Abstract
Innate immune receptors detect microbial pathogens and subsequently activate adaptive immune responses to combat pathogen invasion. MyD88 is a key adaptor molecule in both Toll-like receptor (TLR) and IL-1 receptor superfamily signaling pathways. This is illustrated by the fact that human individuals carrying rare, naturally occurring MYD88 point mutations suffer from reoccurring life-threatening infections. Here we analyzed the functional properties of six reported non-synonymous single nucleotide polymorphisms of MYD88 in an in vitro cellular system. Two variants found in the MyD88 death domain, S34Y and R98C, showed severely reduced NF-κB activation due to reduced homo-oligomerization and IRAK4 interaction. Structural modeling highlights Ser-34 and Arg-98 as residues important for the assembly of the Myddosome, a death domain (DD) post-receptor complex involving the DD of MyD88, IRAK4, and IRAK2 or IRAK1. Using S34Y and R98C as functional probes, our data show that MyD88 homo-oligomerization and IRAK4 interaction is modulated by the MyD88 TIR and IRAK4 kinase domain, demonstrating the functional importance of non-DD regions not observed in a recent Myddosome crystal structure. The differential interference of S34Y and R98C with some (IL-1 receptor, TLR2, TLR4, TLR5, and TLR7) but not all (TLR9) MyD88-dependent signaling pathways also suggests that receptor specificities exist at the level of the Myddosome. Given their detrimental effect on signaling, it is not surprising that our epidemiological analysis in several case-control studies confirms that S34Y and R98C are rare variants that may drastically contribute to susceptibility to infection in only few individuals.
Keywords: Adaptor Proteins, Computer Modeling, Crystal Structure, Death Domain, Genetic Polymorphism, Immunology, Innate Immunity, Interleukin Receptor-associated Kinase (IRAK), MyD88, Pattern Recognition Receptor
Introduction
In mammals, innate immunity relies on Toll-like receptor (TLR)7 pathways for the detection of invading pathogens (1). TLRs are a family of germ line-encoded pattern recognition receptors endowed with the capacity to recognize different pathogen-associated molecular patterns (1, 2). Upon pathogen-associated molecular pattern recognition, TLRs trigger the production of proinflammatory cytokines and interferons by transcriptional regulation and, thus, initiate and shape adaptive immune responses (3). Different TLRs possess specificity for diverse structural classes of pathogen molecules. For example, TLR2 is the receptor for bacterial lipopeptides, and TLR4 detects bacterial lipopolysaccharide (LPS), TLR7 viral single-stranded RNA, and TLR9 bacterial and viral DNA (1). TLRs engage pathogen-associated molecular patterns in the extracellular space or endosomes. Their cytoplasmic Toll/IL-1 receptor (TIR) domains relay signals intracellularly where they are integrated and diversified by four adaptor molecules (4); that is, myeloid differentiation primary response gene 88 (MyD88), MyD88-adaptor-like (Mal), TIR-domain-containing adaptor protein inducing IFN-β (TRIF), and TRIF-related adaptor molecule (TRAM).
MyD88, the first TLR adaptor to be discovered, transduces incoming TLR signals emanating from all TLRs except TLR3 (4). Additionally, MyD88 is required for intracellular signaling in response to IL-1 and IL-18 (5). MyD88-dependent signaling involves three functional domains; that is, a C-terminal, evolutionarily conserved TIR domain (residues 159–296) present in all TLR adaptors that allows for upstream homotypic interactions with stimulated TLR and IL-1 receptor complexes, a central so-called intermediate domain (ID; residues 110–154) of unknown protein architecture that is absent in an inhibitory, shorter splice variant, MyD88s (6), and an N-terminal domain belonging to the death domain (DD; residues 19–109) superfamily required for recruitment and activation of downstream DD-containing kinases of the IL-1 receptor-associated kinase (IRAK)-family. DDs are α-helical and engage in homotypic and heterotypic protein-protein complexes (7–9). In MyD88, ID and DD are required for the recruitment and activation of IRAK4, which subsequently triggers the association and phosphorylation of IRAK1 or IRAK2 (4). Activated IRAK1 has been shown to then dissociate from the MyD88 signaling complex to subsequently activate tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF-6) (10, 11). This process culminates in the activation of the nuclear factor (NF)-κB and activating protein (AP)-1 transcription factors and leads to the expression of a multitude of inflammatory downstream genes (for review, see Ref. 4). In mice, IRAK1 appears to be recruited during early NF-κB responses; IRAK2 plays the same role in late NF-κB responses (12) and is involved in LPS-mediated post-transcriptional control (13). The relative contributions of IRAK1 or -2 in humans have yet to be defined, but it has been suggested that IRAK2 may be the most relevant in humans (14). Upon receptor recruitment of MyD88 via TIR-TIR interactions (15), MyD88 orchestrates the assembly of a post-receptor complex termed the Myddosome (16). A recently reported MyD88-IRAK4-IRAK2 Myddosome crystal structure strikingly elucidated the architecture and stoichiometry of such DD complexes (17). Myddosome assembly is nucleated by the oligomerization of six MyD88 DDs. Through an unknown mechanism, instead of additional MyD88 DD, four IRAK4 DDs are subsequently incorporated into the structure. This forms a docking site for another four IRAK2 DDs. The affinity of the MyD88 DD oligomer for IRAK4 DDs exceeds that for IRAK2 DDs, apparently due to charge complementarity.
The functional importance of MyD88 as a signaling adaptor in innate immune defenses was recently illustrated by rare MYD88 point mutations. Individuals carrying these mutations were found to be highly susceptible to infection with pyogenic bacteria (18). Although single nucleotide polymorphisms (SNPs) in other TLR signaling molecules have been associated with infectious, inflammatory, or autoimmune diseases (for review, see Ref. 19), sequence variants in MyD88 have not been the subject of systematic investigation.
In this study we used naturally occurring non-synonymous (ns) SNPs as probes to study the molecular biology of the adaptor molecule MyD88 in detail with a view of identifying protein phenovariants whose corresponding nsSNPs might be of epidemiological interest due to an interesting functional phenotype. The molecular basis for the observed functional effects were subsequently investigated and led to some novel insights into the molecular interactions of MyD88 in the context of the Myddosome.
