Abstract
Although the trimming of α1,2-mannose residues from precursor N-linked oligosaccharides is an essential step in the delivery of misfolded glycoproteins to endoplasmic reticulum (ER)-associated degradation (ERAD), the exact role of this trimming is unclear. EDEM1 was initially suggested to bind N-glycans after mannose trimming, a role presently ascribed to the lectins OS9 and XTP3-B, because of their in vitro affinities for trimmed oligosaccharides. We have shown before that ER mannosidase I (ERManI) is required for the trimming and concentrates together with the ERAD substrate and ERAD machinery in the pericentriolar ER-derived quality control compartment (ERQC). Inhibition of mannose trimming prevents substrate accumulation in the ERQC. Here, we show that the mannosidase inhibitor kifunensine or ERManI knockdown do not affect binding of an ERAD substrate glycoprotein to EDEM1. In contrast, substrate association with XTP3-B and with the E3 ubiquitin ligases HRD1 and SCFFbs2 was inhibited. Consistently, whereas the ERAD substrate partially colocalized upon proteasomal inhibition with EDEM1, HRD1, and Fbs2 at the ERQC, colocalization was repressed by mannosidase inhibition in the case of the E3 ligases but not for EDEM1. Interestingly, association and colocalization of the substrate with Derlin-1 was independent of mannose trimming. The HRD1 adaptor protein SEL1L had been suggested to play a role in N-glycan-dependent substrate delivery to OS9 and XTP3-B. However, substrate association with XTP3-B was still dependent on mannose trimming upon SEL1L knockdown. Our results suggest that mannose trimming enables delivery of a substrate glycoprotein from EDEM1 to late ERAD steps through association with XTP3-B.
Keywords: Carbohydrate Processing, Glycoprotein Biosynthesis, Glycosylation, Protein Degradation, Protein Folding, Derlin-1, EDEM1, ER Quality Control, HRD1, OS9
Introduction
During their translocation into the ER,3 most polypeptides acquire N-linked glycans, and the cell attempts to fold them to their native state with the help of several resident chaperones (1–3). Calnexin and calreticulin will bind to the sugar chains if they possess only one terminal glucose residue that remains from the original precursor or after its readdition by the folding sensor UDP-Glc: glycoprotein glucosyltransferase (4, 5). If proper folding cannot be achieved in a certain time frame, the glycoprotein molecules are targeted to ERAD by retrotranslocation to the cytosol and degradation by the ubiquitin-proteasome pathway. The decision to send a glycoprotein molecule to ERAD involves differential processing of its sugar chains (6, 7). Three or four mannose residues are excised from misfolded molecules and only one or two from folded molecules that exit to the Golgi. Although it is well established that this trimming of mannose residues is essential for delivery to ERAD (1–3), it is still unclear what events are regulated by this process. Recent studies on affinity of early secretory pathway lectins in vitro show that OS9 binds only after the trimming and cannot bind untrimmed Man9GlcNAc2 or Man8BGlcNAc2 (8–10). The same is true for the OS9 functional homolog XTP3-B (11). Conversely, lectins that support trafficking from the ER to the Golgi (ERGIC53, VIP36, VIPL) cannot bind the trimmed Man5GlcNAc2 and associate with higher affinity with untrimmed molecules (12). This suggests a model whereby extensive excision of mannose residues has a triple function: removal of the glycoprotein from the calnexin cycle (because the acceptor mannose for reglucosylation is lost), prevention of its binding to ER-Golgi lectins, and delivery to OS9 or XTP3-B. OS9 and XTP3-B would thus be the lectin-acceptors for the trimmed glycoprotein ERAD substrates, although evidence for this exists in vivo only for OS9 (8). This role was initially proposed for the ERAD-enhancing factor EDEM1 (ER degradation enhancing mannose-like protein 1) (13, 14), a protein implicated in accepting misfolded substrates released from the calnexin cycle. EDEM1 has a mannosidase-like domain, but a mannosidase activity has not been found yet in vitro, although it accelerates directly or indirectly trimming of mannose residues in vivo (15, 16).
We previously demonstrated that trimming of mannose residues is an obligatory step for ERAD substrate accumulation in the ER quality control compartment (ERQC) (6, 17), a proposed staging ground for ERAD (18, 19). The association of XTP3-B and OS9 with ERAD components (5, 9) that are recruited to the ERQC (19) and the affinity of these lectins for trimmed sugar chains would suggest that the substrate glycoprotein becomes trapped in the ERQC after trimming.
Here, we elucidate steps that require mannose trimming in the targeting of a glycoprotein substrate to ERAD in mammalian cells in vivo. We analyzed the effect of mannose trimming inhibition on association of EDEM1 and downstream ERAD factors with the substrate and the effect on the subcellular localization of these factors in relation to the substrate.
We used as a model an established ERAD substrate, the uncleaved precursor of asialoglycoprotein receptor H2a (20, 21). The precursor of H2a is a type 2 membrane glycoprotein that is expressed endogenously only in hepatocytes and is cleaved next to the transmembrane domain, and its ectodomain is secreted (22). When expressed in other cell types, most H2a precursor molecules remain uncleaved and are targeted to ERAD (18, 20).
Our results suggest that association of EDEM1 with the substrate does not require ERManI and the mannose-trimming event. In contrast, this trimming is essential for association with XTP3-B, substrate sequestration in the ERQC, and delivery to the E3 ubiquitin ligases HRD1 and SCFFbs2.
EXPERIMENTAL PROCEDURES
Materials
Rainbow 14C-labeled methylated protein standards were obtained from GE Healthcare. Promix cell labeling mix ([35S]Met plus [35S]Cys) >1000 Ci/mmol was from PerkinElmer Life Sciences. Protein A-Sepharose was from Repligen (Needham, MA). Lactacystin (Lac) and kifunensine (Kif) were from Cayman Chemicals (Ann Arbor, MI). Streptavidin-agarose-conjugated beads and other common reagents were from Sigma.
