin (4, 5, 7) using RP-HPLC analysis with diode array detection (Fig. 4). A standard of the fourth putative product, trans-6-(4-hydroxy-3-methoxyphenyl)-2,4-dioxo-hexenal (7), was not available. The standards of vanillin and ferulic acid co-eluted at 7.4 min, feruloylmethane eluted at 9.4 min (Fig. 4A). The main oxygenation product of curcumin (1) eluted at 7.2 min (Fig. 4B), about 0.2 min earlier than vanillin and ferulic acid. Co-chromatographic analysis of autoxidized curcumin with the standards confirmed the difference in retention times of 1 and vanillin/ferulic acid (Fig. 4C). Based on the UV spectrum, a minor peak eluting at the corresponding retention time in the autoxidation reaction of curcumin is vanillin (see Fig. 4B) whereas ferulic acid and feruloylmethane were not detected. These analyses provided evidence that the reported cleavage of the heptadienone chain of curcumin into vanillin, ferulic acid, or feruloylmethane is not an abundant reaction (7) and that, in contrast, the almost exclusive transformation of curcumin in aqueous buffer at physiological pH is an autoxidation reaction.
FIGURE 4.
RP-HPLC comparison of the products of curcumin autoxidation with the putative degradation products, vanillin, ferulic acid, and feruloylmethane. A, 500 ng each of vanillin and ferulic acid and 1 μg of feruloylmethane were injected on a Waters Symmetry 5-μm column (4.6 × 250 mm) eluted with a linear gradient of acetonitrile/water/acetic acid (20:80:0.01, by vol) to (80:20:0.01, by vol) in 20 min at a flow rate of 1 ml/min and diode array detection. B, analysis of autoxidized curcumin showing the elution of products 1, 2, and 3. C, co-chromatographic analysis of a mixture of the sample in B and 500 ng each of vanillin, ferulic acid, and feruloylmethane. The chromatograms shown were at recorded at UV 205 nm.
Identification of the Transformation Products
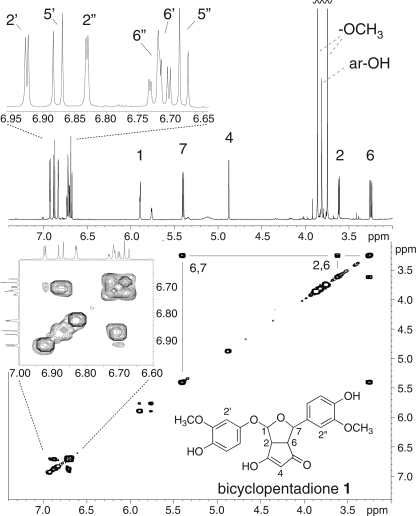
The main product 1 was prepared by autoxidation of 500 μg of curcumin and isolated using RP-HPLC. HRMS analysis gave an exact mass of 401.1225 for the protonated adduct resulting in a molecular formula of C21H20O8 for 1 (calculated: 401.1231) and compatible with the addition of molecular oxygen to curcumin. The 1H NMR and H,H-COSY spectra of 1 recorded in d6-acetone are shown Fig. 5. The proton chemical shifts of product 1 were identical with data reported for the transformation product of curcumin by soybean lipoxygenase (25). Therefore, the main autoxidation product 1 is a bicyclopentadione formed by cyclization and oxygenation of the heptadienone carbon chain of curcumin. Carbons 2 and 6 form the bridge of the two fused five membered rings, and carbons 1 and 7 are connected through an oxygen bridge to form a furan ring. The second atom of oxygen has been inserted between carbon 1 and the phenolic ring (supplemental Fig. S1). Chromatographic, mass-spectrometric, and NMR data were used to identify 2 and 3 as configurational isomers of 1 by comparison to published data (29).
FIGURE 5.
1H and H,H-COSY NMR spectra of product 1 (bicyclopentadione) isolated from autoxidation of curcumin. The 1H spectrum at the top shows five signals corresponding to the hydrogens at carbons 1, 2, 4, 6, and 7 of the original heptadienone chain of curcumin and the signals of the aromatic protons. The H,H-COSY spectrum (below) shows a cross peak between H6 and H7 as well as between H6 and H2. Note the lack of a cross-peak between the neighboring H1 and H2 due to unfavorable arrangement of the two spins at the ring structures. In addition, H1 shows weak coupling (J = 3.5 Hz) with the hydroxyl group at C3 (δ 5.76). H2′ of the aromatic ring showed a cross-peak with C1′ at 148.4 ppm in the HMBC experiments (not shown) that allowed assignment of all six aromatic protons to the corresponding phenolic ring.
