Abstract
Although the mechanisms of generation of signals that control transcriptional activation of Type III IFN (IFNλ)-regulated genes have been identified, very little is known about the mechanisms by which the IFNλ receptor generates signals for mRNA translation of IFNλ-activated genes. We provide evidence that IFNλ activates the p90 ribosomal protein S6 kinase 1 (RSK1) and its downstream effector, initiation factor eIF4B. Prior to its engagement by the IFNλ receptor, the non-active form of RSK1 is present in a complex with the translational repressor 4E-BP1 in IFNλ-sensitive cells. IFNλ-inducible phosphorylation/activation of RSK1 results in its dissociation from 4E-BP1 at the same time that 4E-BP1 dissociates from eIF4E to allow formation of eIF4F and initiation of cap-dependent translation. Our studies demonstrate that such IFNλ-dependent engagement of RSK1 is essential for up-regulation of p21WAF1/CIP1 expression, suggesting a mechanism for generation of growth-inhibitory responses. Altogether, our data provide evidence for a critical role for the activated RSK1 in IFNλ signaling.
Keywords: Interferon, MAP Kinases (MAPKs), S6 Kinase, Serine Threonine Protein Kinase, Signal Transduction, Translation Regulation
Introduction
The recently identified family of Type III IFNs includes IFNλ1, IFNλ2, and IFNλ3, also called interleukin IL-29, IL-28A, and IL-28B, respectively (1, 2). All IFNλs generate their biological effects via binding to a common receptor complex consisting of the specific IFNλR1 subunit and the IL-10Rβ subunit, which is shared with the IL-10 receptor and related cytokine receptors (1, 3–5). Despite engaging entirely different receptor complexes, Type I (α, β, ω) and Type III (λ) IFNs share common Jak-Stat pathways to mediate signals for transcription of target genes. Both Type I IFNs and IFNλ activate receptor-interacting tyrosine kinases Jak1 and Tyk2, which in turn regulate phosphorylation of members of the STAT family (6, 7), leading to formation/assembly of the ISGF3 complex that regulates transcription via binding to interferon-stimulated response elements (6–9). Beyond specific gene expression, IFNλ induces distinct genes but also many genes are known to be IFNα-dependent (1, 3, 10). Notably, IFNλ-induced gene expression has clearly different timing patterns and slower kinetics than IFNα (1, 3, 10).
IFNλ exhibits important biological activities, including antiviral protection, growth-inhibitory and proapoptotic effects, and immunomodulatory and antitumor activities (1, 3, 5, 8, 10, 12–18). Such functional properties of IFNλ have generated substantial interest and raised prospects for utilization of IFNλ in the treatment of human diseases, including viral infections and malignancies. However, despite the advances in the field and the emerging understanding of the mechanisms of Type III IFN-induced transcriptional activation, very little is known about the signals by which the Type III IFN receptor ultimately controls mRNA translation of target genes to induce IFNλ-dependent biological responses.
Over the last 2 decades, our overall understanding of the mechanisms of control of mRNA translation in eukaryotes has evolved, and several important steps have been defined. Control of mRNA translation occurs primarily at the initiation step, in which the small (40 S) ribosomal subunit and the eIF2·tRNAMet·GTP ternary complex form the 43 S complex in association with eIF3. The 43 S complex associates with eIF4F (eukaryotic initiation factor 4F), resulting in formation of the 48 S complex that is recruited to the 5′-end of mRNA and scans the 5′-untranslated region (5′-UTR) for AUG (19). The recruitment of 40 S to mRNA requires the assembly of the eIF4F complex and its binding to the mRNA 5′ cap structure (7-methylguanosine cap structure; m7GpppN). The eIF4F complex is composed of eIF4E, the cap-binding subunit; eIF4G, the scaffolding protein that binds other initiation factors; and eIF4A (an RNA helicase whose function is essential for unwinding of the mRNA 5′ secondary structure) (19–22). Recruitment of eIF4F to the 5′ cap structure is a key step at which mRNA translation in different systems is regulated, via competition between the translational repressor 4E-BP1 and eIF4G for binding to eIF4E (23). Various growth factors, hormones, and mitogens control translation at that level by inducing activation of the mTOR kinase and mTOR-mediated 4E-BP1-phosphorylation, resulting in deactivation of 4E-BP1 and its dissociation from eIF4E (19, 24).
We have previously shown that Type I and II IFNs engage the mTOR pathway and regulate its downstream effectors S6K and 4E-BP1 (25–27). Such engagement of this cascade plays important roles in the ISG mRNA translation process (25, 27–29). Other work has established that the extracellular signal-regulated kinase (ERK) pathway is activated in response to Type I, II, and III IFNs (7, 30). Moreover, there is evidence that RSK mediates Type I IFN-dependent eIF4B phosphorylation in certain cell types (28), but the precise involvement and role of RSK in IFN-dependent translational regulation as well as global mRNA translation in eukaryotes remains to be fully elucidated.
In the present work, we provide evidence for IFNλ-dependent activation of the mTOR and ERK/RSK pathways and demonstrate that both pathways play important roles in IFNλ signaling. Remarkably, our data establish for the first time that the inactive form of RSK1 is constitutively bound to 4E-BP1 and participates in a complex that prevents recruitment of components of eIF4F to the 5′ cap structure. IFNλ treatment results in RSK1 phosphorylation/activation and dissociation from 4E-BP1, and this event is followed by RSK1-mediated eIF4B activation. Interestingly, IFNλ-dependent phosphorylation of 4E-BP1 is only partially blocked by either MEK or mTOR inhibition in IFNλ-sensitive cells, indicating that input from both MEK/ERK and mTOR is necessary for phosphorylation/deactivation of the protein during its engagement by the IFNλ receptor. Our studies also demonstrate that inhibition of RSK1 activity mediates IFNλ-inducible antiproliferative responses, establishing key and essential roles for RSK1 in IFNλ signaling.