EXPERIMENTAL PROCEDURES
Reagents and Cells
Reagents were from Sigma unless otherwise stated. Human embryonic kidney (HEK) 293 cells (A. Dalpke, University of Heidelberg, Germany) were cultured in DMEM supplemented with 10% fetal calf serum, l-glutamine, and penicillin/streptomycin (Invitrogen) at 37 °C and 5% CO2. MyD88-deficient IA3 HEK293 cells (G. Stark, Department of Molecular Genetics, Cleveland, OH; Ref. 20) were cultured as above. Pam3CSK4 and R848 were from Axxora, LPS was from Invivogen, flagellin Salmonella typhimurium was from Biomol, and CpG-ODN was from MWG Biotech. The following expression plasmids were used: TLR2-FLAG, untagged TLR7, and TLR9 (I. Bekeredjian-Ding and A. Dalpke, University of Heidelberg, Infectious Disease Department, Heidelberg, Germany); untagged TLR4, MD-2, and TLR5 (P. Ahmad-Nejad, University of Mannheim, Mannheim, Germany).
Cloning and Site-directed Mutagenesis
A template plasmid containing human MyD88 (NCBI accession no. AAC50954) was a gift from U. Hasan, IARC Lyon. N-terminal hemagglutinin (HA)- and Myc-tagged MyD88 variants were cloned to pcDNA3.1 (+) as described (21). N-terminal tagged MyD88 DD-ID truncations were generated by introducing a stop codon at amino acid position 157. MyD88 and IRAK4 DD constructs for bacterial purification were described in Ref. 16. Mutations corresponding to MyD88 nsSNPs were introduced with a QuikChange II kit (Stratagene). Gateway-compatible entry clones for Mal and IRAK4 full-length (FL; obtained from the German Cancer Research Centre Genomics and Proteomics core facility) or DD-only (16) were transferred via LR reaction (Invitrogen) to a pT-Rex-Dest30-based plasmid containing N-terminal Renilla or Protein A tags. Alternatively, the IRAK 4 DD construct was transferred to a pcDNA5/FRT/TO-based plasmid to add an N-terminal Strep-HA tag (T. Bürckstümmer, CeMM, Vienna. Austria). N-terminal-tagged Mal-Myc was described in George et al. (21). PCR and mutagenesis primers are listed in supplemental Table S2.
Signaling Assays and Quantitative Real-time PCR
HEK293 cells in 24-well plates (7.5 × 104 cells/well) were transfected using the calcium phosphate method with a firefly luciferase reporter (100 ng; Stratagene), pRL-Tk Renilla luciferase control reporter (10 ng; Promega), and 30 ng of pC1-EGFP (Clontech) to monitor transfection, incubated for 48 h, lysed, and measured using the Dual Luciferase Assay system (Promega) on a Fluoskan (Thermo Fisher Scientific) or Fluostar Optima (BMG LabTech) luminometer. Different amounts of MyD88 were transfected as described. For titration experiments the transfected plasmid amount was adjusted to an equal amount with empty pcDNA3.1(+) vector. Luciferase measurements were performed in triplicate (±S.D.). For receptor stimulation experiments, stimuli were added 16 h before lysis. For quantitative real-time PCR, 2.25 × 105 HEK293 cells were transfected with 240 ng of the respective plasmids. 48 h later, total RNA was extracted (RNeasy kit, Qiagen), cDNA was synthesized (Superscript III, Invitrogen), and quantitative PCR was performed in duplicate using the Lightcycler 480 Probes Master Mix (Roche Applied Science). Glycerinaldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene. For primers and probes see supplemental Table S2.
Co-immunoprecipitation and Expression Analysis
Immunoprecipitation experiments were essentially described (21). In brief, for MyD88 interactions cells were lysed in 50 mm HEPES, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 20 mm β-glycerophosphate, 2 mm DTT, 1 mm Na3VO4, and protease inhibitors. For MyD88-IRAK4 DD interactions cells were lysed in 20 mm Tris, pH 7.5, 250 mm NaCl, 1% Triton X-100, 10 mm EDTA, 10 mm DTT, and protease and phosphatase inhibitors (Roche Applied Science). Lysis was performed for 20 min on ice. Immunoprecipitations were performed from cleared cell lysates with monoclonal mouse α-Myc (Sigma) and Protein A/G beads (Pierce). Immunocomplexes were washed three times with lysis buffer and analyzed by immunoblot. For expression analysis epitope tags were detected with rabbit α-Myc or α-HA (Cell Signaling). β-Tubulin expression was monitored with mouse α-β-tubulin (Sigma).
LUMIER was described earlier (22) and performed with Protein A and Renilla luciferase-tagged proteins. In brief this method relies on an affinity purification of Protein A-tagged proteins via magnetic IgG-beads (Dynabeads M-280, sheep anti-rabbit IgG, Invitrogen). Co-purified interacting proteins were detected via Renilla luciferase activity. HEK293 cells were seeded in 96-well plates (10,000 cells/well) and transfected with 20 ng of Protein A- and 20 ng of Renilla-encoding plasmids using Lipofectamine 2000 (Invitrogen). 48 h post-transfection, cells were lysed for 15 min on ice in 10 μl of buffer containing 20 mm Tris, pH 7.5, 250 mm NaCl, 1% Triton X-100, 10 mm EDTA, 10 mm DTT, 0.0125 units/μl Benzonase (Novagen), protease and phosphatase inhibitors (Roche Applied Science), and 1% IgG Dynabeads. The lysates were diluted with 100 μl of PBS. To assess differences in transfection efficiencies, 10 μl of the diluted lysate were measured for raw Renilla activity. The remaining lysate was washed 5 times with ice-cold PBS on a magnetic plate washer (Tecan). The washed samples remained in 20 μl of PBS and were measured for bound luciferase activity. An interaction signal was processed by normalizing the activity of bound Renilla luciferase to the amount of raw Renilla activity. The well established interaction of Protein A-tagged Jun and Renilla-tagged Fos (23) served as a positive control and was included in each experiment. As a negative control Renilla-tagged proteins were co-transfected with plasmids encoding for Protein A alone. The LUMIER interactions are represented as normalized signal to noise (S/N) ratios in which the normalized interaction signal was divided by its respective negative control. Interaction analysis was performed in triplicate in each experiment.
Recombinant Protein Production and Gel Filtration
The DDs of both IRAK4 and MyD88 wild type (WT) and mutants were expressed from pETG-30 (A. Geerlof, EMBL, Heidelberg) expression vectors in BL21 Codon Plus (Novagen; IRAK4 DD) and BL21-DE3 (Novagen; hMyD88) as described in Motshwene et al. (16). Briefly, bacterial cells expressing IRAK4 DDs or MyD88 DDs were resuspended in 20 mm Tris, pH 8.0, and lysed. The lysate was centrifuged at 1050 × g for 1 h, and the supernatant was loaded onto a glutathione-Sepharose column and eluted with 20 mm Tris with or without 50 mm NaCl, pH 8.0, 40 mm reduced glutathione. After cleavage of the GST tag, the solution was loaded onto a nickel-nitrilotriacetic acid column to remove cleaved GST. Protein solutions were concentrated, and the individual DDs were purified using ion exchange followed by Superdex-75 size exclusion column (GE Healthcare) with the same buffer supplemented with 5 mm DTT. The MyD88·IRAK4 complexes were prepared by adding a slight molar excess of IRAK4 DD, and the complex was purified using a Superdex-200 column. Purity at all stages was assessed by SDS-PAGE.
