Abstract
The adaptive response to hyperosmotic stress in yeast, termed the high osmolarity glycerol (HOG) response, is mediated by two independent upstream pathways that converge on the Pbs2 MAP kinase kinase (MAPKK), leading to the activation of the Hog1 MAP kinase. One branch is dependent on the Sho1 transmembrane protein, whose primary role was found to be the binding and translocation of the Pbs2 MAPKK to the plasma membrane, and specifically to sites of polarized growth. The yeast PAK homolog Ste20 is essential for the Sho1-dependent activation of the Hog1 MAP kinase in response to severe osmotic stress. This function of Ste20 in the HOG pathway requires binding of the small GTPase Cdc42. Overexpression of Cdc42 partially complements the osmosensitivity of ste20Δ mutants, perhaps by activating another PAK-like kinase, while a dominant-negative Cdc42 mutant inhibited signaling through the SHO1 branch of the HOG pathway. Since activated Cdc42 translocates Ste20 to sites of polarized growth, the upstream and downstream elements of the HOG pathway are brought together through the membrane targeting function of Sho1 and Cdc42.
Keywords: Cdc42/MAP kinase/signal transduction/Ste20/stress response
Introduction
In eukaryotic cells, a variety of external stimuli are converted to appropriate cellular responses through activation of a small number of signal transduction modules termed mitogen-activated protein (MAP) kinase pathways (for a review see Robinson and Cobb, 1997). In the budding yeast Saccharomyces cerevisiae, five of these MAP kinase cascades have been characterized, which respond to such diverse environmental conditions as the presence of mating pheromone, changes in osmotic pressure, heat stress and nutrient availability (for review see Herskowitz, 1995; Gustin et al., 1998). The MAP kinase cascades are comprised of three sequentially activated kinases: MAP kinase kinase kinase (MAPKKK), which activates MAP kinase kinase (MAPKK), which subsequently activates MAP kinase. Once activated, MAP kinase can in turn activate a set of target proteins, often transcription factors, such that an appropriate cellular response is generated by the external signal.
The yeast S.cerevisiae adapts to growth under conditions of increased external osmolarity through activation of the high osmolarity glycerol (HOG) MAP kinase pathway (for a review see Gustin et al., 1998). The Hog1 MAP kinase is activated upon osmotic stress by two independent upstream mechanisms that converge on Pbs2 MAPKK (Maeda et al., 1995). Activation of the pathway and ultimately phosphorylation of Hog1 at conserved threonine and tyrosine residues initiates the osmo-adaptive response, which includes efflux of water from the cell and intracellular accumulation of glycerol (Gustin et al., 1998). Two transmembrane proteins, Sln1 and Sho1, have been identified that can independently mediate activation of the HOG pathway in response to increased external osmolarity (Ota and Varshavsky, 1993; Maeda et al., 1994, 1995). Thus, we will refer to the two upstream activation mechanisms of the HOG pathway as the SLN1 branch and the SHO1 branch. The Sln1 transmembrane protein, which is a homolog of bacterial two-component signal transducers, is composed of an N-terminal extracellular domain, a cytoplasmic histidine kinase domain and a C-terminal receiver domain. Under low osmotic conditions, the activated Sln1 histidine kinase phosphorylates a histidine residue in Sln1, which is then transferred to an aspartate residue in the Sln1 receiver domain. The phosphate is further transferred, via a phospho-relay mechanism through an intermediary phospho-carrier protein Ypd1, to another receiver domain protein, Ssk1 (Posas et al., 1996). Under high osmolarity conditions, Sln1 histidine kinase activity is suppressed, leading to the accumulation of non-phosphorylated Ssk1 (Maeda et al., 1994; Posas et al., 1996). The unphosphorylated form of Ssk1p in turn activates the MAP kinase cascade composed of the redundant Ssk2 and Ssk22 MAPKKKs, the Pbs2 MAPKK and the Hog1 MAP kinase (see Figure 1).
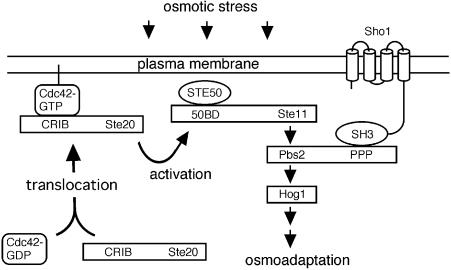
Fig. 1. Schematic model outlining the role of Sho1, Cdc42 and Ste20 in the HOG MAP kinase pathway. The membrane-bound protein Sho1 anchors the Hog1 MAP kinase module to the membrane through its interaction with the Pbs2 MAPKK, which in turn interacts with the Ste11 MAPKKK and Hog1 MAP kinase. In response to osmotic stress, Cdc42-bound, membrane-localized Ste20 phosphorylates Ste11, which leads to the activation of the Pbs2 MAPKK and the Hog1 MAP kinase, and eventually to the osmo-adaptive response. Exactly how osmotic stress induces Ste20 activation is unclear. 50BD, Ste50 binding domain; CRIB, Cdc42/Rac interactive binding domain; PPP, proline-rich sequence.
The SHO1 branch of the HOG pathway has been shown to require Sho1, Ste11 and Ste50 for transmission of the osmotic stress signal (Maeda et al., 1995; Posas and Saito, 1997; Posas et al., 1998). The Sho1 protein comprises four transmembrane domains near its N-terminus, and a cytoplasmic SH3 domain that binds a proline-rich motif near the N-terminus of the Pbs2 MAPKK (Maeda et al., 1995). In the SHO1 branch, activation of Pbs2 is effected by its phosphorylation by the Ste11 MAPKKK (Maeda et al., 1995; Posas and Saito, 1997). Ste11 is also a component of other MAP kinase modules that regulate the mating response, pseudohyphal development and invasive growth response (Liu et al., 1993; Roberts and Fink, 1994). Ste50 is a Ste11-binding protein, and its presence is essential for activation of Pbs2 by Ste11p (O’Rourke and Herskowitz, 1998; Posas et al., 1998; Wu et al., 1999). Ste50 also has roles in the mating response and the pseudohyphal growth response (Ramezani Rad et al., 1992, 1998). Mutation of a single gene that is common to both branches, namely PBS2 or HOG1, confers severe osmosensitivity by itself. In contrast, mutants in either the SLN1 branch (for instance, the ssk2Δ ssk22Δ double mutant) or in the SHO1 branch (sho1Δ, ste11Δ or ste50Δ) can grow normally under hyperosmotic stress conditions, because there are two independent branches leading to the activation of Pbs2 MAPKK. However, mutants that are defective in both branches, for instance ssk2Δ ssk22Δ sho1Δ or ssk2Δ ssk22Δ ste11Δ, are osmosensitive (Maeda et al., 1995; Posas et al., 1998).
Activation of the Ste11 MAPKKK in response to nutrient depletion or mating pheromone requires the p21-activated kinase (PAK) yeast homolog Ste20, which is believed to activate the MAPKKK by direct phosphorylation (Wu et al., 1995; Mösch et al., 1996). The Ste20 kinase is known to function downstream of the heterotrimeric G-protein associated with the mating factor receptors (Leberer et al., 1992; Ramer and Davis, 1993). Two other PAK-like kinases (Cla4 and Skm1) have been identified in yeast and are thought to function upstream of MAP kinase modules that regulate polarized growth and cytokinesis (Cvrcková et al., 1995; Martin et al., 1997). Deletion of the CLA4 gene alone is non-lethal, but does produce morphological abnormalities; deletion of both the CLA4 and STE20 genes, however, is lethal (Cvrcková et al., 1995). This suggests that, in addition to its role in the mating response, Ste20 has a role in vegetative growth that is at least partially redundant with that of Cla4.