MATERIALS AND METHODS
Cells and Reagents
Antibodies against the phosphorylated forms of eIF4B, S6K, 4E-BP1, ERK1/2, PDK1, RSK1, and antibodies against eIF4B, 4E-BP1, eIF4E, GST, eIF4G, and PDK1 were obtained from Cell Signaling Technology (Beverly, MA). Antibodies against p70S6K, ERK1/2, and RSK1 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Human HT-29 cells (colorectal adenocarcinoma) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and antibiotics. Human ARPE-19 cells (retinal pigment epithelial cells) were grown in DMEM/F-12 medium supplemented with 10% fetal bovine serum and antibiotics. Immortalized 4E-BP1+/+ and 4E-BP1−/− mouse embryonic fibroblasts (MEFs)2 (31) were kindly provided by Dr. Nahum Sonenberg (McGill University, Montreal, Canada). The mTOR inhibitor, rapamycin, and the MEK1/2 inhibitor U0126 were obtained from Calbiochem. U0126 was used in the different experiments at a final concentration of 10 μm, whereas rapamycin was used at a final concentration of 20 nm. The RSK inhibitors, SL0101-1 and BI-D1870, were from Tocris Bioscience (Ellisville, MO) and Symansis (Auckland, New Zealand), respectively. SL0101-1 was used at a final concentration of 80 μm, and BI-D1870 was used at a final concentration of 10 μm. GST-4E-BP1, His-tagged 4E-BP1, and active PDK1 were from SignalChem (Richmond, Canada). Activated RSK1 protein and the anti-phospho-Ser221 RSK1 antibodies were purchased from Abcam (Cambridge, MA). Non-active RSK1 was from Abnova (Taipei, Taiwan), whereas activated ERK1 protein was from Milipore (Billerica, MA). The His60 Ni Gravity kit was purchased from Clontech (Mountain View, CA). 7-Methyl-GTP-Sepharose was from GE Healthcare UK Ltd. siRNAs against 4E-BP1 and RSK1 were obtained from Santa Cruz Biotechnology, Inc., and transfections were performed using the siRNA transfection reagent TransIT-TKO from Mirus Bio Corp. (Madison, WI). Protein phosphatase 2A (PP2A) was from Upstate Biotechnology, Inc. (Lake Placid, NY).
Immunoprecipitations and Immunoblotting
Cells were treated with IFNλ1 (10 ng/ml) for the indicated times and lysed in phosphorylation lysis buffer as described previously (32, 33). In some experiments, cells were serum-starved for 24 h prior to the indicated treatments. Immunoprecipitations and immunoblotting using an enhanced chemiluminescence method were performed as described previously (32–36).
Cap Binding Assays
HT-29 cells were incubated for 24 h in serum-free medium and then pretreated for 1 h with U0126 (10 μm) or rapamycin (20 nm) and then treated with IFNλ (10 ng/ml) for the indicated times. Cell lysates were incubated with 7-methyl-GTP-Sepharose (Amersham Biosciences) for 4 h and then washed with lysis buffer. Proteins were resolved by SDS-PAGE electrophoresis, transferred on Immobilon membranes (Millipore), and probed with the indicated antibodies.
Protein Binding Assays
Different GST-proteins (GST-RSK1 protein, activated GST-RSK1 protein, or control GST) were annealed with 0.4 μg of His-4E-BP1 in 100 μl of 50 mm Tris, 100 mm sodium chloride, pH 8.0, for 1 h in 4 °C before they were applied to the 100 μl of His60 Ni superflow resin (Clontech), as described previously (37). After extensive washing, the proteins were eluted using 50 mm sodium phosphate, 300 mm sodium chloride, 300 mm imidazole, pH 7.4. Ten 100-μl fractions were collected from each sample, and equal amounts were used for SDS-polyacrylamide electrophoresis. GST, GST-RSK1 fusion proteins, and 4E-BP1 were detected by Western blot analysis using anti-GST or anti-4E-BP1 antibodies.
In Vitro Phosphatase Assays
These assays were performed according to previously described methodologies (38). 2 μg of active GST-RSK1 was incubated at 37 °C for 30 min in 50 μl of PP2A buffer (50 mm Tris, 1 mm DTT, 1 mm MnCl2, pH 8.0) and 0.1 unit of PP2A.
In Vitro Kinase Assays
Immune complex kinase assays to detect RSK1 kinase activity in anti-RSK1 immunoprecipitates were performed essentially as described previously (39). GST-4E-BP1 or His-4E-BP1 was used as exogenous substrate. Studies to detect in vitro phosphorylation of GST-RSK1 by activated ERK1 or activated PDK1 were performed essentially as described previously (40).
Quantitative RT-PCR (TaqMan)
Cells were treated with 10 ng/ml IFNλ for 6 h, and total RNA was isolated using the RNeasy kit (Qiagen). Real-time RT-PCR was performed as in our previous studies (25, 27).
RESULTS
In initial studies, we examined whether IFNλ treatment of sensitive cell lines induces activation of the MEK/ERK pathway and whether RSK1 is activated by the Type III IFN receptor downstream of ERK. HT-29 cells were incubated for different times in the presence or absence of IFNλ, and cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of RSK1 and ERK1/2. Treatment with IFNλ resulted in phosphorylation of RSK1 on Thr359/Ser363 (Fig. 1A) and ERK1/2 on Thr202/Tyr204 (Fig. 1B), consistent with engagement and activation of these kinases. Such phosphorylation was blocked by pretreatment of cells with the MEK inhibitor U0126, whereas the specific mTOR inhibitor, rapamycin, had no inhibitory effects (Fig. 1, A and B). IFNλ treatment also resulted in phosphorylation of eIF4B (Fig. 1C), a protein that plays an important role in the regulation of mRNA translation in eukaryotes and whose phosphorylation has been previously shown to be mediated by either RSK1 or the mTOR-regulated S6K in other systems (20, 28). eIF4B phosphorylation was blocked by the MEK/ERK inhibitor U0126, but not rapamycin, indicating that IFNλ-dependent engagement of eIF4B on Ser422 in HT-29 cells is a MEK/ERK-dependent signaling event.
FIGURE 1.