Dynamic light scattering was performed using a Zetasizer Nano-S (Malvern) instrument. Protein samples (1.1–2 mg/ml) were centrifuged for 10 min at 13,000 rpm before being loaded on a 12.5 × 12.5 × 45-mm UV-transparent disposable cuvette (Sarstedt) in 20 mm Tris, pH 8.0, 50 mm NaCl. Scattering data were collected at 20 °C at a measurement position of 4.65 mm over a time course of 60–80 s per cycle. The total number of cycles per sample ranged between 13 and 18, and data were averaged across cycles.
Sequence Alignments, Structural Analysis, and Homology Modeling
Protein sequences were from NCBI, alignments were performed with ClustalX and color-formatted according to amino acid physicochemical properties (Joy server). The coordinates for the Myddosome crystal structure (pdb code 3MOP, Ref. 17) were obtained from the Protein Data Bank and analyzed in PyMOL (Schrödinger). Homology models for MyD88 S34Y and R98C were generated using the MODELLER package (24) as described (25, 26) based on the structure of MyD88 M5.
Polymorphism Information, Analysis, and Genotyping
A list of reported SNPs in human MYD88 (Gene ID 4615) was obtained from NCBI (supplemental Table S1). HapMap data were from the International HapMap Project (27), and genotyping was performed using the Sequenom iPLEX assay (Sequenom Inc.) under standard conditions.
Statistical Analysis
p values were determined using Student's t test and designated with p < 0.01 (**) and p < 0.05 (*) throughout.
RESULTS
Two Naturally Occurring Mutations in the MyD88 Death Domain Are Dysfunctional in Downstream Signaling
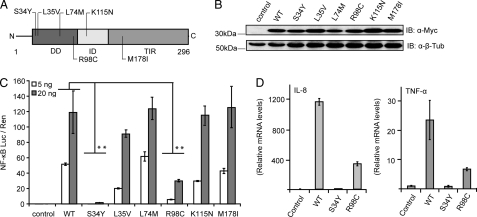
In the human MYD88 gene more than 40 SNPs have been reported, six of which result in amino acid substitutions (nsSNPs) in different MyD88 domains: S34Y, L35V, L74M, R98C (DD), K115N (ID), and M178I (TIR) (see Fig. 1A and supplemental Table S1). Generally, nsSNPs have a high potential impact on protein function (28), and in MyD88 most of the affected amino acid positions are highly conserved in a range of species (supplemental Fig. S1). Although little is known about the frequencies of these naturally occurring variants, we were intrigued to investigate if any of these reported variants were associated with an altered phenotype compared with WT MyD88. To functionally characterize the genetic variants reported for MyD88, Myc-tagged expression constructs corresponding to the nsSNP-associated amino acid changes (“MyD88 mutants”) were generated. The expression levels were similar (Fig. 1B), and the MyD88 mutants were tested for their ability to activate the NF-κB signaling pathway using a dual luciferase assay in HEK293 cells. Fig. 1C shows that overexpression of WT MyD88 results in NF-κB activation and that most MyD88 mutants induce signaling to a level comparable to WT. However, the mutants S34Y and R98C showed a complete loss of function or a reduction to ∼20% of the WT signal, respectively. This reduction was also observed at the level of cytokine induction, as IL-8 and TNF-α transcription levels measured by real-time PCR were reduced for MyD88-S34Y and -R98C (Fig. 1D). To rule out the possibility that our results could be influenced by the presence of endogenous MyD88, we checked NF-κB activation in MyD88-deficient HEK293 IA3 cells (20). In agreement with Fig. 1C, MyD88 S34Y and R98C revealed the same pattern of reduced NF-κB activation (supplemental Fig. S2). In two other cell lines, Huh7 hepatocytes and C33A keratinocytes, NF-κB-driven luciferase production was also strongly reduced for S34Y and R98C compared with WT MyD88 (data not shown). Taken together, these data suggest that the mutants S34Y and R98C interfered with WT MyD88 downstream signaling.
FIGURE 1.
MyD88 S34Y and R98C show reduced NF-κB activation. A, shown is a schematic overview of MyD88 domains with non-synonymous SNPs highlighted. B, expression of all MyD88 mutants is comparable with WT. HEK293 cells were transfected with Myc-tagged expression constructs and analyzed by immunoblot (IB). β-Tubulin served as a loading control. C, S34Y and R98C fail to induce NF-κB activation. HEK293 cells were transiently transfected with the indicated amounts of Myc-tagged MyD88 constructs. NF-κB activation was quantified by dual luciferase assay. D, IL-8 and TNF-α mRNA induction is reduced for S34Y and R98C. HEK293 cells were transiently transfected with MyD88 variants, and induced cytokine mRNA levels were quantified and normalized to GAPDH using LightCycler Realtime PCR. One representative of three independent experiments is shown.