Plasmids and Constructs
H2a was subcloned in pCDNA1 (Invitrogen) (18). The pSUPER vector carrying a shRNA for human ERManI was described previously (17). pSUPER carrying shRNA for human SEL1L was a kind gift from R. Tyler and R. Kopito (Stanford University, Stanford, CA). An insert for pSUPER carrying shRNA for human EDEM1 was constructed as in Ref. 17 using the target sequence AGATTCCACCGTCCAAGTC. EDEM1-HA was a kind gift from K. Nagata (Kyoto University).
Mouse Fbs2 F box deletion mutant (Fbs2ΔF) cloned in pcDNA3-FLAG and human hsHRD1 RING finger mutant cloned in pcDNA3.1-Myc/His-A(−) vector were those used before (23) and were kind gifts from Y. Yoshida (Tokyo Metropolitan Institute of Medical Science) and E. Wiertz (Leiden University), respectively. S-tagged XTP3-B was a kind gift of R. Tyler and R. Kopito. Constructs encoding H2a G78R uncleavable mutant (24) fused through its C terminus to a 38-amino acid streptavidin binding peptide (H2aSBP) or to monomeric red fluorescent protein (H2aRFP) were described before (19, 23).
Primers and RT-PCR
Total cell RNA was extracted with EZ-RNA kit (Biological Industries, Beit Haemek, Israel). ReddyMix (ABgene, Epsom, UK) was used for PCR. Reverse transcription was performed with a VersoTM cDNA kit (Thermo Fisher Scientific), using a mixture of random hexamer and anchored oligo(dT) primers. An aliquot (10%) of the RT product was used for PCR with the following primers: CCTTCAGTGAGTGGTTTGG and GTGGTCCATCTTGGCACTG for ERManI, CAATGAAGGAGAAGGAGAC and CAATGTGTCCCTCTGTTGTG for EDEM1, AAAGCCCTGGAGAGAGTG and TTCCACTGTTCATTCCTG for SEL1L, and CTTTTAACTCTGGTAAAGTGG and TTTTGGCTCCCCCCTGCAAAT for GAPDH.
Antibodies
Rabbit polyclonal anti-H2 carboxyl-terminal and anti-H2 amino-terminal antibodies were the ones used in earlier studies (22, 25). Rabbit polyclonal anti-Derlin-1 was a kind gift from Y. Ye (National Institutes of Health). Mouse monoclonal antibodies were as follows; anti-FLAG (M2) was from Sigma, anti-Myc from Cell Signaling (Beverly, MA), anti-HA from Sigma or Santa Cruz Biotechnology (Santa Cruz, CA) and an anti-S tag from Novagen (Gibbstown, NJ). Goat anti-rabbit IgG antibody conjugated to Cy2, goat anti-mouse IgG conjugated to FITC, and goat anti-rabbit and anti-mouse IgG conjugated to HRP were from Jackson ImmunoResearch Laboratories (West Grove, PA). Goat anti-mouse IgG conjugated to agarose was from Sigma.
Cell Culture and Transfections
HEK 293 cells were grown in DMEM plus 10% FCS and NIH 3T3 cells in DMEM plus 10% new born calf serum. An HEK 293 stable cell line expressing H2aSBP was described previously (23). All cells were grown at 37 °C under an atmosphere of 5% CO2.
Transient transfection of NIH 3T3 cells was performed using FuGENE 6TM reagent (Roche Applied Science) according to the kit protocol. Transient transfection of HEK 293 cells was done according to the calcium phosphate method. The experiments were performed 24–48 h after the transfection.
Metabolic Labeling, Immunoprecipitation, SDS-PAGE, and Quantitation
Subconfluent (90%) cell monolayers in 60-mm dishes were labeled with [35S]Cys, lysed, and immunoprecipitated with anti-H2 antibodies as described previously (20, 22). Kif (100 μm) was added to cells 2 h before the labeling, and it was also present during starvation, labeling, and chase periods. Reducing SDS-PAGE was performed on 10% or 12% Laemmli gels. The gels were analyzed by fluorography using 20% 2,5-diphenyloxazole and were exposed to Biomax MS film using a BioMax TranScreen LE from Kodak (Vancouver, BC). Quantitation was performed in a Fujifilm FLA 5100 phosphorimaging device (Japan).
Coimmunoprecipitation and Immunoblotting
Cell lysis and immunoprecipitation of H2a and related constructs was done as described previously (20), except that an anti-H2a N-terminal antibody was used. Cell lysis was performed in the presence of 2 mm PMSF and 5 μg/ml aprotinin. For precipitation of SBP cell lysis was done in 1%Nonidet P-40, 50 mm Tris/HCl (pH 8), 150 mm NaCl for 30 min on ice, and debris and nuclei were pelleted in a microfuge for 30 min at 4 °C. The samples were immunoprecipitated with streptavidin-agarose-conjugated beads or an appropriate antisera and protein A-agarose. For immunoprecipitation with an anti-HA monoclonal antibody, whole goat anti-mouse IgG antibody immobilized on agarose beads was used. After overnight precipitation, the beads were washed three times with lysis buffer, followed by elution of the bound proteins by boiling with sample buffer containing β-mercaptoethanol at 100 °C for 5 min. Immunoblotting and detection by ECL were done as described previously (18), except for exposure and quantitation in a Bio-Rad ChemiDocXRS Imaging System.
Immunofluorescence Microscopy
The procedures employed were as described previously (17, 18). Whereas for biochemical experiments HEK 293 cells were used for high transfection efficiency, the imaging experiments were done using NIH 3T3 cells for optimal subcellular structure resolution. For treatments with Lac (25 μm) or Kif (100 μm) cells on coverslips were incubated with medium containing the drug at 37 °C in a CO2 incubator for 3–5 h. Confocal microscopy was done on a Zeiss laser scanning confocal microscope (LSM 510; Carl Zeiss, Jena, Germany) as described previously (17). Colocalization analysis (Pearson) was done using ImageJ software.