Members of the PAK family kinases bind the Rho-like GTPase protein Cdc42 or Rac through a conserved Cdc42/Rac interactive binding (CRIB) domain of 16 amino acid residues (Manser et al., 1994). For example, p65PAK was first identified in human cells through its activation by Cdc42 (Manser et al., 1994). In mammalian cells, Cdc42 and Rac regulate cytoskeletal and morphological changes in response to numerous extracellular stimuli (reviewed in Hall, 1994). Furthermore, the mammalian stress-activated MAP kinases, namely JNK1/2 and p38, can be activated by the Cdc42-induced PAK kinase activity (Bagrodia et al., 1995; Polverino et al., 1995; Pombo et al., 1995). In yeast, the GTPase Cdc42, together with its GTP exchange factor Cdc24 and its target Ste20, is required to establish cell polarity during the cell cycle, and is involved in cellular responses to mating pheromone and to nutritional limitation (reviewed in Pringle et al., 1995; Johnson, 1999). Binding of the activated form (i.e. GTP-bound form) of Cdc42 to Ste20 localizes Ste20 to plasma membrane sites at the tip of an emerging bud during vegetative growth, or at the shmoo tip during the mating response (Peter et al., 1996).
We found that Sho1 is localized to the emerging bud, as is Cdc42, and that the main function of Sho1 is to localize the Pbs2 MAPKK to the cell membrane. Thus, we investigated the potential role of Ste20 and Cdc42 in the osmotic stress response, using STE20 alleles that were either kinase-inactive or in which the CRIB domain was deleted. We present evidence that Ste20 is required for osmotic stress signaling via the SHO1 branch of the HOG pathway. In the absence of Ste20 kinase activity, overexpression of either Cdc42 or another PAK family kinase Cla4 can substitute for Ste20 function in activation of the Hog1 kinase. The data support a model whereby Sho1 anchors the Ste11/Ste50/Pbs2/Hog1 MAP kinase module to the cell membrane site where it can be activated by Cdc42-bound membrane-localized Ste20 in response to osmotic stress.
Results
The role of Sho1 in the HOG pathway
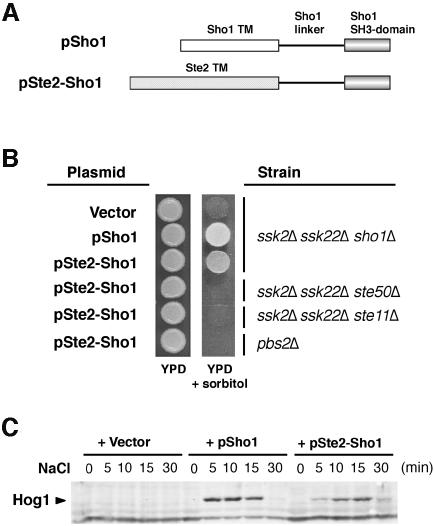
Sho1 is composed of three structural domains: four transmembrane segments near its N-terminus, an SH3 domain at the C-terminus and a linker domain in between. The transmembrane segments are essential for Sho1 function, since deletion of all four transmembrane segments completely abolished the ability of SHO1 to complement the sho1Δ mutation, even if overexpressed (data not shown). We then addressed the question as to whether the specific sequence of Sho1 transmembrane domains was essential for Sho1 function, or whether any transmembrane sequence could fulfill the function of Sho1. To distinguish between these two possibilities, we made a chimeric construct, Ste2–Sho1, in which the four transmembrane segments of Sho1 are replaced by the seven transmembrane segments of the α-factor receptor, Ste2 (see Figure 2A). The Ste2–Sho1 hybrid could complement the osmosensitivity of the ssk2Δ ssk22Δ sho1Δ mutant, but not that of the ssk2Δ ssk22Δ ste50Δ, ssk2Δ ssk22Δ ste11Δ and pbs2Δ mutant strains (Figure 2B). Furthermore, the Ste2–Sho1 chimeric protein could induce tyrosine phosphorylation of Hog1 in response to increased osmotic stress (Figure 2C). These results clearly indicate that neither the number nor the specific sequence of the Sho1 transmembrane segments is essential for its function in the osmotic stress response.
Fig. 2. A chimeric protein composed of the transmembrane domains of Ste2 and the cytoplasmic domain of Sho1 is functional. (A) Schematic representation of the wild-type Sho1 and the Ste2–Sho1 chimeric constructs. A region of the STE2 gene encoding the seven transmembrane (TM) domains was fused to a region of the SHO1 gene encoding the cytoplasmic linker region and the SH3 domain (residues 146–367). (B) Complementation of sho1Δ mutation by the Ste2–Sho1 chimera. Yeast strains MY007 (ssk2Δ ssk22Δ sho1Δ), FP67 (ssk2Δ ssk22Δ ste50Δ), FP50 (ssk2Δ ssk22Δ ste11Δ) and TM260 (pbs2Δ) were transformed with wild-type SHO1 (pSho1), the STE2–SHO1 chimeric construct (pSte2–Sho1), or the empty vector pYES2 (Vector). Rescue of the osmosensitive phenotype of this strain was assessed by spotting transformed cells onto YPD plates containing 1.5 M sorbitol. Although the Ste2–Sho1 chimeric protein could functionally substitute for wild-type Sho1, it failed to complement the osmosensitive defect of ste50Δ, ste11Δ and pbs2Δ strains (ssk2Δ and ssk22Δ mutations are included in some strains to inactivate the SLN1 branch of the HOG pathway). (C) The Ste2–Sho1 chimera can activate the Hog1 MAP kinase with kinetics similar to wild-type Sho1 protein in response to external osmotic stress. The same transformants as in (B) were tested by 4G10 immunoblot assay to detect phosphorylation of the Hog1 MAP kinase before (time 0), and 5, 10, 15 and 30 min after the addition of 0.4 M NaCl.
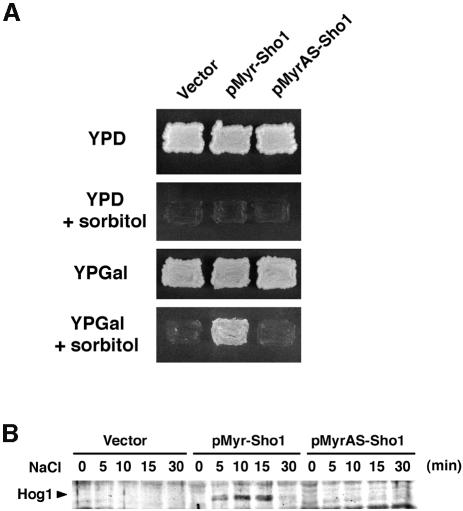
We next tested whether targeting of the cytoplasmic region of Sho1 to the plasma membrane is sufficient for Sho1 function. A Sho1 variant (Myr–Sho1) was constructed in which the four transmembrane segments were replaced with an oligopeptide containing the N-terminal myristoylation site of Gpa1, the Gα subunit of the trimeric G-protein that mediates the mating response in yeast. A mutant version (MyrAS–Sho1) in which the myristoylation site Met-Gly-Cys is altered to Met-Ala-Ser is not membrane-localized and served as a negative control. These constructs were placed under the control of the inducible GAL1 promoter and transformed into an osmosensitive mutant strain, ssk2Δ ssk22Δ sho1Δ. In the presence of glucose, which represses expression of the chimeric Myr–Sho1 protein, the transformed cells are clearly osmosensitive and fail to grow on media containing 1.5 M sorbitol (Figure 3A). However, in the presence of galactose, expression of Myr–Sho1 rescued the osmosensitive phenotype of the host strain; cells expressing the MyrAS–Sho1 construct or vector alone failed to grow on media containing sorbitol. Similarly, in ssk2Δ ssk22Δ sho1Δ cells, expression of the Myr–Sho1 chimera induced tyrosine phosphorylation of Hog1 after the addition of 0.4 M NaCl, whereas the mutant MyrAS–Sho1 protein and vector alone failed to do so (Figure 3B). Thus, this result indicates that membrane targeting of the cytoplasmic domain of Sho1, either through the transmembrane segments or by myristoylation, is necessary and sufficient for its role in the osmotic stress response.