COX-2 Catalyzed Oxidative Transformation of Curcumin
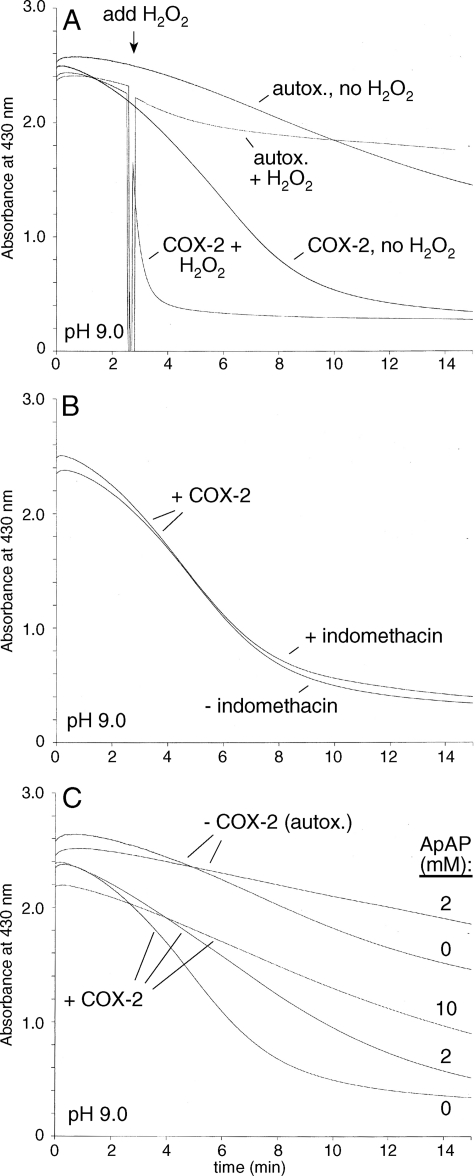
The formation of 1 by autoxidation and by lipoxygenase-catalyzed oxygenation of curcumin (25) prompted the hypothesis that a similar transformation would also be catalyzed by the cyclooxygenase enzymes, COX-1 and COX-2. Reactions with COX-2 were carried out using the purified recombinant human enzyme in the presence of hematin (1 μm) to reconstitute the COX-2 holoenzyme but in the absence of arachidonic acid or a co-substrate for the peroxidase activity (e.g. phenol). The rate of degradation of curcumin increased about 3.5-fold when 50 nm COX-2 was added, and there was a further 10-fold increase by the addition of 300 μm H2O2 (Fig. 6A and Table 1). In the absence of enzyme, addition of 300 μm H2O2 led only to a 2-fold increase of the rate of autoxidative transformation. Product analysis using RP-HPLC with diode array detection or LC-MS confirmed that the same products were formed in the COX-2 catalyzed transformation as in the autoxidative transformation of curcumin. The COX-1 isozyme was active in the presence of H2O2 (300 μm) but inactive when H2O2 was absent.
FIGURE 6.
Spectrophotometric analysis of the COX-2-catalyzed transformation of curcumin. A, transformation of curcumin was accelerated by the presence of 50 nm COX-2 compared with autoxidation. Addition of 300 μm H2O2 further accelerated the rate of COX-2-catalyzed transformation whereas it had only a small effect on autoxidation. B, addition of 10 μm indomethacin did not change the rate of COX-2 (50 nm) catalyzed transformation of curcumin. C, addition of acetaminophen (ApAP) dose-dependently inhibited the COX-2 (50 nm) catalyzed transformation of curcumin. All reactions were conducted using 100 μm curcumin and 1 μm hematin in 100 mm potassium phosphate buffer pH 9 in the absence or presence of 50 nm COX-2 as indicated. The absorbance at 430 nm was recorded in the time-drive mode using a UV/Vis spectrophotometer.
TABLE 1.
Rates of autoxidative and COX-2-catalyzed transformation of curcumin and curcumin derivatives
Rates were determined using 50 μm substrate in 500 μl of 100 mm potassium phosphate buffer, pH 8.0, in a 1-cm cuvette, and in the absence or presence of COX-2, 1 μm hematin, and 300 μm H2O2. Curcumin and its derivatives were monitored at 430 nm (ϵ = 25,000 m−1 cm−1); the reaction with ABTS was monitored at 417 nm (ϵ = 36,000 m−1 cm−1).
| Substrate | Initial rate ± S.D. (μm/min) |
COX-2 turnover number (at 50 μm substrate) | |||
|---|---|---|---|---|---|
| Autoxidation | Autoxidation (hematin + H2O2) | COX-2a | COX-2b + H2O2 | ||
| Curcumin | 2.2 ± 0.4 | 4.4 ± 0.4 | 7.5 ± 0.6 | 88.0 ± 7.0 | 1.76 × 103 ± 1.4 × 102 |
| 4′-Methoxycurcumin | 1.0 ± 0.2c | 3.9 ± 1.0 | 3.9 ± 1.0 | 86.2 ± 5.0 | 1.72 × 103 ± 1.0 × 102 |
| 0.1 ± 0.05 | |||||
| 4′,4″-Dimethoxycurcumin | –d | 0.07 ± 0.01 | –d | 0.06 ± 0.02a | NDe |
| ABTS | ND | ND | ND | 7.2 ± 0.6 | 144 ± 12 |
a The concentration of COX-2 was 50 nm, 1 μm hematin.
b The concentration of COX-2 was 5 nm, 1 μm hematin. The calculated rate is corrected for the dilution of enzyme.
c The reaction proceeded at an initial, faster rate during the first 1–2 min followed by a slower rate over the next 30 min.
d No reaction.
e Not determined.