RESULTS
Mannose Trimming Is Essential for ERAD of Asialoglycoprotein Receptor H2a but Is Not Required for Its Association and Colocalization with EDEM1
We had previously shown that degradation of asialoglycoprotein receptor H2a is strongly dependent on trimming of mannose residues (17, 26). This can be seen in Fig. 1A, that shows the result of a pulse-chase experiment, where the H2a precursor was much stabilized in the presence of the α1,2-mannosidase inhibitor Kif, leading also to an increased amount of the cleaved H2a ectodomain fragment. When mannose trimming is blocked, there is a typical shift to a slower migration of the ERAD substrate (Fig. 1A, lanes 3 and 4 compared with lanes 1 and 2). We tested the requirement of EDEM1 for ERAD of H2a. Anti-EDEM1 shRNA strongly inhibited the degradation of H2a, although it did not lead to a significant change in the migration (Fig. 1B, compare lanes 3 and 4 with 1 and 2 and the graph in Fig. 1D). In contrast, overexpression of EDEM1 accelerated the degradation (Fig. 1C, compare lanes 3 and 4 with 1 and 2, Fig. 1D) and also caused a 30% reduction in the level of the pulse-labeled H2a (Fig. 1C, compare lane 1 with lane 3), probably by reducing the initial lag in degradation (22). Fig. 1E shows the efficiency of the EDEM1 knockdown.
FIGURE 1.
Mannose trimming and EDEM1 are required for ERAD of H2a. A, HEK 293 cells cotransfected with an H2a cDNA encoding vector were pulse-labeled for 20 min with [35S]cysteine and chased for 0 or 5 h in complete medium in the absence (lanes 1 and 2) or in the presence (lanes 3 and 4) of Kif (100 μm). After the pulse (0 h chase) or chase periods, the cells were lysed, H2a was immunoprecipitated, and the immunoprecipitates were separated in 12% SDS-PAGE followed by phosphorimaging. Bands corresponding to the H2a precursor and the naturally occurring cleaved fragment are indicated on the left. For the pulse samples, two bands can be seen for the H2a precursor and also for the fragment, with the lower ones corresponding to underglycosylated species (one of the three glycosylation sites unoccupied). B and C, similar to the pulse-chase experiment in A but performed with untreated cells transiently cotransfected with an H2a cDNA encoding vector together with either a GFP-containing pSUPER-retro vector (B and C, lanes 1 and 2) or with pSUPER encoding anti-EDEM1 shRNA (B, lanes 3 and 4) or with an EDEM1-HA containing pCMVsport2 plasmid (C, lanes 3 and 4). D, the bar graph shows the average percent H2a remaining after chase relative to the corresponding pulse, calculated from phosphorimaging quantitations of all H2a species (precursor and fragment) from three independent experiments similar to each of the ones shown in A–C. Error bars indicate S.D. between experiments. Student's t test renders p < 0.02 for values in all chase samples compared with control chase. E, in parallel with B, HEK 293 cells were transfected with either pSUPER (control, lane 1) or the same plasmid encoding anti-EDEM1 shRNA (lane 2). RNA was extracted 48 h post-transfection and used for RT-PCR with primers for EDEM1 mRNA (upper panel) compared with GAPDH (lower panel). Lane 3 shows a sample with no RNA template.
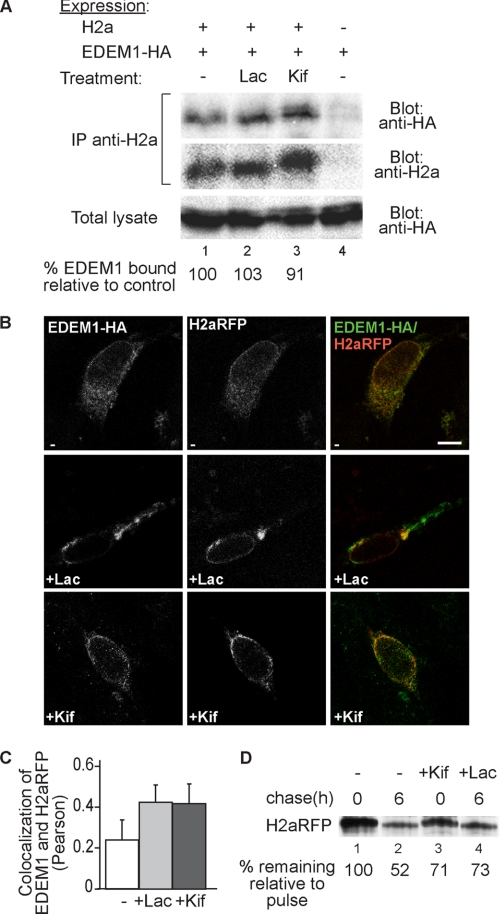
EDEM1 coimmunoprecipitated with H2a to a similar extent in the presence or absence of a proteasomal inhibitor (Lac) and also when inhibiting mannose trimming with Kif (Fig. 2A), similar to what had been observed with another ERAD substrate, Null Hong Kong mutant of α1-antitrypsin (27). We looked at the subcellular localization of EDEM1, which appeared in a somewhat punctate ER pattern (see Ref. 28 for a detailed analysis of EDEM1 localization), colocalizing partially with H2a linked to monomeric red fluorescent protein (H2aRFP) (Fig. 2B, upper panels). H2aRFP is an ERAD substrate (19) and similar to H2a, Kif inhibits its degradation and trimming of its mannose residues (Fig. 2D). Upon proteasomal inhibition, the ERAD substrate accumulates in the ERQC and colocalized to a much higher extent (2-fold) with EDEM1 (Fig. 2C), which partially redistributed to this juxtanuclear region (Fig. 2B, middle panels). Inhibition of mannose trimming causes the ERAD substrate to accumulate in a punctate pattern (6, 17), and also in this case, there was more colocalization with EDEM1 (Fig. 2B, lower panels, and 2C). The behavior of EDEM1 is different from that of ERManI, which is concentrated in the ERQC in all conditions, in untreated cells, after proteasome inhibition and after inhibition of mannose trimming (17).
FIGURE 2.