S34Y and R98C Differently Modify MyD88-dependent Receptor Activation
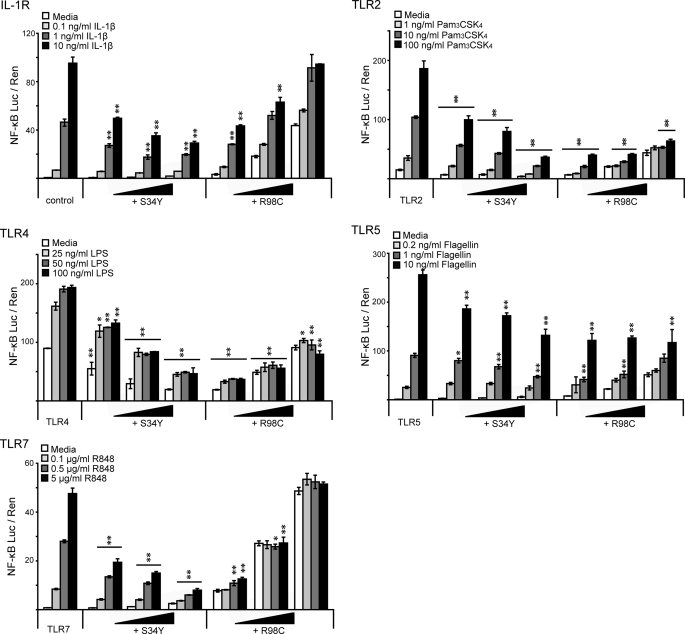
We investigated whether the expression of these mutants interfered with signaling initiated upon activation of TLR2, TLR4, TLR5, TLR7, TLR9, or IL-1R, all of which require MyD88 for downstream signal transduction (1). MyD88 mutants were transiently co-transfected with different TLRs in HEK293 cells, and NF-κB-linked luciferase production was measured upon treatment with the appropriate stimuli. Alternatively, the effect of overexpressed mutants on the stimulation of endogenous IL-1R and TLR5 was analyzed. Whereas overexpression of MyD88 WT increased the overall amount of activated NF-κB as expected (data not shown), both S34Y and R98C exerted a dominant negative effect on most MyD88-dependent receptor signaling pathways in a dose-dependent and stimulation-dependent manner (Fig. 2), with the exception of TLR9 stimulation (supplemental Fig. S3A). S34Y dose-dependently interfered with the induced NF-κB response in the case of all of these receptor pathways when titrating stimulation concentration and the transfected plasmid amount (Fig. 2). However, different TLRs appeared differentially sensitive to this effect; in the case of TLR2, -4, and -7, a dominant negative effect was already observed at the lowest plasmid and stimulation concentrations applied (see Fig. 2). In contrast, in the case of endogenous IL-1R and TLR5 (both endogenous and transfected), signaling was not inhibited in the presence of S34Y at the lowest ligand concentrations (see supplemental Fig. S3B). For R98C, overexpression resulted in a dose-dependent basal level of NF-κB activation (cf. Fig. 1C and see Fig. 4D). In case of IL-1R, TLR2, and TLR5, this signal was further increased in a ligand concentration-dependent way. Nevertheless, the presence of R98C suppressed ligand-induced activation below the level of control-transfected, stimulated cells, especially when low plasmid amounts were transfected and high stimulation concentrations were applied. Interestingly, this effect was even stronger for TLR4 and TLR7 signaling where R98C effectively blocked activation above the level of R98C alone (see Fig. 2, compare white and shaded bars). In summary, our data imply that both S34Y and R98C are able to interfere with MyD88-dependent receptor stimulation in a dominant-negative way across a range of ligand concentrations but with differences in the effectiveness of inhibition between different TLRs (see “Discussion”).
FIGURE 2.
MyD88 S34Y and R98C modulate MyD88-dependent receptor signaling. HEK293 cells were transfected with 10 ng of TLR2-FLAG, 2.5 ng of TLR4 together with 10 ng of MD-2, 50 ng of TLR5, or 100 ng of TLR7. Increasing amounts of MyD88 mutant expression plasmids were co-transfected (5, 10, and 20 ng). Cells were treated with increasing concentrations of receptor agonists accordingly, and NF-κB activation was quantified by dual luciferase assay. p values were calculated referring to the receptor response in the absence of transfected MyD88 variants for each stimulus concentration. One representative of three independent experiments is shown, respectively.
FIGURE 4.
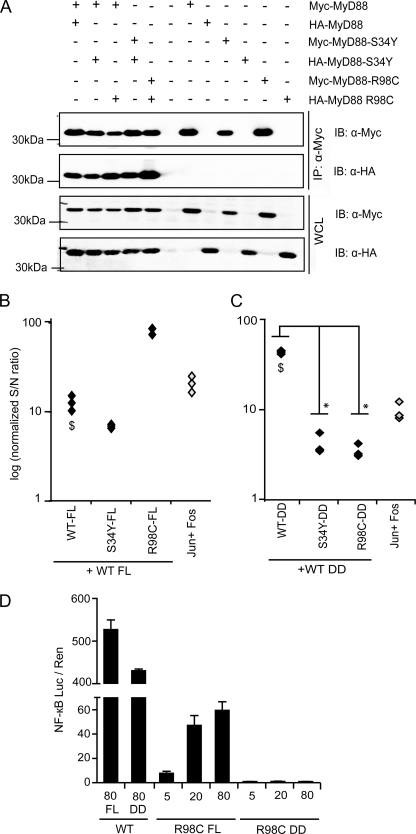
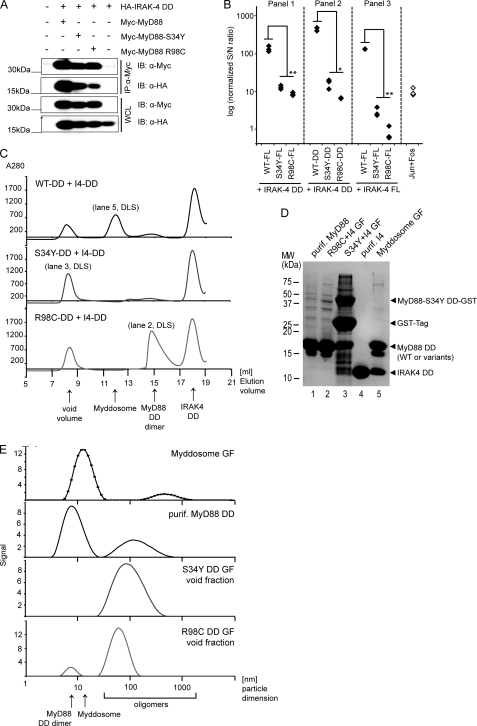
MyD88 homo-oligomerization is not abrogated but reduced in S34Y and R98C. A, S34Y and R98C are able to homo-oligomerize with WT MyD88. Protein complexes were immunoprecipitated (IP) using anti-Myc antibody from HEK293 cells transiently transfected with Myc- and/or HA-tagged MyD88 WT or mutants, respectively. Precipitates and whole cell lysates (WCL) were analyzed by immunoblot (IB). B, in the MyD88 FL context, S34Y-WT interactions are reduced, and R98C-WT interactions increased. LUMIER luciferase readout was from HEK293 cells transfected with Protein A-tagged MyD88 WT or mutants and Renilla luciferase-tagged MyD88 WT. The Jun-Fos interaction served as a positive control. See “Experimental Procedures” for details. C, with regard to DD-DD interactions, S34Y and R98C show drastically reduced homo-oligomerization as assessed by LUMIER. The procedure was as in B with Protein A-tagged MyD88 DD of WT or mutants and Renilla luciferase-tagged MyD88 WT DD. One of at least two independent and representative experiments is shown. The $ symbol denotes a significant difference (p < 0.01) comparing MyD88 FL-FL and DD-DD interactions in respective experiments. D, whereas R98C FL has residual activity, R98C DD does not induce NF-κB activation above background levels. HEK293 cells were transfected with expression constructs for FL or DD constructs for MyD88 WT or R98C, and NF-κB activation was quantified by dual luciferase assay.