The effect of inhibitors of the cyclooxygenase active site (indomethacin) and peroxidase active site (acetaminophen, ApAP) was tested to establish which of the two COX-2 active sites was involved in the transformation of curcumin. Indomethacin at 10 μm concentration had no effect on the COX-2 catalyzed transformation of curcumin (Fig. 6B) whereas acetaminophen dose-dependently decreased the rate of COX-2 catalyzed degradation of curcumin (Fig. 6C). Acetaminophen also had a small inhibitory effect on the autoxidative transformation of curcumin. Neither inhibitor showed an effect on the COX-2-catalyzed transformation of curcumin in the presence of 300 μm H2O2.
Reactions in H218O-Buffer
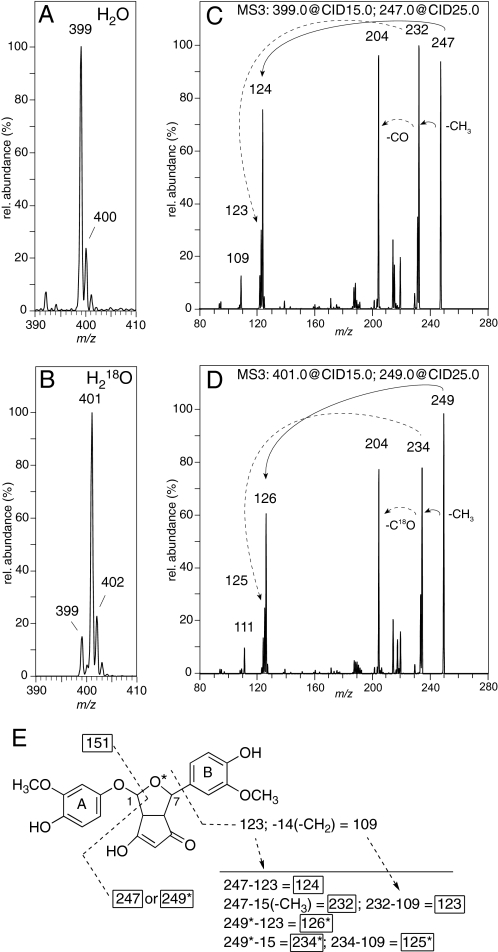
The origin of oxygen in the bicyclopentadione 1 was established by incubations in buffer with H218O or unlabeled water. The reactions were analyzed by LC-ESI-MS using an ion trap mass detector. The calculated concentration of H218O in the labeled buffer was 92%. Product 1 showed an increase of 2 amu from m/z 399 to m/z 401 when the reaction was carried out in H218O-buffer (Fig. 7, A and B). The incorporation of H218O into 1 was 85%, implicating that one of the two oxygen atoms incorporated during autoxidative transformation of curcumin was derived from water. Control experiments showed that 1 did not exchange 18O during incubation (pH 8), extraction (pH 4), or LC-MS analysis.
FIGURE 7.
LC-ESI-MS/MS analysis of the bicyclopentadione 1 obtained by autoxidation of curcumin in regular and H218O-buffer. MS1 spectrum of 1 obtained form the incubation of curcumin in (A) regular buffer and (B) buffer containing 92% H218O. The MS2 spectra obtained by CID of the molecular ions m/z 399 and 401 are shown in supplemental Fig. S2. MS3 spectra obtained by CID of (C) m/z 247 and (D) m/z 249. The solid arrows indicate fragments derived from m/z 247 and m/z 249, respectively. The dashed arrows indicate the fragments obtained by CID of m/z 232 (in panel C) and m/z 234 (in panel D), respectively. E, proposed fragmentation of 1 in the CID experiments. The asterisk denotes the oxygen atom derived from H218O and the fragments that contain 18O. The MSn analyses were performed using an ion trap instrument as described under “Experimental Procedures.”
The site of incorporation of oxygen from water into 1 was determined by CID of the [M-H]− molecular ions at m/z 399 and 401, and by subsequent MSn of the respective fragment ions. The MS2 spectra of the molecular ion showed a fragment ion at m/z 247 or 249, respectively, as the base peak (supplemental Fig. S2). In addition, both spectra contained fragments at m/z 151 and 353, implicating that these ions were derived by loss of a fragment containing the labeled oxygen, the latter m/z 353 likely being derived from the loss of water and CO or C18O, respectively. Further fragmentation (MS3) of m/z 247 or 249 showed consecutive loss of 15 amu (-CH3) to give m/z 232 or 234, respectively, followed by loss of CO or C18O, respectively, to result in m/z 204 in both cases (Fig. 7, C and D). Based on these results, the major fragment ions m/z 247 or 249, respectively, were found to represent the bicyclic ring moiety with one of the phenolic rings attached (ring B) but lacking carbon 1 of the bicyclic ring (Fig. 7E). Carbon 1 was part of the fragment m/z 151 formed by CID of the molecular ions (m/z 399 and 401). Together, these data indicated that the 18O derived from water was incorporated into the substituted furan ring of 1, i.e. between carbons 1 and 7.