Fig. 3. Membrane localization of Sho1 is necessary and sufficient for its role in the osmotic stress response. (A) Osmosensitivity of sho1Δ mutant transformed with the indicated plasmids. pMyr–Sho1 encodes a variant of Sho1 that lacks the four transmembrane domains but does contain a membrane-targeting myristoylation site. pMyrAS–Sho1 is a derivative of pMyr–Sho1 with a myristoylation-defective mutation. These constructs were placed under control of the inducible galactose promoter in the pYES2 vector, and used to transform the yeast strain MY007 (ssk2Δ ssk22Δ sho1Δ). Thus, expression of the Myr–Sho1 constructs is suppressed on glucose media (YPD) and induced on galactose media (YPGal). Expression of the membrane-targeting Myr–Sho1 construct, but not that of the mutant MyrAS–Sho1, complemented the osmosensitive defect of sho1Δ mutation. (B) Tyrosine phosphorylation of Hog1 following the addition of 0.4 M NaCl. The same strains used in (A) were subjected to immunoblot analysis using anti-phosphotyrosine antibody 4G10.
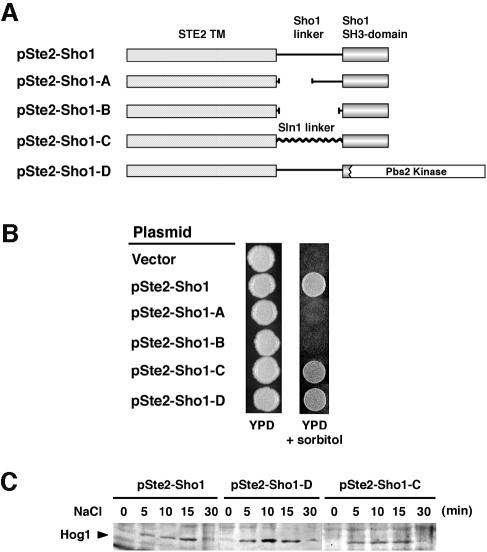
We then tested whether the Sho1 linker segment, which separates the N-terminal transmembrane domains from the C-terminal SH3 domain, is essential for Sho1 function. For this purpose, we constructed two deletion mutants of the Ste2–Sho1 construct, in which one half (Ste2–Sho1-A) or the entire (Ste2–Sho1-B) linker sequence is deleted (Figure 4A). Neither of these deletion mutants complemented the osmosensitivity of ssk2Δ ssk2Δ sho1Δ mutant cells, suggesting that a linker region is essential (Figure 4B). However, the specific linker sequence was not found to be critical, because replacement of Sho1 linker sequence with an unrelated peptide of similar length (Ste2–Sho1-C) fully reconstituted the Sho1 function. These results are consistent with the interpretation that the role of the linker sequence is as a non-specific spacer between the membrane segment and the C-terminal SH3 domain.
Fig. 4. Requirement for the Sho1 cytoplasmic linker region and the SH3 domain for Sho1 function in the osmotic stress response. (A) Schematic representation of the Sho1 constructs used in this experiment. The pSte2–Sho1 chimeric construct is described in Figure 2. Its derivatives, pSte2–Sho1-A and pSte2–Sho1-B, contain deletions within the Sho1 linker region. In pSte2–Sho1-C, the Sho1 linker region is replaced with an unrelated sequence of similar length (150 amino acids) derived from the SLN1 gene. pSte2–Sho1-D contains a truncation of Sho1 in which the SH3 domain is replaced by the PBS2 coding region. (B) Sho1–Ste2 chimeric constructs were transformed into the yeast strain MY007 (ssk2Δ ssk22Δ sho1Δ), and the osmosensitivity of the transformed cells was tested on YPD plates containing 1.5 M sorbitol. (C) Tyrosine phosphorylation of Hog1 at various times after the addition of 0.4 M NaCl to the medium. The same cells used in (A) were subjected to immunoblot analysis using anti-phosphotyrosine antibody 4G10.
Finally, we tested whether the Sho1 SH3 domain has a role other than binding to Pbs2. This was tested by directly targeting Pbs2 to the plasma membrane by fusing the Ste2 transmembrane segments and the Sho1 linker sequence to the N-terminus of Pbs2 (Ste2–Sho1-D). This construct lacks any SH3 domain sequence. As shown in Figure 4B, Ste2–Sho1-D could complement the osmosensitivity of the sho1Δ mutant. It could also induce tyrosine phosphorylation of Hog1 upon hyperosmotic shock (Figure 4C). Thus, the sole function of the Sho1 SH3 domain appears to be binding of Pbs2, and thus target it to the plasma membrane.
Sho1 is localized to regions of polarized cell growth
Although the primary function of the transmembrane domains of Sho1 appears to be targeting of the Pbs2 MAPKK to the plasma membrane, we sought to identify a more subtle, regulatory role for these membrane spanning regions. For this analysis we investigated the localization of Sho1 in the cell. In order to determine the localization of the Sho1 protein, a genomic copy of the SHO1 gene, under its own promoter, was tagged at the C-terminus with the green fluorescent protein (GFP) coding sequence, and placed in a low copy-number (CEN) vector to construct the pSHO1–GFP plasmid. The use of the native SHO1 promoter and a low-copy vector ensures that the level of Sho1–GFP expression is within physiological levels. This Sho1–GFP fusion protein could rescue the osmosensitivity of ssk2Δ ssk22Δ sho1Δ mutant cells, indicating that it is functional (data not shown).
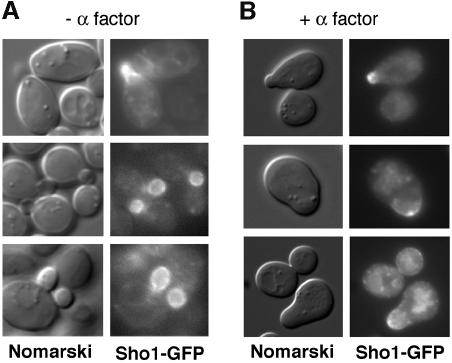
Examination of exponentially growing cultures of sho1Δ cells transformed with the pSHO1–GFP construct revealed a marked asymmetric distribution of the Sho1–GFP protein. Sho1–GFP was clearly localized to the bud in G1–S cells, and was particularly enriched in the plasma membrane of the emerging bud (Figure 5A). The protein appeared more diffusely distributed throughout the cytoplasm of the mother cell, perhaps associated with vacuolar membranes. To determine whether this asymmetric distribution of Sho1–GFP was related to polarization of cell growth, wild-type cells transformed with pSHO1–GFP were incubated in the presence of α mating factor. This produces an altered cell morphology whereby cells of the a mating type (such as TM141) arrest in the G1 phase of the cell cycle and polarize their growth, forming a characteristic shmoo. In the presence of α mating factor, Sho1–GFP was localized to sites of active polarized growth, and specifically to the shmoo tip (Figure 5B). These observations are of particular interest in the light of previous findings that both Ste20 and Cdc42 are also localized to the bud in G1–S cells and to the shmoo tip in cells treated with mating factor (Ziman et al., 1993; Leberer et al., 1997). More recently, the Cdc24 guanine nucleotide exchange factor for the Cdc42 GTPase was also shown to translocate from the nucleus to the bud and shmoo tip (Toenjes et al., 1999; Nern and Arkowitz, 2000; Shimada et al., 2000).