IFNλ-dependent activation of ERK/RSK1 and mTOR signaling cascades in HT-29 cells. A–F, serum-starved HT-29 cells were pretreated for 60 min with U0126 or rapamycin and were either left untreated or treated with IFNλ, in the continuous presence or absence of rapamycin or U0126, as indicated. The cells were lysed, and total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of RSK1 on Thr359/Ser363 or against RSK1 (A); with antibodies against the phosphorylated forms of ERK on Thr202/Tyr204 or against ERK (B); with antibodies against the phosphorylated form of eIF4B on Ser422 or against eIF4B (C); with antibodies against the phosphorylated form of mTOR on Ser2448 or against GAPDH (D); with antibodies against the phosphorylated form of p70S6K on Thr421/Ser424 or against p70S6K (E); or with antibodies against the phosphorylated forms of 4E-BP1 on Thr37/46 or against GAPDH (F), as indicated.
In further studies, we determined whether engagement of the IFNλ receptor results in activation of the mTOR pathway and generation of downstream signals that mediate initiation of mRNA translation. IFNλ treatment of sensitive cells resulted in phosphorylation of mTOR on Ser2448 (Fig. 1D), a site whose phosphorylation correlates with activation of mTORC1 complexes (41). In addition, such treatment resulted in phosphorylation/activation of downstream effector of mTOR, S6K (Fig. 1E). Activation of S6K was blocked by rapamycin, consistent with mTOR-mediated engagement of the pathway. Treatment of cells with IFNλ also resulted in phosphorylation of the translational repressor 4E-BP1 on Thr37/46 (Fig. 1F), a site whose phosphorylation is required for deactivation of this translational repressor and its dissociation from the initiation factor eIF4E (19, 20, 24). Such phosphorylation of 4E-BP1 was only partially blocked by rapamycin, whereas it was also partially inhibited by U0126 (Fig. 1F). This raised the possibility that coordinated engagement of mTOR and ERK/RSK is required for deactivation of 4E-BP1 during IFNλ treatment of cells. Similar results were obtained when studies were performed using the IFNλ-sensitive ARPE-19 cell line. IFNλ also induced phosphorylation of RSK1 (Fig. 2A), eIF4B (Fig. 2A), 4E-BP1 (Fig. 2B), and S6K (Fig. 2C). As for HT-29 cells, phosphorylation of RSK1 and eIF4B was U0126-sensitive and rapamycin-insensitive, whereas engagement of the S6K was blocked by rapamycin (Fig. 2C). On the other hand, phosphorylation of 4E-BP1 on Thr37/46 was partially blocked by either rapamycin or U0126 (Fig. 2B), suggesting dual IFNλ-dependent regulation of the protein by the mTOR and ERK/RSK pathways.
FIGURE 2.
IFNλ-dependent activation of RSK1 and mTOR pathways in ARPE-19 cells. A–C, serum-starved ARPE-19 cells were pretreated with rapamycin or U0126 as indicated and then treated with IFNλ for the indicated times. Total cell lysates were resolved by SDS-PAGE and immunoblotted with anti-phospho-Thr359/Ser363 RSK1, anti-RSK1, anti-phospho-Ser422-eIF4B, or anti-eIF4B antibodies (A); with anti-phospho-Thr37/46-4E-BP1 or anti-GAPDH antibodies (B); or with antibodies against the phosphorylated form of p70S6K on Thr421/Ser424 or against p70S6K or anti-GAPDH (C), as indicated.
Because the recruitment of eIF4F to the 5′ cap structure of mRNA is an important step in the regulation of mRNA translation, we sought to determine the regulatory effects of IFNλ-dependent activated MEK/ERK or mTOR pathways on the binding of translation initiation factors to the 7-methylguanosine cap complex. IFNλ treatment resulted in dissociation of 4E-BP1 and enhanced binding of eIF4E, eIF4G, and eIF4A to the 7-methylguanosine cap complex (Fig. 3A). Such dissociation of 4E-BP1 upon IFNλ-treatment and binding of the different eukaryotic initiation factors was also seen in rapamycin-pretreated cell lysates (Fig. 3A), consistent with the partial resistance of IFNλ-inducible 4E-BP1 phosphorylation to rapamycin seen in these cells (Fig. 1F). Surprisingly, in cells pretreated with U0126, 4E-BP1 did not dissociate from the complex upon IFNλ treatment, raising the possibility that another protein whose activity is inhibited by U0126 is involved in the process (Fig. 3A).
FIGURE 3.
Binding of 4E-BP1 and RSK1 to the 7-methylguanosine cap complex prevents recruitment of eIF4G, eIF4A, and eIF4E to the cap complex. A, serum-starved HT-29 cells were pretreated for 60 min with U0126 or rapamycin and were either left untreated or treated with IFNλ, in the continuous presence or absence of rapamycin or U0126, as indicated. Cell lysates were bound to the cap analog m7GTP conjugated to beads, and bound proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. B, HT-29 cells were treated as indicated and assayed as described in A. Bound proteins were immunoblotted with the indicated antibodies. C, serum-starved HT-29 cells were pretreated for 6 h with SL0101-1 and were subsequently treated with IFNλ, as indicated. D, serum-starved HT-29 cells were pretreated for 60 min with U0126 or rapamycin and then treated with IFNλ, in the continuous presence or absence of rapamycin or U0126, as indicated. Equal amounts of cell lysates were immunoprecipitated (IP) with an anti-4E-BP1 antibody, and immune complexes were resolved by SDS-PAGE and immunoblotted with anti-RSK1 or anti-4E-BP1 antibodies, as indicated. E, HT-29 cells were pretreated for 6 h with SL0101-1 and were left untreated or treated with IFNλ, in the continuous presence or absence of inhibitor, as indicated. Equal amounts of cell lysates were immunoprecipitated with anti-4EBP1 antibodies, and immune complexes were resolved by SDS-PAGE for analysis of 4E-BP1 and RSK1, as indicated. F, HT-29 cells were transfected with either control siRNA or siRNA specifically targeting 4E-BP1 and treated with IFNλ, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies, against 4E-BP1 or GAPDH, as indicated. G, cells were transfected with either control siRNA or siRNA specifically targeting 4E-BP1 and treated with IFNλ, as indicated. Cell lysates were bound to the cap analog m7GTP conjugated to beads, and bound proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Because RSK1 is a known target and effector of the MEK/ERK pathway, we determined whether non-phosphorylated RSK1 binds to the 7-methylguanosine cap complex. As shown in Fig. 3B, RSK1 was present in the complex and, upon IFNλ treatment, dissociated from it (Fig. 3B). However, RSK1 remained in the complex when cells were treated with IFNλ in the presence of U0126 (Fig. 3B). Because U0126, but not rapamycin, inhibits RSK1 phosphorylation in these cells (Fig. 1A), these data suggested that phosphorylation of the protein is an event that ultimately results in its dissociation from the complex. Consistent with this, RSK1 dissociated from the complex in an IFNλ-dependent manner in the presence of rapamycin (Fig. 3B). Notably, RSK1 followed a similar pattern as 4E-BP1, being detectable in the 7-methylguanosine cap complex at base line and dissociating after IFNλ treatment (Fig. 3A). Such dissociation was reversed by U0126 but not rapamycin (Fig. 3B), suggesting that RSK1 and 4E-BP1 associate with each other in a complex that binds to m7GpppN. IFNλ treatment of HT29 cells also resulted in enhanced binding of eIF4G, eIF4A, and eIF4E to the 7-methylguanosine cap complex, and this correlated with 4E-BP1 phosphorylation and dissociation from the complex (Fig. 3C). However, in cells pretreated with SL0101-1, an inhibitor of the RSK N-terminal kinase domain (NTKD) (40, 42, 43), there was no dissociation of 4E-BP1 and RSK1 from the complex upon IFNλ treatment (Fig. 3C).