Inhibition of mannose trimming does not preclude EDEM1 interaction with H2a nor their subcellular colocalization. A, HEK 293 cells were cotransfected with a vector encoding for HA-tagged EDEM1 (EDEM1-HA) with or without an H2a encoding vector. Two days after transfection, the cells were incubated for 3 h in the absence/presence of 25 μm Lac or 100 μm Kif. The cells were lysed in 1% Nonidet P-40, 50 mm Tris/HCl (pH 8), 150 mm NaCl, and 10% of the lysates were run on 10% SDS-PAGE and immunoblotted with anti-HA antibodies (bottom panel). The rest of the lysates were immunoprecipitated (IP) with anti-H2a and protein A-Sepharose. Eluted samples were subjected to 10% SDS-PAGE and immunobloted with anti-HA (upper panel) or anti-H2a (middle panel). H2a (35-kDa fragment) (see Fig. 1) appeared as a very weak band in immunoblots and is therefore not shown. Quantitations of the relative amounts of EDEM1 associated with H2a are shown at the bottom as percent of EDEM1-HA coprecipitated with H2a relative to the mock transfection. The results are normalized by dividing the intensity of the EDEM1 band in the upper panel by that of its corresponding band in the lower panel and by that of H2a in the middle panel. B, NIH 3T3 cells were transiently cotransfected with H2aRFP together with HA-tagged EDEM1. 24 h after transfection, cells were incubated for 3 h in the absence (upper panels) or presence of 25 μm Lac (middle panels) or 100 μm Kif (lower panels), fixed, permeabilized, and incubated with mouse anti-HA followed by probing with FITC conjugated goat-anti mouse IgG. The samples were analyzed in an LSM confocal microscope. Representative optical slices are shown. Colocalization of FITC with RFP appears yellow. Bar, 10 μm. C, the graph shows colocalization analysis (Pearson) performed using the ImageJ program (average of 30 cells from three independent experiments are presented; error bars are S.D.); p < 0.045 for treated samples compared with untreated. D, similar to the pulse-chase experiment in Fig. 1A but performed with cells transiently transfected with an H2aRFP cDNA-encoding vector in the absence or in the presence of Kif (100 μm) or Lac (25 μm).
Because Kif could theoretically compete with substrate binding to EDEM1, we analyzed the effect of inhibition of mannose trimming by knockdown of ERManI, an enzyme that we had shown is required for the extensive mannose trimming and targeting to ERAD (17). Knockdown of ERManI causes a smaller extent of inhibition of mannose trimming compared with Kif (compare band shifts in Figs. 2A and 3A, middle panels), but led to increased coimmunoprecipitation of EDEM1 with H2a (Fig. 3), indicating that association of EDEM1 with the substrate does not depend on the activity or presence of ERManI, the presence of which may actually compete for binding to the substrate. Altogether, the results indicate that EDEM1 is involved in ERAD of H2a and that association and colocalization of EDEM1 and the ERAD substrate do not require mannose trimming or ERManI.
FIGURE 3.
ERManI activity is not required for association of H2a with EDEM1. A, HEK 293 cells were cotransfected with a vector encoding for HA-tagged EDEM1 (EDEM1-HA) with or without an H2a encoding vector and with or without a pSUPER plasmid encoding for anti-ERManI shRNA (shERManI). 24 h post-transfection, cells were lysed and processed, and results were quantitated and normalized as in Fig. 2A. B, in parallel with A, HEK 293 cells were transfected with the plasmid encoding anti-ERManI shRNA used in A or with pSUPER encoding anti-LacZ shRNA (control). RNA was extracted 24 h post-transfection and used for RT-PCR with primers for ERManI mRNA (upper panel) compared with GAPDH (lower panel). IP, immunoprecipitation.
Association of ERAD Substrate to XTP3-B Requires ER Mannosidase I and Mannose Trimming, a Requirement That Is Independent of SEL1L
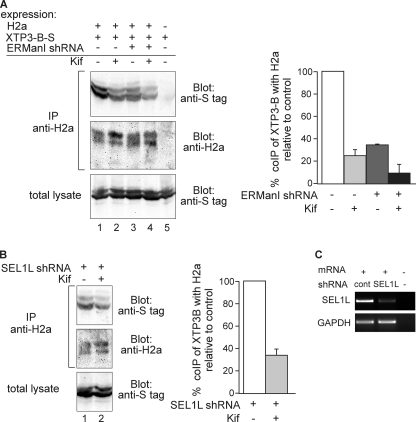
The affinity of the mammalian lectin OS9 and of yeast Yos9 for several oligosaccharides was determined recently in vitro and showed a clear preference for trimmed oligosaccharides and no binding to Man9GlcNAc2 or Man8BGlcNAc2 (missing the middle branch terminal mannose) (8, 10). The same is true for XTP3-B, which has a similar function as OS9 (11). Therefore, the prediction would be that inhibition of α1,2-mannosidases would inhibit ERAD substrate binding to the lectins. Although for some substrates OS9 and XTP3-B seem to be interchangeable (29), other substrates have a preference in associating with one of the lectins (30). Coimmunoprecipitation of H2a with OS9 yielded a very low signal (data not shown), but it showed significant binding to XTP3-B (Fig. 4A, lane 1). Cell treatment with Kif or knockdown of ERManI significantly inhibited the coimmunoprecipitation to 25 and 35% of the control, respectively, even though the total amount of ERAD substrate present increased as its degradation was inhibited (Fig. 4A, lanes 2 and 3 compared with lane 1). Combined knockdown of ERManI and Kif treatment reduced the association even further, to 10% of the control (Fig. 4A, graph).
FIGURE 4.
Association of the ERAD substrate to XTP3-B requires mannose trimming, even after knockdown of SEL1L. A, plasmids encoding for S-tagged XTP3-B and H2a cDNAs were cotransfected into HEK-293 cells with or without ERManI shRNA in a pSUPER plasmid. 24 h after transfection, cells were incubated for 5 h in the absence/presence of 100 μm Kif. Cells were lysed in 1% Nonidet P-40, 50 mm Tris/HCl (pH 8), 150 mm NaCl and 10% of the lysates were run on 10% SDS-PAGE and immunoblotted with anti-S tag antibodies (bottom panel). The rest of the lysates were immunoprecipitated (IP) with anti-H2a and protein A-Sepharose. Eluted samples were subjected to 10% SDS-PAGE and immunoblotted with anti-S tag (upper panel) or anti-H2a (middle panel). Results were quantitated and normalized as in Fig. 2A; values are averages of three independent experiments, error bars are S.D.; p < 0.04 for all coimmunoprecipitations compared with the control. B, experiment similar to that in A, except for SEL1L shRNA that was used instead of ERManI shRNA (p = 0.03). C, in parallel with B, HEK-293 cells were transfected with the plasmid encoding anti-SEL1L shRNA used in B or with pSUPER encoding anti-LacZ shRNA (control, cont). RNA was extracted 24 h post-transfection and used for RT-PCR with primers for SEL1L mRNA (upper panel) compared with GAPDH (lower panel).