Fig. 5. Localization of Sho1 to sites of polarized growth. Localization of the Sho1 protein was determined using a Sho1–GPF fusion construct (pSHO1–GFP). Expression of the Sho1–GFP fusion protein is maintained at physiological levels by using a low copy-number vector (pRS416) and the promoter of the SHO1 gene itself. (A) The majority of the Sho1–GFP protein is localized to the incipient bud (top panel), and to the plasma membrane of the growing bud (lower panels), during vegetative growth. pSHO1–GFP is expressed in the yeast strain FP66 (sho1Δ), and exponentially growing cells in YPD medium were examined by fluorescence microscopy. (B) Sho1–GFP is localized at the shmoo tip during the mating response. pSHO1–GFP is expressed in the wild-type strain TM141 in the presence of 3 µM α mating factor.
PAK-like kinases Ste20 and Cla4 are involved in the SHO1 branch of the HOG pathway
The above finding that the major role of Sho1 is to translocate Pbs2 to specific sites on the plasma membrane suggests that the Sho1-dependent activation of Ste11 and Pbs2 is effected by factors that are localized to similar membrane locations. We will demonstrate that at least one such factor is membrane-associated Cdc42. Furthermore, the yeast PAK homologs Ste20 and Cla4, which bind Cdc42, are involved in this process.
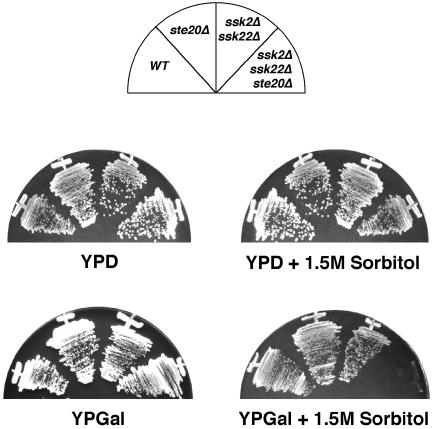
Recently, evidence has been presented for a role of the yeast PAK homolog Ste20 in the Hog1 MAP kinase pathway (O’Rourke and Herskowitz, 1998). It was shown that a ssk1Δ ste20Δ mutant was sensitive to conditions of extreme osmotic stress, such as the presence of 1.2 M NaCl, but not sensitive to less severe conditions, such as 1.0 M NaCl. The ssk1Δ mutation alone did not confer any osmosensitivity. Consistent with these observations, other ste20Δ mutants (such as ssk2Δ ssk22Δ ste20Δ) could grow normally on rich media containing 1.5 M sorbitol (whose osmotic strength is <1.0 M NaCl) (Posas and Saito, 1997). A more systematic examination revealed that ste20Δ cells that have additional mutations in the SLN1 branch exhibited an enhanced osmosensitivity when grown in media containing galactose rather than glucose. Thus, ssk2Δ ssk22Δ ste20Δ mutant cells can grow on YPD media containing 1.5 M sorbitol but fail to grow on YPGal media containing 1.5 M sorbitol (Figure 6). The corresponding ssk2Δ ssk22Δ STE20+ or SSK2+ SSK22+ ste20Δ mutant cells, which are defective in only one of the upstream branches of the HOG pathway, could grow normally on either medium. Possible mechanisms of the galactose effect will be discussed later (see Discussion).
Fig. 6. Deletion of the STE20 gene in cells which are defective for the SLN1 branch of the HOG pathway (in this case, ssk2Δ ssk22Δ double mutant) leads to osmosensitivity with galactose but not dextrose, as the sole carbon source. Wild-type strain TM141 (WT), and mutant strains FP58 (ste20Δ), TM252 (ssk2Δ ssk22Δ) and DR76 (ssk2Δ ssk22Δ ste20Δ) were streaked onto rich media containing 2% glucose (YPD) or 2% galactose (YPGal) with or without 1.5 M sorbitol.
The significantly weaker osmosensitivity of ste20Δ mutants relative to that of the hog1Δ or pbs2Δ mutants suggests that another factor, perhaps a kinase related to Ste20, may partially complement the lack of Ste20. It was of interest, therefore, to determine whether another yeast PAK homolog, Cla4, which also binds Cdc42 (Benton et al., 1997), could rescue the osmosensitivity of the ssk2Δ ssk22Δ ste20Δ triple mutant cells. Expression of CLA4 on a high copy plasmid was found to rescue the osmosensitivity of the mutant strain (Figure 7), indicating that overexpression of Cla4 can complement the osmosensitive defect of a ste20Δ mutation. Because the ste20Δ cla4Δ double mutation is lethal, we could not test the effect of the double mutation on the HOG pathway.
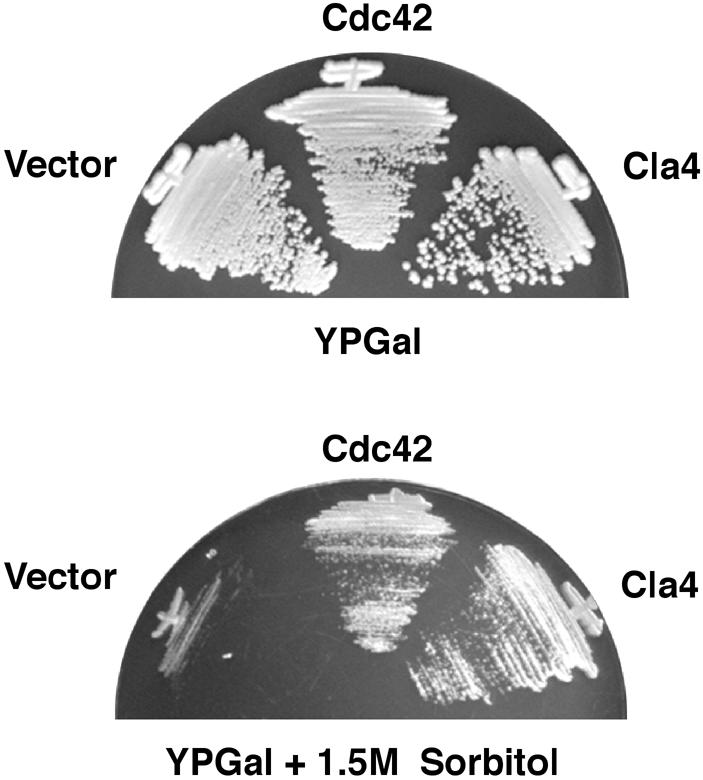
Fig. 7. Overexpression of Cdc42 or the PAK homolog Cla4 rescues the osmosensitive phenotype caused by a ste20Δ mutation. Strain DR76 (ssk2Δ ssk22Δ ste20Δ) was transformed with the galactose promoter containing pYES2 (Vector), wild-type CDC42 inserted into pYES2 (Cdc42) or a high copy-number plasmid, pCC1079 (Chen et al., 1997) containing the wild-type CLA4 gene under its own promoter (Cla4). Transformants were streaked onto YPGal or on YPGal containing 1.5 M sorbitol and incubated at 30°C.