To directly determine whether 4E-BP1·RSK1 complexes are formed and to determine the effects of IFNλ on the formation and binding capacities of such complexes, co-immunoprecipitation experiments were carried out. HT-29 cells were treated with IFNλ in the presence or absence of U0126 or rapamycin, and lysates were immunoprecipitated with an anti-4E-BP1 antibody, followed by immunoblotting with an antibody against RSK1. RSK1 protein was co-immunoprecipitated by the anti-4E-BP1 antibody at base line, but IFNλ treatment resulted in a decrease in the levels of 4E-BP1-associated RSK1 (Fig. 3D). Such IFNλ-dependent dissociation of the RSK1–4E-BP1 complex was not seen when the cells were pretreated with U0126 but was still noticeable in cells treated with rapamycin (Fig. 3D). Similarly, the 4EBP1-RSK1 interaction was reversed by treatment with SL0101-1 (Fig. 3E), suggesting that the NTKD of RSK1 is important for this interaction.
To better define the mechanisms of RSK1 interaction with eIF4E and 4E-BP1 and confirm the findings with SL0101-1, experiments were performed in which 4E-BP1 was knocked down using specific siRNA (Fig. 3F). As expected, such knockdown increased eIF4E and eIF4G binding (Fig. 3G), but at the same time it also reduced RSK1 binding to the 5′ cap complex (Fig. 3G).
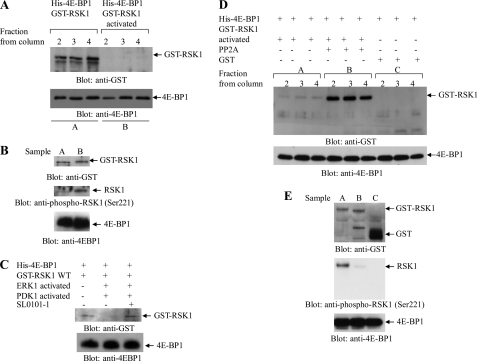
Altogether, our data suggested that 4E-BP1 constitutively associates with RSK1 in HT29 and ARPE-19 cells, but such complexes are only stable when the RSK1 kinase is in the inactive/unphosphorylated state. To further confirm such a hypothesis, protein binding assays were employed. GST-RSK1 or GST-RSK1 (activated) was annealed with His-4E-BP1 and applied to the His60 Ni superflow resin column. Immunoblotting of eluted fractions demonstrated the presence of GST-RSK1 and His-4E-BP1, whereas preactivated GST-RSK1 was not captured in the column together with His-4E-BP1 (Fig. 4A). As shown in Fig. 4B, the inability to bind His-4E-BP1 correlated with phosphorylation of RSK1 at Ser221 in the NTKD (Fig. 4B). To further confirm that the NTKD of RSK1 is required for binding to 4E-BP1, wild type (not activated) RSK1 was used as a substrate for the active ERK1 and active PDK1, in the presence or absence of the SL0101-1 inhibitor. Upon activation in the kinase assay, GST-RSK1 was no longer retained on the column together with His-4E-BP1 (Fig. 4C). However, concomitant treatment with SL0101-1 reversed the binding of RSK1 to 4E-BP1, allowing elution from the column (Fig. 4C). In other studies, we examined whether dephosphorylation of RSK1 enhances its binding to His-4E-BP1. The status of its binding after in vitro dephosphorylation in phosphatase assays using PP2A was examined. As shown in Fig. 4D, in vitro dephosphorylation of preactivated GST-RSK1 resulted in its binding to His-4E-BP1, further demonstrating that only the unphosphorylated form of RSK1 binds to 4E-BP1 (Fig. 4, D and E). The activated form of GST-RSK1 was dephosphorylated after PP2A treatment and had the ability to bind to His-4E-BP1 (Fig. 4E).
FIGURE 4.
Binding of inactive RSK1 to 4E-BP1. A, equal amounts of GST-RSK1 or GST-RSK1 (activated) were annealed with 4E-BP1-His and then bound to the His affinity column. Equal amounts from fractions collected from the affinity column were resolved by SDS-PAGE and immunoblotted with antibodies against GST or 4E-BP1, as indicated. B, input protein levels from the experiment shown in A for GST-RSK1 (sample A) or GST-RSK1 activated (sample B). Immunoblotting with anti-GST or the anti-phospho-Ser221 RSK1 or anti-4E-BP1 (to detect His-4E-BP1) is shown. C, equal amounts of GST-RSK1 were subjected to in vitro kinase assays using active ERK1 and active PDK1, in the presence or absence of the SL0101-1 inhibitor, and then were annealed with 4E-BP1-His and bound to the His affinity column. After extensive washing, proteins were eluted from the column, and equal amounts from each eluted sample were resolved by SDS-PAGE and immunoblotted with antibodies against GST and 4E-BP1, as indicated. D, equal amounts of GST-RSK1 (activated) or GST-RSK1 (activated) that was subjected to an in vitro phosphatase (PP2A) assay were annealed with 4E-BP1-His. After binding to the His affinity column and extensive washing, proteins were eluted from the column, and equal amounts of eluted samples were resolved by SDS-PAGE and immunoblotted with antibodies against GST or 4E-BP1, as indicated. E, input protein levels from the experiment shown in D (panel A) for activated GST-RSK1 (sample A) or activated GST-RSK1 after subjected to in vitro phosphatase (PP2A) assay (sample B) or GST (sample C). Immunoblotting with anti-GST, anti-phospho-Ser221 RSK1 or anti-4E-BP1 (to detect 4E-BP1-His) is shown.