It had been reported that the sugar chains of SEL1L, the HRD1 E3 ligase adaptor (31), are involved in its interactions with EDEM1 (27) and also with OS9 and XTP3-B (30). These findings could suggest an indirect sugar dependence, with a protein-protein interaction of the substrate with SEL1L and sugar-protein interactions of SEL1L with the lectins. In this case, knockdown of SEL1L should abrogate the mannose-trimming dependence of the putative indirect interaction of the ERAD substrate with XTP3-B. Nevertheless, after efficient knockdown of SEL1L (Fig. 4C), the coimmunoprecipitation of H2a with XTP3-B was still inhibited by cell treatment with Kif, to an extent similar to that in the control cells (compare Fig. 4, A and B, lanes 1 and 2).
Targeting of H2a to E3 Ubiquitin Ligases Is Dependent on Mannose Trimming
We analyzed whether delivery of the substrate to factors downstream of XTP3-B in the ERAD pathway is dependent on the mannose-trimming step. We had shown that two E3 ubiquitin ligases participate in the degradation of H2a (23), HRD1 (a transmembrane protein exposing a RING H2 finger domain to the cytosol (32)) and the cytosolic SCFFbs2 (33). This can be seen in Fig. 5, which shows a pulse-chase analysis of H2a and the inhibition of the degradation by overexpression of dominant negative mutants of HRD1 (RING finger mutant) and Fbs2 (F box mutant). In this case, we used SBP-tagged uncleavable H2a, which behaves similarly to the wild type protein, except that it does not yield a cleaved fragment (23).
FIGURE 5.
The E3 ligases HRD1 and SCFFbs2 are involved in ERAD of H2a. HEK 293 cells stably expressing H2aSBP were transiently transfected with plasmids encoding dominant negative mutant Myc-tagged HRD1 (mut HRD1) or FLAG-tagged ΔF box-Fbs2 (Fbs2ΔF). Two days after transfection, cells were labeled with [35S]Cys, and each pool of labeled cells was divided into aliquots that were subjected to 0 or 5 h chase. At the end of the pulse or chase periods, the cells were lysed and immunoprecipitated with anti-H2a. The eluted samples were subjected to SDS-PAGE followed by phosphorimaging detection and quantification. Note that H2aSBP is constructed with an uncleavable H2a variant that does not yield fragment. The plotted data provide quantification analysis of the stability of H2a upon coexpression of Fbs2ΔF or mutant HRD1 (average of three independent experiments); p < 0.03 for expression of both dominant negative proteins compared with control.
We had seen that upon proteasomal inhibition, a large portion of HRD1 colocalizes with H2aRFP at the ERQC (19). This can be seen in Fig. 6A (middle panels). Treatment with Kif redistributed both H2aRFP and HRD1 to a punctate pattern but with much decreased colocalization, similar to that in untreated cells (Fig. 6A, lower panels, and B). Kif also caused a much lower degree of coprecipitation of H2aSBP and HRD1 (Fig. 6C).
FIGURE 6.
Subcellular colocalization of H2a with HRD1 is increased by proteasomal inhibition but not by blocking mannose trimming. The latter treatment reduces their association compared with proteasomal inhibition. A, NIH 3T3 cells were transiently cotransfected with H2aRFP together with Myc-tagged mutant (mut) HRD1. 24 h after transfection, cells were incubated for 3 h in the absence (upper panels) or presence of 25 μm Lac (middle panels) or 100 μm Kif (lower panels), fixed, permeabilized, and incubated with mouse anti-Myc and FITC-conjugated goat anti-mouse IgG. Representative confocal optical slices are shown. Bar, 10 μm. B, colocalization analysis (Pearson) of the fluorescence signals shown in A, performed using the ImageJ program (average of 30 cells from three independent experiments are presented); p = 0.004 for Lac-treated compared with untreated and p = 0.0002 for Lac-treated compared with Kif-treated samples. C, Myc-tagged mutant HRD1 was transfected into the H2aSBP expressing the HEK 293 cell line. Two days after transfection, the cells were incubated for 3 h with 25 μm Lac or 100 μm Kif and then lysed in PBS containing 1% Triton X-100 and 0.5% sodium deoxycholate. 10% of the lysates were run on 10% SDS-PAGE and immunoblotted with anti-Myc (bottom panel). The rest of the lysates were precipitated (ppt) with streptavidin beads. Eluted samples were subjected to 10% SDS-PAGE and immunoblotted with anti-Myc (upper panel) or anti-H2a (middle panel).