Both Ste20 kinase activity and its Cdc42-binding domain are required for activation of the HOG pathway
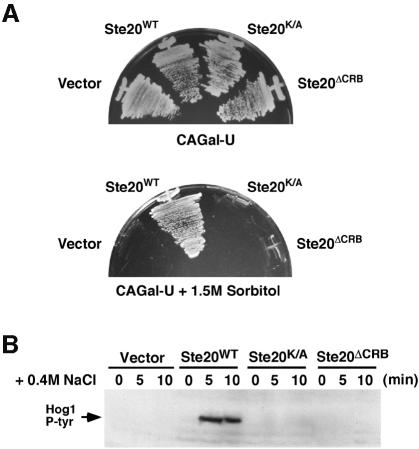
In yeast, the small Rho-type GTPase protein Cdc42 has been shown to directly bind to two PAK-like kinases, Ste20 and Cla4 (Cvrcková et al., 1995; Peter et al., 1996; Benton et al., 1997; Leberer et al., 1997). Yeast cells that are defective in Cdc42 activity show defects in pheromone response, polarized growth and the invasive growth response (Simon et al., 1995; Zhao et al., 1995; Mösch et al., 1996). In order to define more precisely the role of Ste20 in the osmotic stress response, we analyzed the ability of various mutant forms of STE20 to rescue the osmosensitivity caused by a ste20Δ mutation (the mutant also contains the ssk2Δ and ssk22Δ mutations to inactivate the SLN1 branch). In this regard, two specific domains of Ste20 were of particular interest, namely the C-terminal kinase domain and a region of 36 amino acids between residues 334–369, which is the CRIB domain conserved in members of the PAK-family kinases (Manser et al., 1994; Leberer et al., 1997). Yeast strain DR76 (ssk2Δ ssk22Δ ste20Δ) was transformed with high copy-number plasmids expressing either wild-type Ste20, a kinase-inactive mutant Ste20K/A, in which the catalytically essential lysine residue Lys649 is converted to alanine, or a CRIB site deletion mutant Ste20ΔCRB (Leberer et al., 1997). The ability of these STE20 alleles to restore growth of ssk2Δ ssk22Δ ste20Δ cells on galactose medium containing 1.5 M sorbitol was then assessed (Figure 8A). While the expression of wild-type Ste20 clearly rescued the growth defect of DR76 on CAGal + 1.5 M sorbitol medium, both Ste20K/A and Ste20ΔCRB failed to restore growth under these conditions of osmotic stress.
Fig. 8. Both the kinase activity and the CRIB domain of Ste20 are required for the osmotic stress response mediated by the SHO1 branch of the HOG pathway. (A) Strain DR76 (ssk2Δ ssk22Δ ste20Δ) was transformed with a multicopy (2 µm) plasmid containing either wild-type STE20, the kinase inactive STE20K/A allele or the STE20ΔCRB mutant allele lacking the Cdc42-binding domain. Transformants were streaked onto selective media containing galactose as the sole carbon source in the absence (upper panel) or presence (lower panel) of 1.5 M sorbitol. Whereas wild-type STE20 could rescue the osmosensitivity of the ste20Δ strain DR76, both mutant alleles of STE20 and the high copy-number expression vector alone failed to suppress the osmosensitive phenotype. (B) Activation of the Hog1 MAP kinase by osmotic stress in the transformants detailed in (A) was determined by immunoblot analysis using the 4G10 anti-phosphotyrosine antibody. Exponentially growing cultures were harvested before (0), or at the indicated times after, the addition of 0.4 M NaCl, and subjected to immunoblot analysis.
To investigate further the requirement of both Ste20 kinase activity and binding of Cdc42 in the osmotic stress response, we directly assayed Hog1 phosphorylation in response to osmotic stress by immunoblot analysis. Thus, the ssk2Δ ssk22Δ ste20Δ strain was transformed with either vector alone, or a plasmid expressing wild-type Ste20, kinase inactive STE20K/A or Ste20ΔCRB. The ssk2Δ ssk22Δ ste20Δ cells were defective for Hog1 activation in response to the addition of 0.4 M NaCl (Figure 8B, lanes 1–3). Furthermore, tyrosine phosphorylation of Hog1 in response to osmotic shock was dependent on both Ste20 kinase activity and the presence of the Cdc42-binding domain, since Ste20 mutants defective in either kinase activity or Cdc42 binding failed to induce Hog1 phosphorylation (Figure 8B, lanes 4–9). These results therefore suggest an essential role of both Ste20 kinase activity and its ability to bind Cdc42 in the SHO1 branch of the HOG osmosensing pathway. In clear contrast, only Ste20 kinase activity, but not the Cdc42 binding, was implicated in mating pheromone signal transduction (Leberer et al., 1997).
Dominant-negative CDC42A188 inhibits the SHO1 branch of the HOG pathway
As a member of the Ras superfamily of low molecular weight GTP-binding proteins, Cdc42 cycles between the GDP-bound (inactive) form and the GTP-bound (active) form. The activated form of Cdc42 is localized to membrane sites at the bud neck and shmoo tips (Ziman et al., 1993). Membrane association is aided by geranylgeranyl modification of the conserved Cys188 within the consensus Cys-Ala-Ala-Xaa motif at the C-terminus; mutation of this conserved cysteine residue to alanine (A188) blocks the membrane translocation of Cdc42. The CDC42A188 mutant is not only inactive, but also exerts a dominant inhibitory effect, perhaps by abortively interacting with the GTP/GDP exchange factor Cdc24. Other Cdc42 mutants, such as CDC42V12, lock the protein into a GTP-bound conformation and therefore into a constitutively active state. To further determine the potential role of Cdc42 in the HOG pathway, we performed phosphotyrosine immunoblot assays to analyze the effect of overexpressing the dominant-active CDC42V12 and the dominant-negative CDC42A188 alleles (Ziman et al., 1993).