There has been previous evidence from studies using 12-O-tetradecanoylphorbol 13-acetate that, beyond mTOR, engagement of the MEK/ERK pathway can ultimately result in phosphorylation of 4E-BP1 at multiple sites (44). However, the identity of the kinase that directly phosphorylates 4E-BP1 downstream of MEK/ERK has been unknown. We examined whether RSK1 acts as a kinase for 4E-BP1, during its engagement by the IFNλ receptor. Initially, we determined whether RSK1 phosphorylates 4E-BP1 in vitro and in intact cells. Initially, the effects of SL0101-1 on 4E-BP1 phosphorylation on Thr37/46 were examined. As shown in Fig. 5A, IFNλ-dependent phosphorylation of RSK1 on Ser221 and 4E-BP1 on Thr37/46 was blocked by SL0101-1. On the other hand, phosphorylation of RSK1 at Thr573 in the C-terminal kinase domain was not affected by SL0101-1 (Fig. 5A). We also determined the effects of RSK1 knockdown on IFNλ-dependent phosphorylation of 4E-BP1. Knockdown of RSK1 blocked the phosphorylation of 4E-BP1 on Thr37/46 (Fig. 5B), demonstrating that RSK1 is essential for such 4E-BP1 phosphorylation. On the other hand, as expected, RSK1 knockdown did not abrogate IFNλ-dependent phosphorylation/activation of S6K (Fig. 5B). To further establish whether IFNλ-activated RSK1 phosphorylates 4E-BP1, immune complex kinase assays were carried out on anti-RSK1 immunoprecipitates from HT-29 cells using GST-4E-BP1 or 4E-BP1-His as an exogenous substrate. 4E-BP1 was phosphorylated in an IFNλ-activated RSK1 (Fig. 5, C and D), whereas such phosphorylation was selectively blocked by the MEK inhibitor U0126 (Fig. 5C) or RSK inhibitors SL0101-1 and BI-D1870 (Fig. 5D), suggesting that the protein acts as a substrate for RSK1 activity.
FIGURE 5.
RSK1 activity is required for IFNλ-dependent phosphorylation of 4E-BP1 on Thr37/46. A, serum-starved HT-29 cells were pretreated with SL0101-1 or diluent for 6 h and then treated with IFNλ for the indicated times, in the continuous presence or absence of SL0101-1, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Equal cell lysates from the same experiment were analyzed separately by SDS-PAGE and immunoblotted with an anti-phospho-RSK1 (Ser221) and anti-RSK1. B, HT29 cells were transfected with either control siRNA or siRNA targeting RSK1 and treated with IFNλ, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. C, serum-starved HT-29 cells were pretreated with U0126 for 1 h and then treated with IFNλ for 90 min. The cells were lysed, and equal amounts of protein were immunoprecipitated (IP) with an anti-RSK1 antibody. In vitro kinase assays to detect RSK activity were subsequently carried out on the immunoprecipitates, using a 4E-BP1-His protein as an exogenous substrate. D, serum-starved HT-29 cells were pretreated with SL0101-1 for 6 h or BI-D1870 for 1 h and then treated with IFNλ for the indicated times. The cells were lysed, and equal amounts of protein were immunoprecipitated with an anti-RSK1 antibody. In vitro kinase assays to detect RSK activity were subsequently carried out on the immunoprecipitates, using a GST-4E-BP1 protein as an exogenous substrate.
Altogether, our data suggested a model in which the inactive form of RSK1 is associated with 4E-BP1 in a complex that negatively controls eIF4F formation and in which, upon treatment with IFNλ, the kinase is activated and phosphorylates 4E-BP1, resulting in its deactivation and dissociation from eIF4E. To further establish the validity of such a model, experiments were performed using MEFs with targeted disruption of the 4e-bp1 gene (25, 31). Due to the cell type specificity of IFNλ receptor expression, such studies were performed using mouse IFNα. 4E-BP1+/+ and 4E-BP1−/− MEFs were treated with mouse IFNα, and lysates were used in a 7-methylguanosine cap binding assay (Fig. 6A). Abundant amounts of non-phosphorylated RSK1 were detectable in the 7-methylguanosine cap complex in untreated 4E-BP1+/+ but not in 4E-BP1−/− MEFs (Fig. 6A). Consistent with our findings in IFNλ-sensitive cells, IFNα treatment resulted in dissociation of RSK1 from the complex in parental 4E-BP1+/+ MEFs (Fig. 6A). Moreover, binding of eIF4E to the 7-methylguanosine cap complex increased in an IFNα-dependent manner and correlated with phosphorylation and release of 4E-BP1 (Fig. 6A). There were no significant differences in the overall eIF4E levels before or after IFNα treatment in either 4E-BP1 knockouts or parental MEFs, whereas IFNα-inducible phosphorylation of RSK1 was detectable in the presence or absence of 4E-BP1 (Fig. 6B).
FIGURE 6.