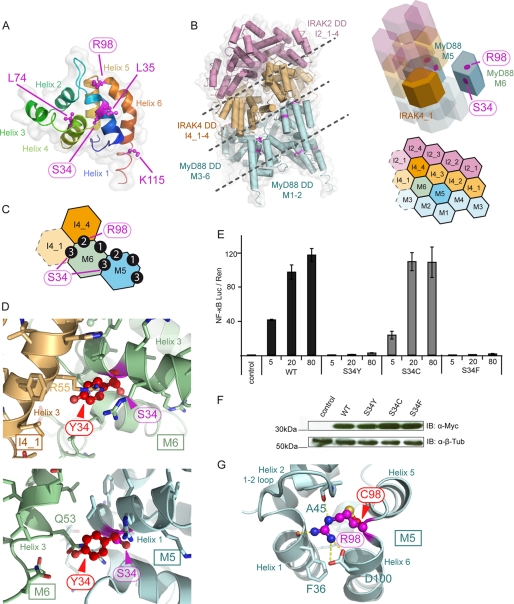
Ser-34 and Arg-98 Play Important Structural Roles in the Myddosome
We next sought to determine the molecular basis for the effects observed for MyD88 S34Y and R98C. Before investigating which protein-protein interactions might be affected by MyD88 S34Y and R98C, their location was analyzed in the three-dimensional framework in which MyD88 has been proposed to operate, the Myddosome (16). Ser-34 is found on helix 1, and Arg-98 is located in the loop connecting helix 5 and 6 (Fig. 3A) in a recently published MyD88-IRAK4-IRAK2 DD Myddosome crystal structure (17) (Fig. 3B). Ser-34 is part of a type 3 helix → helix 3 interaction interface of consecutive MyD88 molecules (e.g. M5 and M6) and of MyD88 M6 with the first IRAK4 DD (I4_1′, see Fig. 3C and Ref. 17). Using homology modeling, a Tyr-34 structure of MyD88 DD was generated and superimposed onto M5 to assess its impact on MyD88-MyD88 interactions (Fig. 3D, lower panel) and on M6 for MyD88-IRAK4 DD interactions (Fig. 3D, upper panel). Evidently, due to its larger size, Tyr-34 would protrude from helix 1 and, thus, clash with the neighboring M6 (glutamine 63) or I4_1 (arginine 55) amino acids. To assess whether steric clashes provide a structural explanation for the observed loss of function, serine 34 was also mutated to the serine-isosteric residue cysteine (S34C) and to phenylalanine (S34F), which is of a similar size as tyrosine. NF-κB activation was subsequently assessed. Fig. 3E shows that S34C signals normally, whereas S34F is also severely impaired in signaling with comparable expression levels (Fig. 3F). This suggests that the introduction of a bulky amino acid like tyrosine or phenylalanine appears to compromise MyD88-MyD88 and MyD88-IRAK4 interactions and, thus, downstream signaling. The fact that S34C signals normally rules out the possibility that loss of serine 34 phosphorylation contributes to the S34Y defect in NF-κB activation. A closer inspection of the Myddosome structure revealed that Arg-98 maps to the helix 5–6 loop near a type 2 interface and that the arginine 98 side chain coordinates the interface with the IRAK4 ring above (Fig. 3, A–C). For example, Arg-98 is involved in hydrogen bonds with the backbone carbonyls of alanine 45 (helix 1–2 loop) and phenylalanine 36 (helix 1) and with the side-chain carboxyl of aspartic 100 at the top of helix 6 (Fig. 3G). A superimposed Cys-98 model clearly illustrates that a cysteine residue at position 98 would fail in “stapling” this surface together due to the loss of hydrogen bonding options. This would most likely lead to a perturbation of the MyD88-IRAK4 interface and potentially also of the MyD88 assembly.
FIGURE 3.
S34Y and R98C impact on the structure of the Myddosome. A, shown is the structure of a human MyD88 monomer in schematic representation with Ser-34, Arg-98, and other nsSNPs highlighted in magenta throughout. Helices are numbered and colored from the N (blue) to the C termini (red). Coordinates were from the pdb file 3MOP (17). B, shown is the structure of the MyD88-IRAK4-IRAK2 DD Myddosome (17). Left, shown is a schematic representation with IRAK2 DDs in pink, IRAK4 DDs are in light orange, and MyD88 DDs is in light cyan. Right upper, shown is a schematic helical assembly structure of the Myddosome with those subunits whose interactions were subjected to detailed analysis shown non-transparently. Right lower, shown is a planar arrangement of the Myddosome complex. C, DD interaction types in the Myddosome are shown. The positions of Ser-34 (type 3 interface) and Arg-98 (type 2 interface) are shown. D, exchange of serine 34 to tyrosine leads to steric clashes with residues of helix 3 in IRAK4 I4_1 (upper) and MyD88 M6 (lower). Upper, the interface of helix 3 of I4_1 and M6 helix 1 was analyzed, and a homology model for the M6 S34Y mutant was superimposed. Evidently, Tyr-34 (red, ball-and-stick) clashed with the side chain of Arg-55 of I4_1 helix 1. Ser-34 is buried. Lower, the S34Y mutant model was also superimposed on M5 in the context of the M5:M6 interaction. Tyr-34 clashes with Gln-53. E, bulky amino acids at position 34 in MyD88 impair downstream signaling, whereas the isosteric cysteine signals normally. HEK293 cells were transfected with Myc-tagged expression constructs for WT MyD88 (Ser-34), S34Y, S34C, and S34F, and NF-κB activation was quantified by dual luciferase assay. One of at least two independent and representative experiments is shown. F, expression of S34Y, S34C, and S34F MyD88 mutants is comparable with WT. HEK293 cells were transfected with Myc-tagged expression constructs and analyzed by immunoblot (IB). β-Tubulin expression served as a loading control. G, Arg-98 plays an important structural role in coordinating the interface of MyD88 with the IRAK4 ring above. As evident, Arg-98 structurally stabilizes helix 5, helix 1, helix 2, and the 1–2 loop (which interdigitates with IRAK4) through hydrogen bonds (yellow dashes) with the Asp-100 side-chain and Phe-36 and Ala-45 backbone oxygens. A R98C model was superimposed onto the elucidated MyD88 structure (M5). Cys-98 (red, ball-and-stick) would fail to engage in these hydrogen bonds.