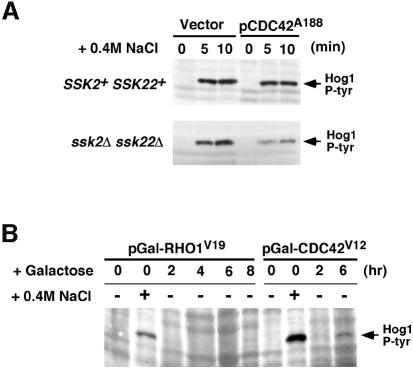
The effect of overexpression of dominant-negative Cdc42A188 on Hog1 activation in wild-type cells, in which both the SLN1 and SHO1 branches are intact, was compared with that in ssk2Δ ssk22Δ mutant cells, in which only the SHO1 branch is functional. Galactose-induced overexpression of the dominant-negative CDC42A188 allele substantially reduced Sho1-dependent Hog1 phosphorylation in response to osmotic stress (Figure 9A, lower panel). In contrast, overexpression of CDC42A188 in wild-type cells, with a fully functional SLN1 branch of the pathway, did not significantly affect Hog1 activation in response to stress (Figure 9A, upper panel). Conversely, overexpression of the dominant-active CDC42V12 induced, albeit weakly, Hog1 phosphorylation in the absence of osmotic stress (Figure 9B). This effect seemed to be specific to the Cdc42 GTPase, since galactose-induced expression of the dominant-active form of the Rho GTPase (RHO1G19) had no effect on Hog1 phosphorylation (Figure 9B). These data, however, do not exclude the possibility that this weak Hog1 activation is an indirect effect of non-specific cellular changes caused by Cdc42V12 overexpression. Finally, we tested whether overexpression of wild-type Cdc42 could rescue the osmosensitivity of ssk2Δ ssk22Δ ste20Δ cells. Expression of wild-type CDC42 under the control of the galactose promoter on a high copy-number plasmid was found to rescue the osmosensitivity of the triple deletion mutant strain (Figure 7). This effect is likely to be mediated by the enhanced activation of Cla4 by overexpressed Cdc42. These data presented above suggest a direct and specific role for the Cdc42 GTPase in the activation of the Sho1–Ste20–Ste11 branch of the HOG MAP kinase pathway.
Fig. 9. Effect of dominant-active and dominant-negative CDC42 on Hog1 activation. (A) The dominant-negative CDC42A188 allele inhibits the SHO1 branch of the HOG pathway. Wild-type strain TM141 (SSK2+ SSK22+) and a mutant strain TM252 (ssk2Δ ssk22Δ) were transformed with an expression plasmid pYES2 with a galactose-inducible promoter (Vector), or pYES2 containing the dominant-negative CDC42A188 allele (pCDC42A188). Expression of the CDC42A188 allele was induced by the addition of 2% galactose to exponentially growing cultures containing non-repressing raffinose as carbon source. Two-and-a-half hours after the addition of galactose, the ability of the cells to activate the HOG pathway was determined by monitoring Hog1 tyrosine phosphorylation following osmotic shock. Samples were taken before (time 0), and at 5 and 10 min after the addition of 0.4 M NaCl for 4G10 immunoblot analysis. Expression of the dominant-negative CDC42A188 allele significantly inhibited signaling through the SHO1 branch of the HOG pathway in ssk2Δ ssk22Δ cells. In contrast, activation of Hog1 was unaffected in wild-type cells (SSK2+ SSK22+), in which the SLN1 branch is intact. (B) Expression of the dominant-active CDC42V12 allele activates the HOG pathway in the absence of osmotic stress. Strain TM252 (ssk2Δ ssk22Δ) was transformed with galactose-inducible constructs of dominant-active CDC42V12 or dominant-active RHO1V19. Hog1 activation was assessed as above by 4G10 immunoblot analysis. Exponentially growing cells in non-inducing raffinose media (time 0) were either untreated (–) or treated with NaCl for 5 min (+) as controls for baseline and maximal Hog1 activation, respectively. At the indicated times after addition of 2% galactose to induce expression of RHO1V19 or CDC42V12, samples were harvested (without osmotic shock) for 4G10 analysis. At 6 h of CDC42V12 expression, weak, but significant, phosphorylation of the Hog1 kinase was detected in the absence of osmotic stress.
Discussion
Activation of the Hog1 MAP kinase pathway is initiated by either of two independent upstream pathways; the Sln1 transmembrane histidine kinase is required for activation of the Ssk2 and Ssk22 MAPKKKs, while the Sho1 transmembrane protein mediates activation of the Ste11 MAPKKK. Here, we have demonstrated that targeting of the Pbs2 MAPKK to appropriate membrane sites is the major role of the Sho1 transmembrane protein. Through the use of chimeric proteins containing the cytoplasmic domain of Sho1 fused to unrelated membrane localization domains, we showed that membrane localization of the Sho1 cytoplasmic domain was necessary and sufficient for its role in the osmotic stress response. Furthermore, it was found that once Pbs2 MAPKK is targeted to the membrane, no specific part of Sho1 is required for HOG pathway activation. Using a Sho1–GFP fusion protein we found that Sho1 was enriched in the emerging bud of vegetatively growing cells and the shmoo tip during the mating response. Thus, we propose that the primary role of Sho1 is to localize the Pbs2 MAPKK to the plasma membrane, specifically to sites of polarized growth. Because Pbs2 also binds Ste11 and Hog1 (Posas and Saito, 1997), and Ste11 constitutively binds Ste50 (Posas et al., 1998), it is possible that the entire complex composed of Ste11, Ste50, Pbs2 and Hog1 is localized to the plasma membrane site by Sho1 (see Figure 1). The function of Sho1 as a membrane-bound anchor of the HOG1 MAP kinase pathway, although integral to the osmosensing mechanism, does not constitute an osmosensor per se. Clearly, activation of other as yet unidentified elements of the pathway, in addition to the appropriate localization of the Ste11–Pbs2–Hog1 module, is required for cellular adaptation to osmotic stress.
The pseudohyphal growth pathway and the HOG pathway share several upstream elements, namely Cdc42, Ste20, Ste11, Ste50 and, interestingly, Sho1 (Roberts and Fink, 1994; Mösch et al., 1996; O’Rourke and Herskowitz, 1998). Their signal specificity appears to derive from the activation of distinct MAPKKs and MAPKs, namely Ste7 and Kss1, in the case of pseudohyphal growth (Madhani et al., 1997), and Pbs2 and Hog1 in the HOG pathway. There does not appear to be any requirement for Pbs2 in the pseudohyphal pathway, however, suggesting that Sho1 may have an additional role in pseudohyphal development distinct from that in the osmotic stress response.
In this paper, we have shown that activation of Hog1 by the SHO1 branch requires the yeast PAK homologs Ste20 and Cla4, and the small GTPase Cdc42. The PAK-like Ste20 kinase is involved in a number of signal transduction pathways in yeast, for instance the pseudohyphal and invasive growth response, as well as in the mating response (Herskowitz, 1995). Evidence for a requirement for Ste20 for growth under conditions of extreme hyperosmolarity was also presented recently (O’Rourke and Herskowitz, 1998). Here we show that cells with a defect in the SLN1 branch of the HOG pathway combined with a deletion of STE20 (for example ssk2Δ ssk22Δ ste20Δ) fail to grow on media containing 1.5 M sorbitol with galactose as the carbon source. However, these cells can grow in the presence of 1.5 M sorbitol with glucose as the carbon source (Figure 6). Since this strain can grow on galactose-containing media in the absence of sorbitol, there appears to be no defect in galactose metabolism per se. This differential osmosensitivity could be accounted for by a number of possible factors. First, altered Ras2 activity could affect the activation of Cdc42 and Ste20, as was found for the pseudohyphal pathway (Mösch et al., 1996; Lorenz and Heitman, 1997). Alternatively the reduced levels of cAMP in galactose media might affect cellular stress responses through altered protein kinase A activity or localization (Gancedo, 1998; Griffioen et al., 2000). Growth on galactose rather than glucose may in addition affect the expression or activity of a gene redundant with STE20. Finally, it is also possible that the galactose effect is simply caused by the combination of a poorer carbon source and elevated osmotic stress. Under conditions of moderate stress, for instance on YPD containing 1.5 M sorbitol, ste20Δ cells can survive, perhaps through activation of another Cdc42-binding PAK-like kinase, Cla4. This activity may provide sufficient activation of the Hog1 pathway to allow growth under moderate conditions, but not under more severe conditions of osmotic stress (O’Rourke and Herskowitz, 1998). Indeed, if overexpressed, Cla4 can fully complement the osmosensitive defect of ste20Δ mutants.