RSK1 associates with 4E-BP1 in the 7-methylguanosine cap complex. A, serum-starved 4E-BP1+/+ and 4E-BP1−/− cells were treated with mouse IFNα for the indicated times. Equal amounts of cell lysates were incubated with cap analog beads, and after intensive washing, the retained proteins were resolved by SDS-PAGE and immunoblotted with antibodies against RSK1, 4E-BP1, or eIF4E. B, total cell lysates from the same experiment shown in A were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
We subsequently sought to directly determine the functional relevance of IFNλ-dependent engagement of RSK1, as it relates to generation of growth-inhibitory responses. Several studies have previously established that IFNλ suppresses the growth of several tumor cell lines and intestinal epithelial cells (13, 16, 45). It has been previously shown that growth-inhibitory activity of type I IFN toward HT29 results from the prolongation of cell cycle via the induction of p21WAF1/CIP1, and this effect of IFN was suppressed when p21 expression was down-regulated (46). Recent studies have also demonstrated that the growth-suppressive activity of IFNλ1 in human esophageal carcinoma cell lines is accompanied by up-regulation of p21 (45). We sought to determine whether activation of RSK1 is required for p21WAF1/CIP1 protein expression. IFNλ treatment resulted in induction of p21WAF1 protein expression (Fig. 7A), and this expression was suppressed by treatment of cells with the RSK inhibitor SL0101-1 (Fig. 7A). On the other hand, the transcriptional activation of p21WAF1/CIP1 was not inhibited by SL0101-1 treatment (Fig. 7B). Thus, engagement of RSK1 by IFNλ is required for p21WAF1/CIP1 protein expression but not p21WAF1/CIP1 gene transcription, suggesting a mechanism by which RSK1 may mediate generation of IFNλ-dependent growth-inhibitory responses via control of mRNA translation of p21WAF1/CIP1.
FIGURE 7.
Requirement of RSK1 activity in IFNλ-dependent p21WAF1 expression. A, serum-starved HT29 cells were either not pretreated or pretreated with SL0101-1 and then treated with IFNλ for 24 h. Cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against p21WAF1 or GAPDH, as indicated. B, HT29 cells were either not pretreated or were pretreated with SL0101-1 and then treated with IFNλ for 6 h. Expression of mRNA for the p21 gene was assessed by quantitative real-time RT-PCR. The GAPDH transcript was used for normalization. Data are expressed as -fold increase over IFNλ-untreated samples and represent means ± S.E. (error bars) from three experiments.
DISCUSSION
The family of RSK kinases includes four isoforms, all of which share significant structural homology with each other (47–49). All kinases have in their structure two unique kinase domains, an NTKD that is responsible for phosphorylation of RSK substrates and an autophosphorylation C-terminal kinase domain (47–59). The regulation of RSK activation is complex and involves a series of signaling events that lead to phosphorylation of six conserved sites in the structure of RSKs, including Ser221, Thr359, Ser363, Ser380, Thr573, and Ser749 (48). The phosphorylation of such sites requires the coordinated functions of both the MEK/ERK and the phosphoinositide-dependent protein kinase 1 (PDK1) pathways (43, 48–54).
Several substrates for RSK activity have been previously identified. A major substrate is eIF4B, which regulates the helicase activity of eIF4A (22, 48, 55). RSK-mediated eIF4B phosphorylation also enhances its association with eIF3, resulting in enhanced cap-dependent mRNA translation (48, 56, 57). There is also evidence that RSK1 phosphorylates TSC2 on Ser1798, resulting in its inactivation and enhanced mTOR signaling to S6K (58). RSK1/2 activity is also required for phosphorylation of Raptor at the RXRXXp(S/T) motif (where p(S/T) represents phosphoserine/phosphothreonine) that results in enhanced mTORC1 activity (59), providing further evidence for coordinated regulation and action of the Ras/MAPK and mTOR pathways.
There is extensive interest in defining the mechanisms of signaling by the Type III IFN (IFNλ) receptor. The specific cellular patterns of expression of IFNλ receptors in different cell types have raised the possibility that this recently discovered family of cytokines may provide a unique approach for the treatment of certain viral infections and malignancies with minimal toxicities. Recent work has demonstrated that a key mechanism for transcriptional activation of IFN-regulated genes is engagement of Jak-Stat pathways, but the mechanisms that regulate mRNA translation of IFNλ-regulated genes have been largely unknown. In the present study, we examined whether IFNλ engages the mTOR and MEK/ERK pathways and whether the coordinated activities of these signaling cascades regulate downstream events that may ultimately mediate initiation of mRNA translation of IFNλ-sensitive genes. In initial studies, we found that treatment of sensitive cells with IFNλ results in phosphorylation of mTOR at Ser2448 and downstream engagement of effectors known to be regulated by mTORC1 complexes in other systems, including S6K and its effector S6 ribosomal protein. Moreover, IFNλ was found to induce phosphorylation of the translational repressor 4E-BP1 on sites required for its deactivation and dissociation from the eukaryotic initiation factor 4E (eIF4E). In parallel studies, we found that IFNλ stimulation results in activation of the MEK/ERK pathway and downstream engagement of RSK1. As expected, phosphorylation of S6K was found to be rapamycin-sensitive, consistent with regulation by mTORC1 complexes. On the other hand, the phosphorylation of eIF4B was mediated by RSK1 downstream of ERK, consistent with previous work from our laboratory in the Type I IFN system that had demonstrated that eIF4B can be a substrate for the kinase activities of either RSK1 or S6K, depending on the cellular context (28). Taken together, these studies suggest that IFNλ engages both the mTOR and MEK/ERK pathways for the regulation of events required for the initiation of mRNA translation.
Beyond regulation of eIF4B phosphorylation, RSK activity, downstream of ERK1/2 and PDK1, has previously been shown to regulate, under certain circumstances, rpS6 phosphorylation (11), suggesting a mechanism by which RSK activity regulates mRNA translation. Our current studies in the IFNλ system identify and define another important link between RSK and the translation machinery. Our findings demonstrate that IFNλ-dependent RSK1 activity is required for recruitment of translation initiation factors to the 7-methylguanosine cap complex. Surprisingly, inactive RSK1 was found to be associated with the active form of 4E-BP1 that binds to eIF4E and prevents the recruitment of eIF4G, eIF4E, and eIF4A to the 7-methylguanosine cap complex. Upon IFNλ treatment of sensitive cells, RSK1 is activated and phosphorylates 4E-BP1, and such kinase activity is required for optimal phosphorylation and deactivation of 4E-BP1. Notably, we found that the RSK1 is not present in the 7-methylguanosine cap complex in 4E-BP1 knock-out MEFs. Similarly, the levels of RSK1 present in the complex were diminished in HT29 cells in which 4E-BP1 expression was partially knocked down by specific siRNAs.