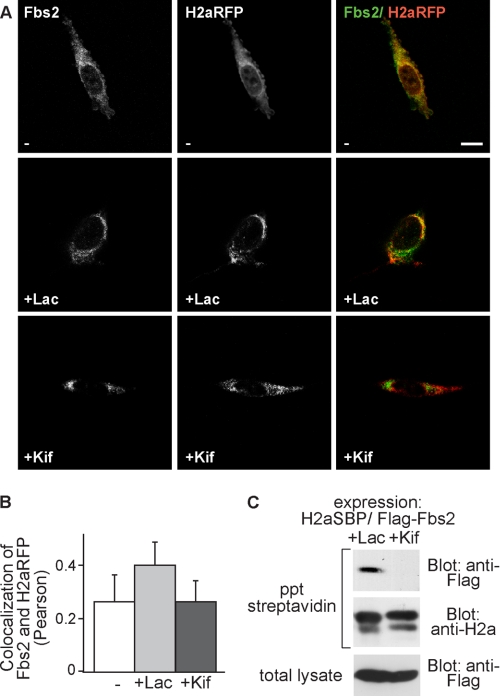
For SCFFbs2, Fbs2, the substrate recognition component of this E3 ligase presents additional interest. Fbs2 is a lectin that binds substrate N-glycans (34) and for sugar chain recognition, the glycoprotein must undergo retrotranslocation to the cytosol where the E3 ligase is located. Thus, Fbs2 associates with the substrate at a very late ERAD stage. We had shown that Fbs2 targets the sugar moiety of H2a, whereas the protein moiety is the target of HRD1 (23). Fbs2 colocalized partially with H2aRFP at the ERQC in Lac-treated cells (Fig. 7A, middle panels) and also in this case, there was a strong reduction of the colocalization upon Kif treatment of the cells, resulting in similar levels to those in untreated cells (Fig. 7A, lower panels, and B). In vitro Fbs2 binds a large range of trimmed and untrimmed high mannose oligosaccharides (33), indicating that lack of trimming of the ERAD substrate should not abolish Fbs2 binding. However, the Fbs2-H2aSBP interaction was drastically reduced by incubation of cells with Kif, showing no detectable coprecipitation (Fig. 7C). Altogether, the results suggest that mannose trimming is necessary for delivery to late ERAD stages, including the retrotranslocation step. Therefore, we looked at the protein Derlin-1, implicated in a putative retrotranslocation complex (35). Derlin-1 also coprecipitated with H2aSBP and colocalized with the ERAD substrate at the ERQC upon proteasomal inhibition. Surprisingly, inhibition of mannose trimming did not affect the extent of colocalization nor of coprecipitation compared with proteasomal inhibition (Fig. 8). Similar to H2aRFP and EDEM1, there also was a large increase in the colocalization of H2aRFP with Derlin-1 comparing untreated cells with Lac- or Kif-treated cells. This suggests that the association with Derlin-1 takes place before the mannose-trimming event.
FIGURE 7.
Association and subcellular colocalization of H2a with Fbs2 also is much reduced by inhibition of mannose trimming compared with the effect of proteasomal inhibition. A and B, experiment similar to that in Fig. 6 (A and B) but with H2aRFP and FLAG-tagged Fbs2ΔF. Mouse anti-FLAG antibodies and FITC-conjugated goat anti-mouse IgG were used to visualize Fbs2. Bar, 10 μm. p = 0.045 for Lac-treated compared with untreated and p = 0.07 for Lac-treated compared with Kif-treated samples. C, FLAG-tagged Fbs2ΔF was transfected into an H2aSBP-expressing HEK 293 cell line. Two days after transfection, the cells were incubated for 3 h with 25 μm Lac or 100 μm Kif. The cells were lysed in 1% Nonidet P-40, 50 mm Tris/HCl (pH 8), 150 mm NaCl, and the same procedure as in Fig. 6C was performed, except that anti-FLAG antibody was used instead of anti-Myc. ppt, precipitation.
FIGURE 8.
Association of H2a with Derlin-1 is not affected by inhibition of mannose trimming. A, experiment similar to that in Fig. 6A, except that endogenous Derlin-1 was visualized using rabbit polyclonal anti-Derlin-1 and Cy2-conjugated goat anti-rabbit IgG. B, experiment similar to that in Fig. 7C, except that Derlin-1 was detected on the blot using rabbit polyclonal anti-Derlin-1. ppt, precipitation.
DISCUSSION
When being targeted to ERAD, a misfolded glycoprotein undergoes ERManI-dependent trimming to yield the N-linked oligosaccharide structures Man5–6GlcNAc2 (6, 17). These are the best binders to OS9 (10) and XTP3-B (11). Substrate binding to XTP3-B in cells in vivo requires this trimming and ERManI (Fig. 4). This requirement is not dependent on the presence of SEL1L. Altogether, our results suggest that the substrate associates directly with XTP3-B after excision of the α1,2-mannose residues from its sugar chains. Reported sugar-dependent interactions of SEL1L with EDEM1 (27), OS9 and XTP3-B (30) might be nonproductive for ERAD of the substrate. For example, Kif affected EDEM1-SEL1L binding but 1-deoxymannojirimycin had no effect on the EDEM1-SEL1L association (27). However, 1-deoxymannojirimycin is known to inhibit strongly ERAD of many glycoprotein substrates, including H2a (17). Alternatively, EDEM1, OS9 and XTP3-B may each exist as oligomers, one subunit associating with the substrate and another with SEL1L. The HRD1 complex, to which both OS9 and XTP3-B are associated, was shown to require oligomerization to be functional in yeast (36). In any case, our data favors a model whereby mannose trimming of the N-glycans of the ERAD substrate glycoprotein regulates its binding to XTP3-B, in a way similar to that reported for OS9 (8) (see model in Fig. 9). It was shown that XTP3-B and OS9 can bind nonglycosylated proteins (37). However, XTP3-B still seems to bind glycoprotein substrates through their sugar chains (Fig. 4) (11). Substrates can bind to OS9 and XTP3-B through both protein and sugar moieties (8, 30, 38, 39), but an initial lectin-sugar recognition event may normally be required.
FIGURE 9.