A role of Cdc42 in the SHO1 branch of the Hog1 pathway was implicit in the failure of the Ste20 CRIB deletion mutant to complement a ste20Δ strain, and was more directly demonstrated by the effect of expressing dominant-active and dominant-negative forms of Cdc42 on Hog1 activation. Overexpression of the constitutively activated, membrane-localized form of Cdc42 (CDC42V12) weakly activates the Hog1 pathway in the absence of external stress, whereas expression of the dominant inhibitory form (CDC42A188), which cannot be membrane localized, significantly reduced activation of Hog1 in response to osmotic stress. While these results suggest that Cdc42 is involved in Hog1 activation, it also suggests that active Cdc42 alone is not sufficient for robust Hog1 activation. Thus, involvement of additional factors seems likely.
Cdc42-binding is not required for Ste20 kinase activity itself. Indeed, signaling in response to α mating factor is not affected by deletion of the Cdc42-binding domain in Ste20 (Peter et al., 1996). However, the Cdc42-binding domain does appear essential for Ste20 activity in the pseudohyphal/invasive growth response and for the suppression of the cell cycle defect of a ste20Δ cla4Δ mutant (Peter et al., 1996; Leberer et al., 1997). Similarly, our results indicate that the function of Ste20 in the Hog1 MAP kinase pathway requires Cdc42 binding (Figure 8). Cdc42 binding is required for localization of the Ste20 kinase to the emerging bud during the G1–S phase of the cell cycle and to the shmoo tip during the mating response (Peter et al., 1996; Leberer et al., 1997). Thus, we propose that appropriate localization of Ste20, as mediated by binding of Cdc42–GTP, is essential for the function of the Ste20 kinase in the Hog1 MAP kinase pathway.
The data are consistent with a model whereby activation of Cdc42 brings Ste20 into close proximity to the Hog1 MAP kinase module and thereby facilitates its activation, presumably through phosphorylation of the Ste11 MAPKKK (see Figure 1). Furthermore, our data suggest a unique role for the SHO1 branch of the HOG pathway during periods of polarized growth. During shmoo formation towards a mating partner or during bud emergence, Sho1 may localize the Hog1 MAP kinase cascade to the site of polarized growth. This will allow the cell to monitor increased osmotic stress in those vulnerable regions of the cell undergoing morphological change and new cell wall deposition.
Materials and methods
Yeast strains
The following yeast strains, which are all isogenic, were used: TM141 (MATa ura3 leu2 trp1 his3); TM252 (MATa ura3 leu2 trp1 ssk2::LEU2 ssk22::LEU2); TM257 (MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2); TM260 (MATa ura3 leu2 trp1 pbs2::LEU2); DR76 (MATa leu2 ura3 trp1 ssk2::ura3::Neo ssk22::ura3::Neo ste20::TRP1); FP50 (MATa ura3 leu2 his3 ssk2::LEU2 ssk22::LEU2 ste11::HIS3); FP58 (MATa ura3 leu2 trp1 his3 ste20::HIS3); FP66 (MATa ura3 leu2 trp1 his3 ste50::HIS3); FP67 (MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste50::HIS3); and MY007 (MATa ura3 leu2 his3 ssk2::LEU2 ssk22::LEU2 sho1::HIS3).
Media
Standard yeast media and genetic procedures were as described previously (Rose et al., 1990). YPD contains 10 g/l yeast extract; 20 g/l tryptone and 2% glucose; YPGal contains 10 g/l yeast extract; 20 g/l tryptone and 2% galactose; YPD- and YPGal-sorbitol media contain the indicated concentrations of sorbitol; CAGal-Ura contains 5 g/l casamino acids, 6.7 g/l yeast nitrogen base, 40 µg/ml tryptophan, 40 µg/ml adenine and 2% galactose (casamino acids lack uracil).
Plasmids
The following STE20 plasmids used in this study were kind gifts from E.Leberer (Leberer et al., 1997): pVTU-STE20 contains wild-type STE20 coding region on the 2 µm URA3+ plasmid pVTU (Vernet et al., 1987); pVTU-STE20Δ42 contains a deletion between residues 334–369 spanning the Cdc42-binding domain; and pCW1 is a high copy-number vector containing the entire STE20 coding region with a lysine-to-alanine (K649A) mutation in the kinase domain. The coding region of wild-type CDC42, dominant-negative CDC42A188 and dominant-active CDC42V12 (Ziman et al., 1991) was placed in the galactose-inducible expression vector pYES2 (Invitrogen). The high copy CLA4 expression vector, pCC1079, was a gift from C.Chan (Chen et al., 1997). pRS416–GFP was constructed by cloning the GFP coding sequence and the NUF2 3′ untranslated region from pPS934 (J.Kahana and P.Silver, unpublished) into the XhoI and KpnI sites of pRS416 (URA3+, CEN) (Sikorski and Hieter, 1989). A PCR fragment containing the SHO1 promoter region and the SHO1 ORF was then inserted at the XhoI site 5′ of the GFP coding sequence in pRS416–GFP to construct pSHO1–GFP. The dominant-active form of the RHO1 gene, RHO1V19, was isolated from the plasmid pRHO1V19 (a gift from D.Levin) and cloned into pYES2.
The plasmid vector pMyr–Sho1 for membrane targeting of the C-terminal domain of Sho1 was constructed by fusing the myristoylation signal sequence of Gpa1 (MGCTVSTQIG) to residue Asp146 of Sho1, and the hemagglutinin epitope coding sequence to the 3′ end of the SHO1 ORF, using appropriate PCR primers. The Myc–Sho1 sequence is then cloned into pYES2 downstream of the galactose-inducible promoter. The non-myristoylation mutant (MyrAS–Sho1) was generated similarly, except the mutant myristoylation sequence (MASTVSTQIG) was used.
Using a similar PCR-based strategy, Ste2–Sho1 fusion constructs between the Ste2 transmembrane domains and the cytoplasmic domain of Sho1 were generated. A region of the STE2 gene encoding the seven transmembrane domains (amino acid positions 1–301) together with the preceding promoter region (535 nucleotides upstream from the initiating methionine codon) was fused to the following coding segments of the SHO1 gene. The prototype pSte2–Sho1 construct contains the entire C-terminal region of Sho1 (residues 146–367); pSte2–Sho1-A contains residues 215–367 of Sho1, which spans half of the Sho1 linker region and the entire SH3 domain; pSte2–Sho1-B contains residues 294–367 comprising only the SH3 domain of Sho1. Construct pSte2–Sho1-C contains only the Sho1 SH3 domain fused to a region of 150 amino acids between residues 637 and 787 of the SLN1 coding sequence, approximately the same length as that of the Sho1 linker region. The construct pSte2–Sho1-D contains the Ste2 transmembrane region and only the Sho1 linker region (residues Asp146–Tyr303) fused to full-length PBS2 ORF. All pSte2–Sho1 plasmids were based on the plasmid vector pRS416 (Stratagene).
Microscopy
Yeast cells were observed using a Nikon Optiphot-2 epifluorescence microscope equipped with a MicroMax charge-coupled device camera (Princeton Instruments). GFP signal was detected using a Chroma filter #41018 (Chroma Technology Corp., Brattleboro, VT). Digital images were converted to PhotoShop format (version 4.0, Adobe Systems).