Altogether, our studies propose a model in which the inactive form of RSK1 and 4E-BP1 form a complex that binds to eIF4E and prevents initiation of mRNA translation in some cell types. Upon activation of RSK1 by the IFN-regulated MEK/ERK pathway, 4E-BP1 is phosphorylated on Thr37/46, an event required for its deactivation, and, at least in some cell types, such phosphorylation is RSK1-dependent. Thus, RSK1 may play a key role in the initiation of IFNλ-inducible cap-dependent mRNA translation by its presence in 7-methylguanosine cap complex and its ability to phosphorylate and deactivate associated 4E-BP1, possibly in a cell type-specific manner. Although RSK1 is known to be involved in growth factor pathways, it is possible that IFNλ engages the pathway for selective translation of growth-inhibitory genes whose transcription is induced during engagement of the IFNλ receptor. At the same time, engagement and utilization of RSK1 by the IFNλ receptor may result in diminished amounts of RSK1 that are available for growth factor receptor pathways that utilize it for the generation of mitogenic responses.
Although additional studies will be required in the future to precisely define the mechanisms of RSK1–4E-BP1 interaction and the motifs in the structures of the proteins that mediate such complex formation, such effects of RSK1 appear to have important functional implications. Based on our studies, RSK1 activity appears to be required for IFNλ-dependent p21WAF1/CIP1 expression, an event implicated in the generation of antiproliferative responses by IFNλ (46), providing a direct link between the coordinated functions of the mTOR and ERK/RSK pathways and the generation of growth-regulatory and antineoplastic activities by this cytokine.
This work was supported, in whole or in part, by National Institutes of Health Grants CA77816, AG029138, CA121192, HL082946, and CA100579. This work was also supported by a Merit Review Grant from the Department of Veterans Affairs.
- MEF
- mouse embryo fibroblast
- NTKD
- N-terminal kinase domain
- PP2A
- protein phosphatase 2A.
REFERENCES
- 1. Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., Langer J. A., Sheikh F., Dickensheets H., Donnelly R. P. (2003) Nat. Immunol. 4, 69–77 [DOI] [PubMed] [Google Scholar]
- 2. Donnelly R. P., Kotenko S. V. (2010) J. Interferon Cytokine Res. 30, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T. E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F. J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K. M. (2003) Nat. Immunol. 4, 63–68 [DOI] [PubMed] [Google Scholar]
- 4. Kotenko S. V., Langer J. A. (2004) Int. Immunopharmacol. 4, 593–608 [DOI] [PubMed] [Google Scholar]
- 5. Lasfar A., Lewis-Antes A., Smirnov S. V., Anantha S., Abushahba W., Tian B., Reuhl K., Dickensheets H., Sheikh F., Donnelly R. P., Raveche E., Kotenko S. V. (2006) Cancer Res. 66, 4468–4477 [DOI] [PubMed] [Google Scholar]
- 6. Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 7. Zhou Z., Hamming O. J., Ank N., Paludan S. R., Nielsen A. L., Hartmann R. (2007) J. Virol. 81, 7749–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dumoutier L., Tounsi A., Michiels T., Sommereyns C., Kotenko S. V., Renauld J. C. (2004) J. Biol. Chem. 279, 32269–32274 [DOI] [PubMed] [Google Scholar]
- 9. Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 10. Marcello T., Grakoui A., Barba-Spaeth G., Machlin E. S., Kotenko S. V., MacDonald M. R., Rice C. M. (2006) Gastroenterology 131, 1887–1898 [DOI] [PubMed] [Google Scholar]
- 11. Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., Sonenberg N., Blenis J. (2007) J. Biol. Chem. 282, 14056–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartlett N. W., Buttigieg K., Kotenko S. V., Smith G. L. (2005) J. Gen. Virol. 86, 1589–1596 [DOI] [PubMed] [Google Scholar]
- 13. Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S. T., Otte J. M., Diebold J., Diepolder H., Adler B., Auernhammer C. J., Göke B., Dambacher J. (2005) Am. J. Physiol. Gastrointest. Liver Physiol. 289, G960–G968 [DOI] [PubMed] [Google Scholar]
- 14. Hong S. H., Cho O., Kim K., Shin H. J., Kotenko S. V., Park S. (2007) Virus Res. 126, 245–249 [DOI] [PubMed] [Google Scholar]
- 15. Li W., Lewis-Antes A., Huang J., Balan M., Kotenko S. V. (2008) Cell Prolif. 41, 960–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meager A., Visvalingam K., Dilger P., Bryan D., Wadhwa M. (2005) Cytokine 31, 109–118 [DOI] [PubMed] [Google Scholar]
- 17. Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. (2007) J. Biol. Chem. 282, 7576–7581 [DOI] [PubMed] [Google Scholar]
- 18. Zitzmann K., Brand S., Baehs S., Göke B., Meinecke J., Spöttl G., Meyer H., Auernhammer C. J. (2006) Biochem. Biophys. Res. Commun. 344, 1334–1341 [DOI] [PubMed] [Google Scholar]
- 19. Gingras A. C., Raught B., Sonenberg N. (1999) Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 20. Hay N., Sonenberg N. (2004) Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 21. Hernández G., Vazquez-Pianzola P. (2005) Mech. Dev. 122, 865–876 [DOI] [PubMed] [Google Scholar]
- 22. Jaramillo M., Dever T. E., Merrick W. C., Sonenberg N. (1991) Mol. Cell Biol. 11, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C., Jr., Sonenberg N. (1994) Nature 371, 762–767 [DOI] [PubMed] [Google Scholar]