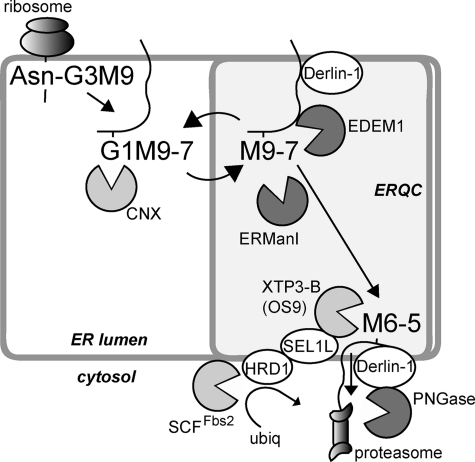
Model for mannose trimming-mediated delivery of the ERAD substrate from EDEM1 to XTP3-B and to a retrotranslocation complex. Cotranslational glycosylation is followed by trimming of two glucose residues and association with the chaperones/lectins calnexin (CNX) or calreticulin. The glycoprotein is transported to the ERQC (18, 21) and can recycle back to the peripheral ER, undergoing during this process cycles of deglucosylation, reglucosylation by UDP-Glc:glycoprotein glucosyltransferase (4) and trimming of up to 2 α1,2-mannose residues by ERManI (17). Association to Derlin-1 and EDEM1 is followed by trimming of one or two more α1,2-mannose residues, which removes the glycoprotein from the calnexin folding cycle, determining its targeting to ERAD by association with XTP3-B or OS9 and delivery to the ubiquitination and retrotranslocation machinery. All proteins with a lectin activity are in light gray, including the cytosolic SCF E3 ligase component Fbs2, and glycosidases are in dark gray, including EDEM1, for which mannosidase activity has still not been proven in vitro, and peptide N-glycanase (PNGase), which cleaves the sugar chain before proteasomal degradation of the protein.
After association of the substrate with XTP3-B (or OS9) upon mannose trimming, the lectin would deliver the glycoprotein to downstream ERAD factors such as HRD1 and SCFFbs2. HRD1 was recently reported to be mandatory for soluble luminal ERAD substrates but not for membrane-bound ones (29). However, membrane-bound H2a (Fig. 5) (23) as well as other membrane-bound ERAD substrates (TCRα, CD3δ) (32) are targets of HRD1. As these proteins are substrates of more than one ligase, this may explain why down-regulation or knock-out of HRD1 does not significantly affect their rate of degradation (29) but overexpression of a dominant negative HRD1 mutant does affect their rate of degradation (Fig. 5) (32).
There was a strong inhibition by Kif of the coimmunoprecipitation of H2a with XTP3-B and HRD1 and a total block in the association of H2a with cytosolic Fbs2. This is probably due to the inability of the ERAD substrate to reach the retrotranslocation complex and to be transported to the cytosol. This result is consistent with an observation that proteasomal inhibition but not mannosidase inhibition led to the accumulation of another ERAD substrate, mutant hERG in the cytosol (40). This suggests that the retrotranslocation to the cytosolic proteasomes is highly dependent on the excision of α1,2-mannose residues.
Interestingly, binding to Derlin-1 is independent of the mannose-trimming event (Fig. 8). Therefore, although Derlin-1 has been implicated in retrotranslocation, it probably associates with the substrate at an early stage, before excision of mannose residues. This is consistent with a study of the involvement of Derlin-1 in both ER retention of mutant CFTRΔF508 and its delivery to the ubiquitination machinery, which suggested that Derlin-1 functions at several steps in the ERAD pathway (41). Indeed, Derlin-1 associates with Bap31 (42), a protein that participates in early quality control events, as it associates with calnexin and cycles between the peripheral ER and the ERQC (43), but also in late events in substrate retrotranslocation (42). Derlin-1 also associates with late ERAD factors, E3 ubiquitin ligases (44, 45), the retrotranslocation factor p97 (35, 46, 47), and cytosolic N-glycanase (48). Interestingly, Derlin-1 forms hetero-oligomers with Derlin-2 (47). Derlin-2 and -3 were shown to interact with EDEM1 (49) that, as we show here, associates with the substrate independently of the excision of mannose residues. Recognition of substrates by EDEM1 does not require mannose trimming (Fig. 2) (27) or the activity of ERManI (Fig. 3). Mannose trimming is not required either for the appearance of EDEM1 and the glycoprotein in the same region of the cell (Fig. 2). These events would take place before the trimming step. Therefore, it can be suggested that N-glycans with trimmed mannose residues are not the signal for substrate recognition by EDEM1. Consistently, EDEM1 was not found in complex with E3 ubiquitin ligases or with OS9 (30, 50–52). EDEM1 was also suggested to act as a chaperone binding to misfolded polypeptides (53, 54).4 Therefore, in addition to its direct or indirect participation in the excision of mannose residues (15, 16), a possible role of EDEM1 is in chaperoning or shuttling the substrate together with Derlin-1 through the ERQC, where it is delivered to XTP3-B and OS9, after the mannose-trimming event.
Fig. 9 shows a model for the role of mannose trimming, based on our results and those of others, where during the calnexin cycle, the misfolded glycoprotein is delivered to the ERQC. There, it binds Derlin-1 and EDEM1. ERManI, concentrated in the ERQC compartment, trims the α1,2-mannose residues with the help of EDEM1. At this stage, the glycoprotein can be released and recycled to the peripheral ER where, if still not properly folded, it is reglucosylated and reassociates with calnexin and is targeted again to the ERQC. If during these cycles, there is trimming to M6–5, and loss of the mannose acceptor for reglucosylation, this releases the glycoprotein from the calnexin cycle and delivers it to XTP3-B (or OS9), which traps the substrate in the ERQC and targets it to the ubiquitination and retrotranslocation machinery.
Acknowledgments
We are grateful to Ron Kopito, Kazuhiro Nagata, Emmanuel Wiertz, Yukiko Yoshida and Yihong Ye for reagents.
This work was supported by grants from the Israel Science Foundation (1229/07) and German-Israeli Project Cooperation (DIP).
B. Groisman, M. Shenkman, E. Ron, E. Avezov, Y. Izenshtein, and G. Z. Lederkremer, unpublished results.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- ERQC
- ER-derived quality control compartment
- ERManI
- ER mannosidase I
- Lac
- lactacystin
- Kif
- kifunensine
- RFP
- red fluorescent protein
- SBP
- streptavidin binding peptide.