S34Y and R98C Modulate MyD88 Oligomerization
To verify these hypotheses and test which interactions of the MyD88 mutants were influenced by the amino acid changes, we initially analyzed MyD88 homotypic interactions. Although it is unclear how this property relates to the interactions of endogenous MyD88 upon native TLR stimulation, it has been shown that MyD88 is able to homo-oligomerize using purified MyD88 or overexpression (16, 29). In agreement with these data we were able to detect homo-oligomerization of overexpressed MyD88 WT in HEK293 cells (see Fig. 4A). Furthermore, Fig. 4A shows that MyD88 variants were also able to interact with themselves as well as with MyD88 WT. To accurately quantify the strength of interaction, we applied LUMIER, a novel technique to quantitatively measure protein-protein interactions within minutes of cell lysis (Ref. 22 and “Experimental Procedures”). For this purpose, MyD88 WT and mutants were tagged with Protein A or Renilla luciferase. These fusion constructs showed the same pattern of NF-κB activation as that determined for Myc-tagged MyD88 variants in Fig. 1C (data not shown). The interaction of the AP-1 transcription factor subunits Jun and Fos served as an internal control for a positive and measurable interaction (23). These LUMIER measurements confirmed that upon overexpression in HEK293 cells, the variants can interact with MyD88 WT (Fig. 4B). The levels of interaction between WT-WT and S34Y-WT were similar to that of the Jun-Fos positive control. Interestingly, the R98C-WT interaction was considerably stronger. To exclude the possibility that the TIR domain interferes with these homotypic interactions, we also conducted the experiments with MyD88 constructs consisting only of DD-ID (residues 1–156). The LUMIER technique revealed an interaction for all DD mutants (Fig. 4C). S34Y and R98C DDs showed a significantly decreased affinity for WT DDs. Interestingly, WT-WT interactions were considerably stronger when the DDs alone were transfected rather than the FL MyD88 (cf. Fig. 4, B and C). Of note, whereas R98C FL shows residual activity, R98C DD does not induce NF-κB activation (Fig. 4D). These data imply that S34Y and R98C retain the ability to form homo-oligomers in both the MyD88 FL and DD context. However, the mutant proteins appear to mediate these homotypic DD interactions with reduced affinity. Furthermore, DD oligomerization is significantly influenced by the TIR domain (see “Discussion”).
S34Y and R98C Fail to Recruit IRAK4
MyD88 homo-oligomerization has been proposed to nucleate Myddosome assembly (17). Because S34Y and R98C impact on this step, our modeling results prompted us to determine whether the association of the MyD88 variants with downstream signaling components of the IRAK family is altered. We investigated the association of FL and DD MyD88 WT and the mutants with FL and DD IRAK4 (residues 1–106) by co-immunoprecipitation and LUMIER. We initially generated an N-terminal Strep-HA-tagged construct of IRAK4 solely comprising the DD, a construct known to be sufficient for MyD88 interactions (16, 17). Fig. 5A shows that all FL MyD88 mutants were still able to interact with IRAK4 DDs (an interaction found to be highly dependent on the presence of EDTA; data not shown). However, the band intensities implied reduced affinities of S34Y and R98C for IRAK4 DDs. This was confirmed using LUMIER (Fig. 5B, Panel 1). We also analyzed the association of MyD88 DD-only constructs with IRAK4 DDs. In agreement with the previous experiment, the DDs of S34Y and R98C showed more than a 10-fold reduced binding but nevertheless weak binding to the IRAK4 DD (Fig. 5B, Panel 2). We sought to confirm this finding using the FL constructs of both MyD88 and IRAK4. As the association of IRAK4 with MyD88 is a transient event, it cannot be captured by co-immunoprecipitation (30). In view of this, we conducted LUMIER experiments, and this showed that the interaction of MyD88 WT FL with IRAK4 FL was comparable with MyD88 FL or DD interactions with IRAK4 DDs. By contrast, no interaction of IRAK4 FL could be detected with the S34Y and R98C mutants (Fig. 5B, Panel 3), implying that the presence of the kinase domain influences the assembly of the Myddosome in vitro. To confirm these findings by an alternative method, we purified MyD88 WT, S34Y, and R98C DDs from bacteria (16) and tested their ability to interact with purified IRAK4 DD. We analyzed the elution profiles of mixtures of the MyD88 mutants in the presence of a molar excess of IRAK4 DDs by size exclusion chromatography (Fig. 5C). Individual fractions or purified reference proteins were analyzed by SDS-PAGE (Fig. 5D). IRAK4 is recruited into a characteristic higher Mr Myddosome peak by WT MyD88 (elution volume Ve = 11.95 ml corresponding to 165 kDa, lane 5) but not by S34Y or R98C. In these mixtures, the IRAK4 DD instead eluted exclusively on its own (Ve = 18 ml, corresponding to an IRAK4 DD monomer). S34Y (lane 3) and R98C DDs (not shown) were present in the void volume (Ve = 8.38 ml), but the peaks did not contain any IRAK4 DDs. In the case of R98C, a second peak (Ve = 14.7 ml, lane 2) eluted at a position characteristic of a MyD88 WT dimer (not shown, see Ref. 16). Individual peaks or samples (as indicated in Fig. 5, C and D) were also analyzed by dynamic light scattering, which provides an assessment of the size of complexes formed by MyD88 mutants and/or IRAK4 DDs. As found previously (16), MyD88 WT DD formed two species, one corresponding to a MyD88 DD dimer (with a hydrodynamic diameter, d(H) of 8.6 nm), the other to a higher oligomer (d(H) of 135 nm; Fig. 5E). By contrast, the Myddosome complex fraction (Ve = 11.95 ml, lane 5 in Fig. 5D) is detected as one predominant species of d(H) 13 nm (16). Void volume fractions of S34Y and R98C contained mainly higher order oligomers only, suggesting they that they have an enhanced tendency to aggregate (d(H) 110.02 and 64.05 nm, respectively). This is especially evident with the S34Y DD, which does not appear to form a stable dimeric species and is highly resistant to proteolysis from its GST tag (Fig. 5D, cf. lane 1 and 2 with lane 3), indicating that the cleavage site is sequestered by aggregation. R98C DD aggregation appears partially reversible, as the R98C DD void volume fraction also gave a minor signal corresponding to a dimer (d(H) 7.43 nm). Although there are likely to be subtle differences between MyD88 and IRAK4 DDs from bacteria and in mammalian cells, our data conclusively illustrate the failure of MyD88 S34Y and R98C to associate with IRAK4 and identify this as the defect in signal transduction.
FIGURE 5.
S34Y and R98C fail to interact with IRAK4. A, the interaction of S34Y and R98C with the IRAK4 DD is reduced. Protein complexes were immunoprecipitated using anti-Myc antibody from HEK293 cells transiently transfected with Myc-MyD88 WT or mutants and Strep-HA-tagged IRAK4 DD, respectively. Precipitates and whole cell lysates (WCL) were analyzed by immunoblot (IB). B, MyD88 S34Y-and R98C-IRAK4 interactions are strongly reduced in the context of MyD88 FL and IRAK4 DD (Panel 1). Similar results were obtained in the DD-DD interactions (Panel 2). Interactions are reduced to a level of no interaction in the context of mutant FL MyD88-FL IRAK4 interactions (Panel 3). Protein A-tagged MyD88 WT and mutants in FL or DD context and Renilla luciferase-tagged IRAK4 FL or DD constructs were transfected into HEK293 cells, and LUMIER measurements were conducted as described under “Experimental Procedures.” One representative of at least two independent experiments is shown. C, S34Y and R98C are unable to assemble into a Myddosome on size exclusion chromatography of mixtures of bacterially purified MyD88 DDs and IRAK4 DDs (added in excess). WT MyD88+IRAK4 (upper graph) mixtures elute in a discrete Myddosome peak. This peak is absent in S34Y (middle graph) and R98C (lower graph) mixtures. Both mutants elute in the void volume, containing higher order oligomers only and no IRAK4 DD. R98C also shows a dimer peak. Individual peak fractions from gel filtration (labeled GF), purified MyD88 DD or IRAK4 DD alone (for size comparison) were analyzed by reducing SDS-PAGE (D) or dynamic light scattering (marked DLS) to measure the size of protein species found in these fractions or samples (E). One representative of two independent experiments is shown respectively.