To provide additional evidence for the assignment of the fragments observed in the MSn experiments, the asymmetric curcumin analog 4′-methoxycurcumin was reacted with COX-2 in unlabeled and in H218O-buffer. The proposed mechanism of autoxidative and COX-2 catalyzed transformation of curcumin implies that only one of two isomeric oxygenation products will be formed, and that, therefore, the increase in 14 amu due to the additional methyl group at one of the phenolic rings can be used to assign the mass spectrometric fragment ions (supplemental Fig. S3). The reaction of COX-2 with 4′-methoxycurcumin proceeded at similar initial rate but less total conversion was observed (Table 1). As expected, the oxygenated product had a MW of 414 or 416, respectively, when formed in H218O-enriched buffer. The incorporation of 18O into the product was calculated to be about 25%. MS2 analyses of the molecular ions at m/z 413 and 415, respectively, gave m/z 247 or 249 as the base peak (supplemental Fig. S3). This indicated that the additional 4′-methyl group was not part of the bicyclic ring structure that contained the 18O but was located on the phenolic ring (ring A) that is attached to carbon 1 through the ether bridge.
Additional Mechanistic Experiments
We hypothesized that 4′,4″-dimethoxycurcumin would be protected from autoxidative as well as COX-2 catalyzed transformation due to blocking of both phenolic hydroxyl groups. 4′,4″-Dimethoxycurcumin at 50 μm was stable in potassium phosphate buffer pH 8 showing no measurable decrease of the chromophore at 430 nm over the course of 15 min. Addition of 300 μm H2O2 and/or 50 nm COX-2 induced only a minimal loss of the chromophore of 4′,4″-dimethoxycurcumin (Table 1).
Catalysis by peroxidase and lack of reaction of 4′,4″-dimethoxycurcumin implicated formation of a phenoxyl radical as a likely initial step of the transformation of curcumin. We hypothesized that the phenoxyl radical is formed by oxidation of the phenolate anion with molecular oxygen serving as the oxidant (electron acceptor) that would be reduced to superoxide. The formation of superoxide was determined as the reduction of cyctochrome c (12.5 μm) measured at 550 nm, and its inhibition by superoxide dismutase (28). The rate of superoxide formed was 0.1 μm/min over the course of autoxidation of 50 μm curcumin (20 min). Addition of catalase in order to prevent the possibility of re-oxidation of reduced cytochrome c by H2O2 (derived from dismutation of superoxide) did not change the rate of reduction of cytochrome c.
Attempts to trap a putative hydroperoxide intermediate of oxidative transformation by reduction were not successful. When the autoxidative and COX-2 catalyzed transformation of curcumin were extracted and treated immediately with an excess of triphenylphosphine no change in the product profile was observed upon analysis by RP-HPLC with diode array detection. Similarly, inclusion of 1 mm SnCl2 in the reaction of curcumin with COX-2 did not change the product profile but reduced the extent of transformation.
Transformation of Curcumin in RAW264.7 Cells
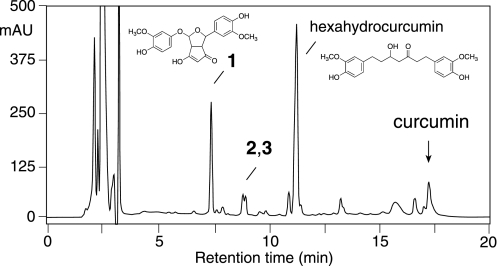
Oxidative transformation of curcumin was also analyzed upon addition to RAW264.7 murine macrophage cells. Cells were treated for 5 h without or with LPS (100 ng/ml) to induce expression of COX-2 and then treated with 20 μm curcumin for 2 h. RP-HPLC analyses with UV detection showed that 1 was one of two major transformation products of curcumin detectable (Fig. 8). A second prominent peak at 11.4 min retention time consisted of hexahydrocurcumin and an unidentified co-eluting product. Hexahydrocurcumin was identified by comparison of its retention time and UV spectrum to an authentic standard prepared by partial hydrogenation of curcumin.
FIGURE 8.
RP-HPLC analysis of the incubation of curcumin with LPS-activated RAW264.7 cells. RAW264.7 cells were activated with 100 ng/ml LPS for 5 h in DMEM adjusted to pH 7.6 using 20 mm sodium phosphate buffer in the absence of FBS. Curcumin (20 μm) was added to the RAW264.7 cells and incubated for additional 2 h. The medium was removed, acidified to pH 4, and extracted twice with 2.5 ml of ethyl acetate/isopropanol (90:10, by vol). An aliquot was injected on RP-HPLC with diode array detection using the same conditions as in Fig. 4. The peak identified as hexahydrocurcumin (structure sown) contained an unidentified product that eluted as a front shoulder. The chromatogram was recorded at 205 nm.
The metabolic profile of curcumin was similar in RAW264.7 cells activated with LPS and in unactivated cells. In unstimulated cells, however, 83.0 ± 0.9% of curcumin was degraded after 2 h incubation whereas in activated cells 92.7 ± 0.4% of curcumin was degraded. The peak area of the bicyclopentadione product was increased 1.8-fold in LPS-activated RAW264.7 cells compared with unactivated cells.