- 24. Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. (2001) Genes Dev. 15, 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaur S., Lal L., Sassano A., Majchrzak-Kita B., Srikanth M., Baker D. P., Petroulakis E., Hay N., Sonenberg N., Fish E. N., Platanias L. C. (2007) J. Biol. Chem. 282, 1757–1768 [DOI] [PubMed] [Google Scholar]
- 26. Kaur S., Katsoulidis E., Platanias L. C. (2008) Cell Cycle 7, 2112–2116 [DOI] [PubMed] [Google Scholar]
- 27. Kaur S., Sassano A., Dolniak B., Joshi S., Majchrzak-Kita B., Baker D. P., Hay N., Fish E. N., Platanias L. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kroczynska B., Kaur S., Katsoulidis E., Majchrzak-Kita B., Sassano A., Kozma S. C., Fish E. N., Platanias L. C. (2009) Mol. Cell Biol. 29, 2865–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thyrell L., Hjortsberg L., Arulampalam V., Panaretakis T., Uhles S., Dagnell M., Zhivotovsky B., Leibiger I., Grandér D., Pokrovskaja K. (2004) J. Biol. Chem. 279, 24152–24162 [DOI] [PubMed] [Google Scholar]
- 30. David M., Petricoin E., 3rd, Benjamin C., Pine R., Weber M. J., Larner A. C. (1995) Science 269, 1721–1723 [DOI] [PubMed] [Google Scholar]
- 31. Tsukiyama-Kohara K., Poulin F., Kohara M., DeMaria C. T., Cheng A., Wu Z., Gingras A. C., Katsume A., Elchebly M., Spiegelman B. M., Harper M. E., Tremblay M. L., Sonenberg N. (2001) Nat. Med. 7, 1128–1132 [DOI] [PubMed] [Google Scholar]
- 32. Uddin S., Majchrzak B., Woodson J., Arunkumar P., Alsayed Y., Pine R., Young P. R., Fish E. N., Platanias L. C. (1999) J. Biol. Chem. 274, 30127–30131 [DOI] [PubMed] [Google Scholar]
- 33. Uddin S., Yenush L., Sun X. J., Sweet M. E., White M. F., Platanias L. C. (1995) J. Biol. Chem. 270, 15938–15941 [DOI] [PubMed] [Google Scholar]
- 34. Giafis N., Katsoulidis E., Sassano A., Tallman M. S., Higgins L. S., Nebreda A. R., Davis R. J., Platanias L. C. (2006) Cancer Res. 66, 6763–6771 [DOI] [PubMed] [Google Scholar]
- 35. Kannan-Thulasiraman P., Katsoulidis E., Tallman M. S., Arthur J. S., Platanias L. C. (2006) J. Biol. Chem. 281, 22446–22452 [DOI] [PubMed] [Google Scholar]
- 36. Altman J. K., Yoon P., Katsoulidis E., Kroczynska B., Sassano A., Redig A. J., Glaser H., Jordan A., Tallman M. S., Hay N., Platanias L. C. (2008) J. Biol. Chem. 283, 1992–2001 [DOI] [PubMed] [Google Scholar]
- 37. Krajcíková D., Lukácová M., Müllerová D., Cutting S. M., Barák I. (2009) J. Bacteriol. 191, 3212–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ito A., Kataoka T. R., Watanabe M., Nishiyama K., Mazaki Y., Sabe H., Kitamura Y., Nojima H. (2000) EMBO J. 19, 562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Itoh S., Ding B., Bains C. P., Wang N., Takeishi Y., Jalili T., King G. L., Walsh R. A., Yan C., Abe J. (2005) J. Biol. Chem. 280, 24135–24142 [DOI] [PubMed] [Google Scholar]
- 40. Sapkota G. P., Cummings L., Newell F. S., Armstrong C., Bain J., Frodin M., Grauert M., Hoffmann M., Schnapp G., Steegmaier M., Cohen P., Alessi D. R. (2007) Biochem. J. 401, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Copp J., Manning G., Hunter T. (2009) Cancer Res. 69, 1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nguyen T. L. (2008) Anticancer Agents Med. Chem. 8, 710–716 [DOI] [PubMed] [Google Scholar]
- 43. Smith J. A., Poteet-Smith C. E., Xu Y., Errington T. M., Hecht S. M., Lannigan D. A. (2005) Cancer Res. 65, 1027–1034 [PubMed] [Google Scholar]
- 44. Herbert T. P., Tee A. R., Proud C. G. (2002) J. Biol. Chem. 277, 11591–11596 [DOI] [PubMed] [Google Scholar]
- 45. Li Q., Kawamura K., Ma G., Iwata F., Numasaki M., Suzuki N., Shimada H., Tagawa M. (2010) Eur. J. Cancer 46, 180–190 [DOI] [PubMed] [Google Scholar]
- 46. Katayama T., Nakanishi K., Nishihara H., Kamiyama N., Nakagawa T., Kamiyama T., Iseki K., Tanaka S., Todo S. (2007) Int. J. Oncol. 31, 613–620 [PubMed] [Google Scholar]
- 47. Fisher T. L., Blenis J. (1996) Mol. Cell Biol. 16, 1212–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anjum R., Blenis J. (2008) Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 49. Jones S. W., Erikson E., Blenis J., Maller J. L., Erikson R. L. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3377–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gavin A. C., Nebreda A. R. (1999) Curr. Biol. 9, 281–284 [DOI] [PubMed] [Google Scholar]
- 51. Smith J. A., Poteet-Smith C. E., Malarkey K., Sturgill T. W. (1999) J. Biol. Chem. 274, 2893–2898 [DOI] [PubMed] [Google Scholar]
- 52. Dalby K. N., Morrice N., Caudwell F. B., Avruch J., Cohen P. (1998) J. Biol. Chem. 273, 1496–1505 [DOI] [PubMed] [Google Scholar]
- 53. Frödin M., Jensen C. J., Merienne K., Gammeltoft S. (2000) EMBO J. 19, 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jensen C. J., Buch M. B., Krag T. O., Hemmings B. A., Gammeltoft S., Frödin M. (1999) J. Biol. Chem. 274, 27168–27176 [DOI] [PubMed] [Google Scholar]
- 55. Shahbazian D., Roux P. P., Mieulet V., Cohen M. S., Raught B., Taunton J., Hershey J. W., Blenis J., Pende M., Sonenberg N. (2006) EMBO J. 25, 2781–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005) Cell 123, 569–580 [DOI] [PubMed] [Google Scholar]
- 57. Etchison D., Milburn S. C., Edery I., Sonenberg N., Hershey J. W. (1982) J. Biol. Chem. 257, 14806–14810 [PubMed] [Google Scholar]
- 58. Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]