Electrophoresis and immunoblot analysis
Activated, tyrosine phosphorylated Hog1 was detected by use of the 4G10 anti-phosphotyrosine antibody (a gift from T.Roberts). Cell samples were harvested at the times indicated and quickly frozen on dry ice; after the addition of 2× SDS–PAGE sample buffer, samples were boiled for 10 min, pelleted in a microfuge and supernatants loaded onto a 7.5% polyacrylamide gel for PAGE analysis. Proteins were transferred to nitrocellulose, probed with antibody, and visualized by ECL (Amersham).
Acknowledgments
Acknowledgements
We thank E.A.Witten and David Luyimbazi for their excellent technical assistance, E.Leberer and P.Silver for plasmids, P.Ferrigno for help with microscopy, and Q.Ge and P.O’Grady for valuable comments on the manuscript. This work was supported by NIH grants (GM50909 and GM56699) and a grant from Novartis/DFCI Drug Discovery Program to H.S., and by a postdoctoral fellowship to F.P. from la Dirección General de Investigación Científica y Técnica of the Spanish Government.
References
- Bagrodia S., Dérijard,B., Davis,R.J. and Cerione,R.A. (1995) Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J. Biol. Chem., 270, 27995–27998. [DOI] [PubMed] [Google Scholar]
- Benton B.K., Tinkelenberg,A., Gonzalez,I. and Cross,F. (1997) Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol., 17, 5067–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.-C., Kim,Y.-J. and Chan,C.S.M. (1997) The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev., 11, 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrcková F., De Virgilio,C., Manser,E., Pringle,J.R. and Nasmyth,K. (1995) Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev., 9, 1817–1830. [DOI] [PubMed] [Google Scholar]
- Gancedo J.M. (1998) Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev., 62, 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen G., Anghileri,P., Imre,E., Baroni,M.D. and Ruis,H. (2000) Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J. Biol. Chem., 275, 1449–1456. [DOI] [PubMed] [Google Scholar]
- Gustin M.C., Albertyn,J., Alexander,M. and Davenport,K. (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 62, 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. (1994) Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu. Rev. Cell Biol., 10, 31–54. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. (1995) MAP kinase pathways in yeast: for mating and more. Cell, 80, 187–197. [DOI] [PubMed] [Google Scholar]
- Johnson D.I. (1999) Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev., 63, 54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard,D., Harcus,D., Thomas,D.Y. and Whiteway,M. (1992) The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J., 11, 4815–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Wu,C., Leeuw,T., Fourest-Lieuvin,A., Segall,J.E. and Thomas,D.Y. (1997) Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J., 16, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles,C.A. and Fink,G.R. (1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science, 262, 1741–1744. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C. and Heitman,J. (1997) Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J., 16, 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D., Styles,C.A. and Fink,G.R. (1997) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell, 91, 673–684. [DOI] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy,S.M. and Saito,H. (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature, 369, 242–245. [DOI] [PubMed] [Google Scholar]
- Maeda T., Takekawa,M. and Saito,H. (1995) Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science, 269, 554–558. [DOI] [PubMed] [Google Scholar]
- Manser E., Leung,T., Salihuddin,H., Zhao,Z. and Lim,L. (1994) A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature, 367, 40–46. [DOI] [PubMed] [Google Scholar]
- Martin H., Mendoza,A., Rodriguez-Pachon,J.M., Molina,M. and Nombela,C. (1997) Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol. Microbiol., 23, 431–444. [DOI] [PubMed] [Google Scholar]
- Mösch H.-U., Roberts,R.L. and Fink,G.R. (1996) Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 93, 5352–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A. and Arkowitz,R.A. (2000) Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J. Cell Biol., 148, 1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M. and Herskowitz,I. (1998) The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev., 12, 2874–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota I.M. and Varshavsky,A. (1993) A yeast protein similar to bacterial two-component regulators. Science, 262, 566–569. [DOI] [PubMed] [Google Scholar]
- Peter M., Neiman,A.M., Park,H.-O., van Lohuizen,M. and Herskowitz,I. (1996) Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J., 15, 7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Polverino A., Frost,J., Yang,P., Hutchison,M., Neiman,A.M., Cobb,M.H. and Marcus,S. (1995) Activation of mitogen-activated protein kinase cascades by p21-activated protein kinases in cell-free extracts of Xenopus oocytes. J. Biol. Chem., 270, 26067–26070. [DOI] [PubMed] [Google Scholar]
- Pombo C.M., Kehrl,J.H., Sánchez,I., Katz,P., Avruch,J., Zon,L.I., Woodgett,J.R., Force,T. and Kyriakis,J.M. (1995) Activation of the SAPK pathway by the human STE20 homologue germinal centre kinase. Nature, 377, 750–754. [DOI] [PubMed] [Google Scholar]
- Posas F. and Saito,H. (1997) Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science, 276, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Posas F., Wurgler-Murphy,S.M., Maeda,T., Witten,E.A., Thai,T.C. and Saito,H. (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 ‘two-component’ osmosensor. Cell, 86, 865–875. [DOI] [PubMed] [Google Scholar]
- Posas F., Witten,E.A. and Saito,H. (1998) Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol., 18, 5788–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J.R., Bi,E., Harkins,H.A., Zahner,J.E., De Virgilio,C., Chant,J., Corrado,K. and Fares,H. (1995) Establishment of cell polarity in yeast. Cold Spring Harb. Symp. Quant. Biol., 60, 729–744. [DOI] [PubMed] [Google Scholar]
- Ramer S.W. and Davis,R.W. (1993) A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani Rad M., Xu,G. and Hollenberg,C.P. (1992) STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol. Gen. Genet., 236, 145–154. [DOI] [PubMed] [Google Scholar]
- Ramezani Rad M., Jansen,G., Bühring,F. and Hollenberg,C.P. (1998) Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol. Gen. Genet., 259, 29–38. [DOI] [PubMed] [Google Scholar]
- Roberts R.L. and Fink,G.R. (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev., 8, 2974–2985. [DOI] [PubMed] [Google Scholar]
- Robinson M.J. and Cobb,M.H. (1997) Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol., 9, 180–186. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shimada Y., Gulli,M.-P. and Peter,M. (2000) Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nature Cell Biol., 2, 117–123. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.N., De Virgilio,C., Souza,B., Pringle,J.R., Abo,A. and Reed,S.I. (1995) Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature, 376, 702–705. [DOI] [PubMed] [Google Scholar]
- Toenjes K.A., Sawyer,M.M. and Johnson,D.I. (1999) The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr. Biol., 9, 1183–1186. [DOI] [PubMed] [Google Scholar]
- Vernet T., Dignard,D. and Thomas,D.Y. (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene, 52, 225–233. [DOI] [PubMed] [Google Scholar]
- Wu C., Whiteway,M., Thomas,D.Y. and Leberer,E. (1995) Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem., 270, 15984–15992. [DOI] [PubMed] [Google Scholar]
- Wu C., Leberer,E., Thomas,D.Y. and Whiteway,M. (1999) Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell, 10, 2425–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.-S., Leung,T., Manser,E. and Lim,L. (1995) Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol. Cell. Biol., 15, 5246–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., O’Brien,J.M., Ouellette,L.A., Church,W.R. and Johnson,D.I. (1991) Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol., 11, 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Preuss,D., Mulholland,J., O’Brien,J.M., Botstein,D. and Johnson,D.I. (1993) Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell, 4, 1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]