DISCUSSION
The obvious structural differences of curcumin and the bicyclopentadione product prevented a ready explanation of the mechanism of oxidative transformation. The net increase of 32 amu of the bicyclopentadione initially suggested that both oxygen atoms were derived from incorporation of molecular oxygen, similar to the addition of O2 to a polyunsaturated fatty acid during autoxidation, and likewise, the incorporation of O2 into a fatty acid substrate in COX-2 and lipoxygenase catalysis (30, 31). Important clues about the mechanism came from analysis of the reaction catalyzed by COX-2. In the case of enzymatic transformation of curcumin, binding in the fatty acid binding site and catalytic reaction with O2 appeared a straightforward possibility. Certainly for COX-2 catalysis this is not the case since indomethacin, an inhibitor that blocks access of substrate to the oxygenase active site, did not inhibit COX-2 catalyzed transformation of curcumin. Acetaminophen, on the other hand, which acts at the peroxidase active site of COX-2 (32) inhibited the reaction, implicating that the peroxidase rather than the oxygenase activity of the enzyme was crucial.
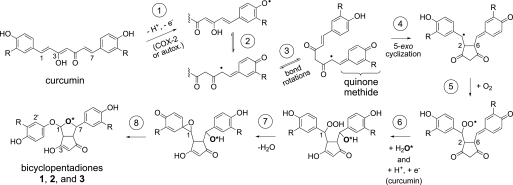
A mechanism for autoxidative and COX-2 catalyzed transformation of curcumin that is consistent with the experimental observations is proposed in Fig. 9. In both cases, reaction is initiated by the formal hydrogen atom abstraction from one of the phenolic hydroxyl groups (step 1 in Fig. 9). Given the pH of the medium (pH 8) and the first pKa of curcumin (≈8.5), it is likely that the reaction is initiated by an electron transfer from the phenolate anion, akin to the concept of “sequential proton loss electron transfer (SPLET)” advanced by Litwinienko and Ingold (33). The peroxidase active site of the COX enzyme is promiscuous with regard to the H-atom donor and accepts a large variety of phenols, aromatic amines, β-dicarbonyls, and naturally occurring compounds (e.g. ascorbic acid, uric acid, glutathione) (34). As expected for catalysis by the peroxidase active site, addition of H2O2 greatly accelerated the rate of reaction (Table 1). The initial oxidation is presumably the only true enzymatic reaction in the COX-2 catalyzed transformation of curcumin, all of the following steps being non-enzymatic. The resulting phenoxyl radical has significant radical character at C6 of the heptadienone chain, which allows it to undergo 5-exo cyclization onto C2 to yield the cyclopenta-1,3-dione moiety of 1 upon rotation about the C4-C5 bond (steps 2–4). Addition of O2 to the newly formed C1 radical is presumably diffusion controlled (step 5), and the resultant peroxyl radical can serve as the autoxidation chain-carrying radical through reaction with another molecule of curcumin. It is not clear whether hydration of the quinone methide moiety or reduction of the peroxyl radical to the hydroperoxide by curcumin would occur next (step 6). While the rate constants for hydration of analogous o-methoxy p-quinone methides have been determined (pseudo first order rate constant of ≈0.12 s−1) (35), the rate constant for the curcumin peroxyl reaction has only been determined in chlorobenzene (3.4 × 105 m−1 s−1) (36) where the mechanism is presumably H-atom transfer rather than a SPLET mechanism, making comparison of the rates difficult. The tetrahydrofuran ring of the bicyclic moiety is formed when the C1-hydroperoxide dehydrates to undergo a 1,2-carbon-to-oxygen aryl migration whereby the electron rich phenolic ring attacks the electrophilic oxygen of the hydroperoxide to form a spiro cyclohexadienone epoxide intermediate (step 7). The unstable epoxide is opened by the neighboring alcohol to yield the bicyclopentadione 1 (step 8).
FIGURE 9.
Proposed mechanism of autoxidative and COX-2 catalyzed transformation of curcumin. The reaction steps are explained in the “Discussion.” Products 2 and 3 are isomers of 1 differing in the configuration of carbons 1 and 7, respectively. R = -OCH3.
Incorporation of water during the transformation was evident from reactions carried out in H218O-buffer. MSn analyses proved that the oxygen derived from water is the one forming the ether bridge linking carbons 1 and 7 of the tetrahydrofuran ring. This was confirmed by analysis of the main reaction product formed from the asymmetrical analog 4′-methoxycurcumin. The second oxygen connecting the distal aromatic ring with the newly formed bicyclic moiety is derived from molecular oxygen.