Myddosome Function Is Regulated by Non-DD Regions
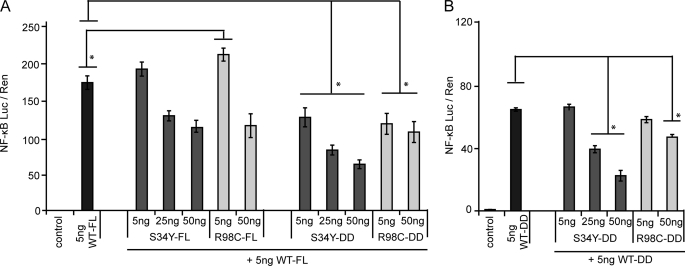
The difference observed in the interaction strengths between different MyD88 and IRAK4 constructs (cf. Fig. 5B) suggests a significant influence of non-DD regions on Myddosome assembly. To investigate this further, we tested S34Y and R98C for dominant negative effects on MyD88 signaling. As shown in Fig. 6A, NF-κB activation mediated by FL MyD88 WT was only significantly inhibited by an excess of S34Y or R98C FL plasmid DNA. On the other hand, S34Y and R98C DD influenced MyD88 FL activation already at equimolar amounts of transfected plasmid DNA. When, instead, MyD88 WT DD was used for NF-κB activation, S34Y DD failed to significantly impair NF-κB activation at a 1:1 ratio (Fig. 6B). This suggests that non-DD regions influence Myddosome function not only at the interaction (cf. Fig. 4) but also at the functional level (see “Discussion”).
FIGURE 6.
DD and non-DD regions in MyD88 differentially modulate dominant-negative activity of S34Y and R98C. A, signaling induced by MyD88 WT plasmid is reduced by S34Y and R98C. Whereas S34Y and R98C FL exhibit only a weak dominant-negative effect, DD-only constructs impair signaling at equimolar plasmid ratio. B, when a constant amount of MyD88 DD plasmid is used to induce NF-κB signaling, DD-only mutant constructs do not reduce signaling at a 1:1 level, albeit at higher molar ratios. In both A and B HEK293 cells were transfected with 5 ng of MyD88 WT constructs (DD or FL) and increasing amounts of MyD88 mutants. NF-κB activation was quantified by dual luciferase assay. One representative of at least three independent experiments is shown, respectively.
S34Y and R98C Are Relatively Rare MyD88 Variants
Having discovered that two naturally occurring variants of MyD88 affect the signaling properties of this universal TLR adaptor on the molecular level, we became interested in the epidemiological significance of the associated nsSNPs. Unfortunately, according to the NCBI dbSNP data base, few details on the frequency of S34Y (rs1319438) and R98C (rs199396) are available. In one study in 184 samples conducted by the Centre d'Etude du Polymorphisme Human comprising genomic DNA samples from individuals with UTAH (93%), French (4%), and Venezuelan (3%) background, both variants were monomorphic. Additionally, rs199396 was also monomorphic in a Japanese study (n = 183; Ref. 31). To verify these results, we genotyped rs1319438 and rs199396 in case-control study collectives for several infectious diseases from different ethnic groups, namely invasive pneumococcal disease (IPD) from the United Kingdom (222 cases, 282 controls), bacteremia from Kenya (149 cases, 220 controls), and leprosy from India (227 cases, 166 controls). We did not detect a single carrier for rs1319438 or rs199396, suggesting that these variants are rare in the human population (<1%). Of note, in some databases the C allele of R98C has been listed as WT. Our genotyping studies clearly define the R as the major allele.
DISCUSSION
S34Y and R98C Are Rare Mutations in Humans
The phenotype of MyD88-deficient mice clearly illustrates the pivotal role of MyD88 in host defense against a broad range of pathogens (32, 33). Similar findings were obtained in human patients with apparent MyD88 deficiency, who display a high susceptibility to pyogenic bacteria. Cytokine responses from patient cells were impaired and transcriptional profiles found to be drastically altered (18). The responsible mutations E52del, L93P and R196C, mapped to the MyD88 DD and TIR, respectively. In this paper we have studied the function of additional variants reported for MyD88. Our data on the mutations S34Y (rs1319438) and R98C (rs199396) show an effect similar to the previously characterized E52del in terms of reduced NF-κB activation (Ref. 18 and our data in the HEK293 system, not shown). It is interesting to note that even though E52del was found in two unrelated kindreds according to the report by von Bernuth et al. (18), the mutation was not identified in more than 1800 unrelated individuals, indicating that it represents a very rare mutation in humans. This suggests that loss of function mutations in a pivotal adaptor molecule like MyD88 encounter strong negative selective pressure. It is to be expected that in the absence of antibiotics these two mutations would be associated with high lethality during childhood and, hence, would not be maintained in the human population. Interestingly, the TLR adaptor Mal loss of function alleles D96N and S180L have been identified in human study populations (21, 34). This may be due to the fact that Mal is only required for TLR2 and TLR4 signaling or that Mal deficiency may be associated with an attenuated cytokine response beneficial to the host (34). Our in vitro data regarding the effect of S34Y and R98C (cf. Fig. 6) on a MyD88 WT allele and on several TLR pathways (cf. Fig. 2) are an indication that these mutations would affect TLR signaling to a similar level as observed for E52del. It was, therefore, not surprising that our preliminary studies suggest that S34Y (rs1319438) and R98C (rs199396) are equally rare. There are additional examples for rare sequence variants in other components of the innate immune system such as IRAK4 (35), TLR3 (36), or RIG-I (37). Although more frequent variants may have a wider yet more subtle effect on disease susceptibility in the population at large, rare variants may contribute drastically to disease susceptibility on an individual level (38).