REFERENCES
- 1. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 2. Hebert D. N., Bernasconi R., Molinari M. (2010) Semin. Cell Dev. Biol. 21, 526–532 [DOI] [PubMed] [Google Scholar]
- 3. Lederkremer G. Z. (2009) Curr. Opin. Struct. Biol. 19, 515–523 [DOI] [PubMed] [Google Scholar]
- 4. D'Alessio C., Caramelo J. J., Parodi A. J. (2010) Semin. Cell Dev. Biol. 21, 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Määttänen P., Gehring K., Bergeron J. J., Thomas D. Y. (2010) Semin. Cell Dev. Biol. 21, 500–511 [DOI] [PubMed] [Google Scholar]
- 6. Frenkel Z., Gregory W., Kornfeld S., Lederkremer G. Z. (2003) J. Biol. Chem. 278, 34119–34124 [DOI] [PubMed] [Google Scholar]
- 7. Lederkremer G. Z., Glickman M. H. (2005) Trends Biochem. Sci. 30, 297–303 [DOI] [PubMed] [Google Scholar]
- 8. Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. (2009) J. Biol. Chem. 284, 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hosokawa N., Kamiya Y., Kato K. (2010) Glycobiology 20, 651–660 [DOI] [PubMed] [Google Scholar]
- 10. Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. (2008) Mol. Cell 32, 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamaguchi D., Hu D., Matsumoto N., Yamamoto K. (2010) Glycobiology 20, 348–355 [DOI] [PubMed] [Google Scholar]
- 12. Kamiya Y., Kamiya D., Yamamoto K., Nyfeler B., Hauri H. P., Kato K. (2008) J. Biol. Chem. 283, 1857–1861 [DOI] [PubMed] [Google Scholar]
- 13. Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. (2001) EMBO Rep. 2, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. (2001) EMBO Rep. 2, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hosokawa N., Tremblay L. O., Sleno B., Kamiya Y., Wada I., Nagata K., Kato K., Herscovics A. (2010) Glycobiology 20, 567–575 [DOI] [PubMed] [Google Scholar]
- 16. Olivari S., Molinari M. (2007) FEBS Lett. 581, 3658–3664 [DOI] [PubMed] [Google Scholar]
- 17. Avezov E., Frenkel Z., Ehrlich M., Herscovics A., Lederkremer G. Z. (2008) Mol. Biol. Cell 19, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S. V., Ehrlich R., Lederkremer G. Z. (2001) Mol. Biol. Cell 12, 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kondratyev M., Avezov E., Shenkman M., Groisman B., Lederkremer G. Z. (2007) Exp. Cell Res. 313, 3395–3407 [DOI] [PubMed] [Google Scholar]
- 20. Shenkman M., Ayalon M., Lederkremer G. Z. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11363–11368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frenkel Z., Shenkman M., Kondratyev M., Lederkremer G. Z. (2004) Mol. Biol. Cell 15, 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tolchinsky S., Yuk M. H., Ayalon M., Lodish H. F., Lederkremer G. Z. (1996) J. Biol. Chem. 271, 14496–14503 [DOI] [PubMed] [Google Scholar]
- 23. Groisman B., Avezov E., Lederkremer G. Z. (2006) Isr. J. Chem. 46, 189–196 [Google Scholar]
- 24. Yuk M. H., Lodish H. F. (1993) J. Cell Biol. 123, 1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shenkman M., Ehrlich M., Lederkremer G. Z. (2000) J. Biol. Chem. 275, 2845–2851 [DOI] [PubMed] [Google Scholar]
- 26. Ayalon-Soffer M., Shenkman M., Lederkremer G. Z. (1999) J. Cell Sci. 112, 3309–3318 [DOI] [PubMed] [Google Scholar]
- 27. Cormier J. H., Tamura T., Sunryd J. C., Hebert D. N. (2009) Mol. Cell 34, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuber C., Cormier J. H., Guhl B., Santimaria R., Hebert D. N., Roth J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. (2010) J. Cell Biol. 188, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mueller B., Lilley B. N., Ploegh H. L. (2006) J. Cell Biol. 175, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 33. Yoshida Y., Tokunaga F., Chiba T., Iwai K., Tanaka K., Tai T. (2003) J. Biol. Chem. 278, 43877–43884 [DOI] [PubMed] [Google Scholar]
- 34. Yoshida Y. (2007) Biosci. Biotechnol. Biochem. 71, 2623–2631 [DOI] [PubMed] [Google Scholar]
- 35. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 36. Horn S. C., Hanna J., Hirsch C., Volkwein C., Schütz A., Heinemann U., Sommer T., Jarosch E. (2009) Mol. Cell 36, 782–793 [DOI] [PubMed] [Google Scholar]
- 37. Alcock F., Swanton E. (2009) J. Mol. Biol. 385, 1032–1042 [DOI] [PubMed] [Google Scholar]
- 38. Bernasconi R., Pertel T., Luban J., Molinari M. (2008) J. Biol. Chem. 283, 16446–16454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. (2008) J. Biol. Chem. 283, 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong Q., Keeney D. R., Molinari M., Zhou Z. (2005) J. Biol. Chem. 280, 19419–19425 [DOI] [PubMed] [Google Scholar]
- 41. Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006) Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 42. Wang B., Heath-Engel H., Zhang D., Nguyen N., Thomas D. Y., Hanrahan J. W., Shore G. C. (2008) Cell 133, 1080–1092 [DOI] [PubMed] [Google Scholar]
- 43. Wakana Y., Takai S., Nakajima K., Tani K., Yamamoto A., Watson P., Stephens D. J., Hauri H. P., Tagaya M. (2008) Mol. Biol. Cell 19, 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 45. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hegde R. S., Ploegh H. L. (2010) Curr. Opin. Cell Biol. 22, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lilley B. N., Ploegh H. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Katiyar S., Joshi S., Lennarz W. J. (2005) Mol. Biol. Cell 16, 4584–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006) J. Cell Biol. 172, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 51. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 52. Gauss R., Jarosch E., Sommer T., Hirsch C. (2006) Nat. Cell Biol. 8, 849–854 [DOI] [PubMed] [Google Scholar]
- 53. Hosokawa N., Wada I., Natsuka Y., Nagata K. (2006) Genes Cells 11, 465–476 [DOI] [PubMed] [Google Scholar]
- 54. Olivari S., Cali T., Salo K. E., Paganetti P., Ruddock L. W., Molinari M. (2006) Biochem. Biophys. Res. Commun. 349, 1278–1284 [DOI] [PubMed] [Google Scholar]