The antioxidant mechanism of curcumin has been the subject of a controversial debate for more than a decade, e.g. Ref. 33, 36–38. What is clear is that the antioxidant reaction of curcumin (as with other phenols) is the donation of a hydrogen atom to a lipid peroxyl radical, thereby forming a lipid hydroperoxide and interrupting the radical chain reaction. What is less clear is at which of the two possible sites hydrogen abstraction takes place, i.e. at the phenolic OH or the central CH2 in the β-diketo form of curcumin, and also whether a classical hydrogen atom transfer (“HAT”) takes place or rather what has been described as a sequential proton loss electron transfer (SPLET) reaction (33, 38). The essence of the SPLET reaction is that first loss of a proton occurs and then an electron is donated from the phenolate anion. This is the most likely mechanism in polar solvents such as water as we have used here. Nevertheless, our results contribute little to this discussion since they are concerned with what happens to curcumin after the antioxidant reaction, i.e. after the hydrogen atom has been abstracted from the phenolic hydroxyl.
Leaving detailed mechanistic considerations aside, there is the unexpected finding that in the COX-2 catalyzed transformation of curcumin the peroxidase rather than the cyclooxygenase active site is involved in the oxygenation reaction. The insertion of oxygen, however, is non-enzymatic and dependent on the particular structure of curcumin that enables a radical intermediate to undergo reaction with molecular oxygen and to further rearrange the peroxide into a stable product. Other natural phenolic compounds that react at the peroxidase site of the COX enzymes, for example eugenol and resveratrol, undergo a similar initial hydrogen abstraction to yield a phenoxyl radical but fail to incorporate molecular oxygen as part of the transformation since phenoxyl radicals are unreactive to O2 (39, 40). By donating a hydrogen, many flavonoids and other plant phenolic compounds have been shown to inhibit the activity of various LOX enzymes (41). Hydrogen abstraction from the phenolic hydroxyl keeps the catalytic non-heme iron of LOX enzymes in its reduced, inactive state. In the transformation of curcumin, however, the peroxyl radical can regenerate the oxidized enzyme allowing for catalytic turnover, and thus for the use of curcumin as a substrate in LOX catalysis (25). This hypothesis is supported by crystallization studies that suggest binding of curcumin in or near the fatty acid-binding site of a soybean LOX isozyme (42).
The degradation of curcumin in buffer at alkaline pH and upon addition to cultured cells is well documented (7). Our studies implicate that oxidative transformation could also contribute to the degradation of curcumin in cultured cells. Using RAW264.7 mouse macrophage-like cells we found that the cyclopentadione metabolite is almost equally abundant to hexahydrocurcumin, the main reductive metabolite found in animals and humans (43). Furthermore, since LPS-activated RAW264.7 cells showed greater degradation of curcumin and almost 2-fold increased formation of the bicyclopentadione product there is the possibility of enzymatic contribution to the oxidative transformation of curcumin. Enzymatic oxidation of curcumin is likely to occur not only by COX-2 but also by other peroxidases like myeloperoxidase and NADPH oxidase.
Several studies have addressed the question as to whether degradation products of curcumin could be responsible for some of the biological effects described for curcumin. The reduced metabolites, tetra-, hexa-, and octahydrocurcumin, have either greatly decreased or different biological activity compared with curcumin (43–45). In a few instances the biological activities of the putative degradation products vanillin, ferulic acid, and feruloylmethane were investigated, and these were also found to be only weakly active, at best, when compared with the same effects of curcumin (10, 12, 46, 47). There is the possibility, however, that the quinone methide intermediate or the bicyclopentadione product are biologically active and responsible for some of the activities of curcumin. Quinone methides derived from natural phenolic compounds readily form adducts with glutathione and cellular macromolecules, e.g. protein and DNA (48–51), and the antitumor activity of many chemotherapeutic drugs, e.g. etoposide or mitomycin C, is mediated by the formation of a reactive quinone methide (52–54). In line with this argument, the formation of covalent protein adducts by curcumin has been observed, for example, with thioredoxin reductase (55) and in the cross-linking of cystic fibrosis transmembrane conductance regulator polypeptides (56). These adducts could have been formed by the quinone methide rather than by curcumin itself. The possibility of oxidative activation should be considered as a potential mechanism of the therapeutic activity of curcumin.
Supplementary Material
Acknowledgment
We thank Dr. M. Wade Calcutt for help with HRMS.
This work was supported, in whole or in part, by National Institutes of Health Pilot and Feasibility Grant P30 ES000267 from the Center of Molecular Toxicology and Grant R01GM076592 from the NIGMS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- COX
- cyclooxygenase
- ABTS
- [2,2′-azinobis-(3-ethyl-benzothiazoline-6-sulphonic acid
- ApAP
- acetaminophen (N-acetyl-p-aminophenol)
- CID
- collision-induced dissociation
- ESI
- electrospray ionization
- HRMS
- high resolution mass spectrometry
- LC-MS
- liquid chromatography-mass spectrometry
- LOX
- lipoxygenase
- LPS
- lipopolysaccharide
- PG
- prostaglandin
- SPLET
- sequential proton loss electron transfer.
REFERENCES
- 1. Aggarwal B. B., Sung B. (2009) Trends Pharmacol. Sci. 30, 85–94 [DOI] [PubMed] [Google Scholar]
- 2. Aggarwal B. B., Shishodia S. (2006) Biochem. Pharmacol. 71, 1397–1421 [DOI] [PubMed] [Google Scholar]
- 3. Hatcher H., Planalp R., Cho J., Torti F. M., Torti S. V. (2008) Cell. Mol. Life Sci. 65, 1631–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tønnesen H. H., Karlsen J., van Henegouwen G. B. (1986) Z. Lebensm. Unters. Forsch. 183, 116–122 [DOI] [PubMed] [Google Scholar]
- 5. Khurana A., Ho C. T. (1988) J. Liquid Chromatogr. 11, 2295–2304 [Google Scholar]
- 6. Tonnesen H. H., Karlsen J. (1985) Z. Lebensm. Unters. Forsch. 180, 132–134 [DOI] [PubMed] [Google Scholar]
- 7. Wang Y. J., Pan M. H., Cheng A. L., Lin L. I., Ho Y. S., Hsieh C. Y., Lin J. K. (1997) J. Pharm. Biomed. Anal. 15, 1867–1876 [DOI] [PubMed] [Google Scholar]
- 8. Pfeiffer E., Hoehle S. I., Solyom A. M., Metzler M. (2003) J. Food Eng. 56, 257–259 [Google Scholar]
- 9. Dempe J. S., Pfeiffer E., Grimm A. S., Metzler M. (2008) Mol. Nutr. Food Res. 52, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 10. Appiah-Opong R., Commandeur J. N., van Vugt-Lussenburg B., Vermeulen N. P. (2007) Toxicology 235, 83–91 [DOI] [PubMed] [Google Scholar]
- 11. Shen L., Ji H. F. (2009) Clin. Cancer Res. 15, 7108–7109 [DOI] [PubMed] [Google Scholar]
- 12. Dhillon N., Sung B., Kurzrock R., Aggarwal B. B. (2009) Clin. Cancer Res. 15, 7108–710919903776 [Google Scholar]
- 13. Reddy B. S., Rao C. V. (2002) J. Environ. Pathol. Toxicol. Oncol. 21, 155–164 [PubMed] [Google Scholar]
- 14. Murakami A., Ohigashi H. (2007) Int. J. Cancer 121, 2357–2363 [DOI] [PubMed] [Google Scholar]
- 15. Marnett L. J., DuBois R. N. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 55–80 [DOI] [PubMed] [Google Scholar]
- 16. Sharma R. A., McLelland H. R., Hill K. A., Ireson C. R., Euden S. A., Manson M. M., Pirmohamed M., Marnett L. J., Gescher A. J., Steward W. P. (2001) Clin. Cancer Res. 7, 1894–1900 [PubMed] [Google Scholar]
- 17. Garcea G., Berry D. P., Jones D. J., Singh R., Dennison A. R., Farmer P. B., Sharma R. A., Steward W. P., Gescher A. J. (2005) Cancer Epidemiol. Biomarkers Prev. 14, 120–125 [PubMed] [Google Scholar]
- 18. Plummer S. M., Holloway K. A., Manson M. M., Munks R. J., Kaptein A., Farrow S., Howells L. (1999) Oncogene 18, 6013–6020 [DOI] [PubMed] [Google Scholar]
- 19. Goel A., Boland C. R., Chauhan D. P. (2001) Cancer Lett. 172, 111–118 [DOI] [PubMed] [Google Scholar]
- 20. Ramsewak R. S., DeWitt D. L., Nair M. G. (2000) Phytomedicine 7, 303–308 [DOI] [PubMed] [Google Scholar]
- 21. Koeberle A., Northoff H., Werz O. (2009) Mol. Cancer Ther. 8, 2348–2355 [DOI] [PubMed] [Google Scholar]
- 22. Huang M. T., Lysz T., Ferraro T., Abidi T. F., Laskin J. D., Conney A. H. (1991) Cancer Res. 51, 813–819 [PubMed] [Google Scholar]
- 23. Prasad N. S., Raghavendra R., Lokesh B. R., Naidu K. A. (2004) Prostaglandins Leukot. Essent. Fatty Acids 70, 521–528 [DOI] [PubMed] [Google Scholar]
- 24. Rao C. V. (2007) Adv. Exp. Med. Biol. 595, 213–226 [DOI] [PubMed] [Google Scholar]
- 25. Schneider C., Amberg A., Feurle J., Ross A., Roth M., Tóth G., Schreier P. (1998) J. Mol. Catalysis B: Enzymatic 4, 219–227 [Google Scholar]
- 26. Pabon H. J. (1964) Recl. Trav. Chim. Pays Bas. 83, 379–386 [Google Scholar]
- 27. Schneider C., Boeglin W. E., Brash A. R. (2004) J. Biol. Chem. 279, 4404–4414 [DOI] [PubMed] [Google Scholar]
- 28. Klegeris A., Korkina L. G., Greenfield S. A. (1995) Free Radic. Biol. Med. 18, 215–222 [DOI] [PubMed] [Google Scholar]
- 29. Toth G., Roth M., Weckerle B., Schreier P. (2000) Magn. Res. Chem. 38, 51–54 [Google Scholar]
- 30. Porter N. A., Caldwell S. E., Mills K. A. (1995) Lipids 30, 277–290 [DOI] [PubMed] [Google Scholar]
- 31. Schneider C., Pratt D. A., Porter N. A., Brash A. R. (2007) Chem. Biol. 14, 473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aronoff D. M., Oates J. A., Boutaud O. (2006) Clin. Pharmacol. Ther. 79, 9–19 [DOI] [PubMed] [Google Scholar]
- 33. Litwinienko G., Ingold K. U. (2004) J. Org. Chem. 69, 5888–5896 [DOI] [PubMed] [Google Scholar]
- 34. Markey C. M., Alward A., Weller P. E., Marnett L. J. (1987) J. Biol. Chem. 262, 6266–6279 [PubMed] [Google Scholar]
- 35. Bolton J. L., Comeau E., Vukomanovic V. (1995) Chem. Biol. Interact. 95, 279–290 [DOI] [PubMed] [Google Scholar]
- 36. Barclay L. R., Vinqvist M. R., Mukai K., Goto H., Hashimoto Y., Tokunaga A., Uno H. (2000) Org. Lett. 2, 2841–2843 [DOI] [PubMed] [Google Scholar]
- 37. Jovanovic S. V., Steenken S., Boone C. W., Simic M. G. (1999) J. Am. Chem. Soc. 121, 9677–9681 [Google Scholar]
- 38. Galano A., Alvarez-Diduk R., Ramirez-Silva M. T., Alarcon-Angeles G., Rojas-Hernandez A. (2009) Chem. Phys. 363, 13–23 [Google Scholar]
- 39. Thompson D., Constantin-Teodosiu D., Norbeck K., Svensson B., Moldéus P. (1989) Chem Res Toxicol 2, 186–192 [DOI] [PubMed] [Google Scholar]
- 40. Szewczuk L. M., Lee S. H., Blair I. A., Penning T. M. (2005) J. Nat. Prod. 68, 36–42 [DOI] [PubMed] [Google Scholar]
- 41. Werz O. (2007) Planta Med. 73, 1331–1357 [DOI] [PubMed] [Google Scholar]
- 42. Skrzypczak-Jankun E., Zhou K., McCabe N. P., Selman S. H., Jankun J. (2003) Int. J. Mol. Med. 12, 17–24 [PubMed] [Google Scholar]
- 43. Ireson C., Orr S., Jones D. J., Verschoyle R., Lim C. K., Luo J. L., Howells L., Plummer S., Jukes R., Williams M., Steward W. P., Gescher A. (2001) Cancer Res. 61, 1058–1064 [PubMed] [Google Scholar]
- 44. Pan M. H., Lin-Shiau S. Y., Lin J. K. (2000) Biochem. Pharmacol. 60, 1665–1676 [DOI] [PubMed] [Google Scholar]
- 45. Liu Z., Xie Z., Jones W., Pavlovicz R. E., Liu S., Yu J., Li P. K., Lin J., Fuchs J. R., Marcucci G., Li C., Chan K. K. (2009) Bioorg. Med. Chem. Lett. 19, 706–709 [DOI] [PubMed] [Google Scholar]
- 46. Klasing S. A., Mora M. I., Wilson W. C., Fahey G. C., Jr., Garst J. E. (1985) Proc. Soc. Exp. Biol. Med. 179, 529–538 [DOI] [PubMed] [Google Scholar]
- 47. Deters M., Knochenwefel H., Lindhorst D., Koal T., Meyer H. H., Hänsel W., Resch K., Kaever V. (2008) Pharm. Res. 25, 1822–1827 [DOI] [PubMed] [Google Scholar]
- 48. Bodell W. J., Ye Q., Pathak D. N., Pongracz K. (1998) Carcinogenesis 19, 437–443 [DOI] [PubMed] [Google Scholar]
- 49. Thompson D. C., Thompson J. A., Sugumaran M., Moldéus P. (1993) Chem. Biol. Interact. 86, 129–162 [DOI] [PubMed] [Google Scholar]
- 50. Thompson D. C., Perera K., Krol E. S., Bolton J. L. (1995) Chem. Res. Toxicol. 8, 323–327 [DOI] [PubMed] [Google Scholar]
- 51. van der Woude H., Alink G. M., van Rossum B. E., Walle K., van Steeg H., Walle T., Rietjens I. M. (2005) Chem. Res. Toxicol. 18, 1907–1916 [DOI] [PubMed] [Google Scholar]
- 52. Haim N., Nemec J., Roman J., Sinha B. K. (1987) Cancer Res. 47, 5835–5840 [PubMed] [Google Scholar]
- 53. Peterson D. M., Fisher J. (1986) Biochemistry 25, 4077–4084 [DOI] [PubMed] [Google Scholar]
- 54. Moore H. W., Czerniak R., Hamdan A. (1986) Drugs Exp Clin Res 12, 475–494 [PubMed] [Google Scholar]
- 55. Fang J., Lu J., Holmgren A. (2005) J. Biol. Chem. 280, 25284–25290 [DOI] [PubMed] [Google Scholar]
- 56. Bernard K., Wang W., Narlawar R., Schmidt B., Kirk K. L. (2009) J. Biol. Chem. 284, 30754–30765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.