Structural Insights into Myddosome Assembly
Our findings are of special interest in the context of the recently published MyD88-IRAK4-IRAK2 Myddosome crystal structure (17). Our data show that not only directly interacting residues at MyD88-MyD88 or MyD88-IRAK4 interfaces impact on Myddosome assembly but also distal residues like serine 34 and arginine 98, which are essentially required for the internal structural integrity of MyD88 DDs (see below). Although it is directly adjacent to Ser-34, L35V does not alter NF-κB activation. We attribute this to the fact that a small and similarly hydrophobic amino acid can be accommodated at this position. Most other nsSNPs in MYD88 also result in relatively conservative amino acid substitutions. Of interest is that K115N maps to the functionally important ID (39). As evident from the Myddosome structure, parts of the intermediate domain are included in the DD structure as an elongated helix 6, which lies at the outside of the Myddosome. Lys-115 has no direct intra- or intermolecular contacts so that amino acid changes in the ID remain functionally silent, as observed for the mutations E110A and D112A (40).
Our modeling data provide a molecular explanation for the defects seen in the S34Y and R98C mutants. In the Myddosome, the DDs of MyD88, IRAK4, and IRAK1 interact via type 1, type 2, and type 3 interfaces to form a helical superstructure (Ref. 17; cf. Fig. 3, B and C) in which consecutive DD along the superhelix are stabilized by type 3 DD-DD interactions. Although there are many heterotypic type 1 and type 2 DD-DD interactions between the constituent subunits of the Myddosome, there is only one type 3 interaction between the MyD88 and the first IRAK4 protomer to be incorporated (Fig. 3C). This type 3 interface occurs between helix 1 residues in M6 and helix 3 of I4_1 (Fig. 3D, upper panel). A very similar structural configuration is observed at the type 3 interface between M5 and M6 (Fig. 3D, lower panel). It is clear from the modeling study we present that substitution of serine 34 in helix 1 by tyrosine would cause steric hindrance in both the homotypic (M5:M6) and the single heterotypic type 3 interfaces (M6:I4_1), leading to reduced affinity of homotypic (Fig. 4C) and heterotypic (Fig. 5B, panel 2) interactions. It is conceivable that a weakened type 3 interface additionally decreases the stability of type 1 and/or 2 interactions and, thus, helical assembly. The reason why MyD88 dimers are seen for R98C (Fig. 5C) and WT (not shown, Ref. 16) but not S34Y may be due to the fact that initial MyD88 dimer formation depends exclusively on a type 3 interface. For the R98C mutant the molecular basis of the defect appears to be different. Arg-98 is located close to (but not part of) a type 2 interface. Several hydrogen bonds formed by the Arg-98 side-chain coordinate helix 1, 5, 6, and the 1–2 loop, which contacts any subsequent ring (MyD88 or IRAK4) above. Substitution of Arg-98 for cysteine would cause perturbations in this type 2 interface. Whereas dimer formation would not be affected (as evidenced by gel filtration of purified R98C, cf. Fig. 5C), full Myddosome assembly is probably compromised. The residual level of R98C activity observed in the FL but not DD context (Fig. 4D) may be due to the fact that the TIR domain recovers interaction with WT MyD88 (Fig. 4B) through an unknown mechanism (see also below). In conclusion, this identifies the formation of the heterotypic type 2 and 3 interfaces between IRAK4 and MyD88 and, by implication, helical assembly of the Myddosome complex as essential steps in signal transduction.
Influence of Non-DD Regions on Myddosome Function
Another important insight from our work is the influence of non-DD regions on Myddosome assembly. The crystal structure of the Myddosome obviously does not include non-DD regions like the MyD88 TIR domain or the IRAK kinase domains (17). We found that DD-DD interactions in general are stronger than looking at the same interactions in an FL context as assessed by LUMIER, a technique providing a more quantitative measurement of protein-protein interactions than co-immunoprecipitation. For example, the presence of the MyD88 TIR domain significantly modulated MyD88 homo-oligomerization (cf. Fig. 4, B versus C, WT FL versus WT DD) and heterotypic interaction with IRAK4 DDs (cf. Fig. 5B, Panel 1 versus Panel 2, WT FL versus WT DD). This is in keeping with earlier immunoprecipitation studies which found that the TIR domain plays a role in MyD88 homodimer formation independently of the DD. Unfortunately, the strengths of interaction for different MyD88 (FL, DD or TIR-only) constructs was not quantified (29). The MyD88 TIR domain also seems to play an important role in the dominant-negative effect of MyD88 (cf. Fig. 6). For example, MyD88 FL is only inhibited significantly by DD-only S34Y and R98C at equimolar ratios of transfected plasmid (right part of Fig. 6A). Based on our LUMIER data, this may be due to a stronger interaction upon removal of the TIR domain of S34Y or R98C. Surprisingly, when TIR domains are absent in both MyD88 WT and mutants, this inhibition is again lost (left part of Fig. 6B). Due to the fact that S34Y DD and R98C DD interact with WT DD far less strongly than WT DD; cf. Fig. 4C), inactive S34Y or R98C DD would be excluded from active WT DD Myddosomes. In the case of WT FL-WT FL versus WT FL-mutant DD interactions, the difference in affinity may be less, so that signaling-deficient mutant DDs are also incorporated into Myddosomes, leading to an overall decrease in downstream signal activation. We also found that the IRAK4 kinase domain influences MyD88-IRAK4 DD interactions (cf. (Fig. 5B): for instance, S34Y and R98C FL interacted, albeit weakly, with the IRAK4 DD (Panel 1). The interaction with IRAK4 FL for the same MyD88 constructs was however abrogated (Panel 3). Because the structural assembly shown in the crystal structure appears to form spontaneously from purified DDs (16, 17), in a cellular context TIR domain and kinase domains may, thus, modulate Myddosome assembly. Although at this stage the precise mechanisms of Myddosome regulation in vivo remain elusive, our data suggest that additional work both structural and biochemical is required at the level of FL proteins. We introduce LUMIER as a technique able to assess even transient IRAK4 FL interactions. Additionally, the spatiotemporal framework of Myddosome assembly in living cells should be addressed to complement the information derived from isolated crystallized proteins.
Receptor-specific MyD88 Post-receptor Complexes
Our data suggest that the mutants S34Y and R98C negatively influence MyD88-dependent receptor stimulation and that the MyD88 requirement for different TLR pathways may vary. We would have expected that, if at all, a loss of function mutation in the MyD88 DD would interfere with all MyD88-dependent signaling pathways in a similar way, as receptor-specific coupling is assumed to be mediated exclusively by the respective TIR domains. Notwithstanding the limitations of the HEK293 system, according to the data shown in Figs. 2 and supplemental Fig. S3, S34Y and R98C only affected IL-1R, TLR2, TLR4, TLR5, and TLR7 (not TLR9), and differences in the effectiveness of inhibition between the different receptors were repeatedly observed: whereas S34Y inhibited TLR2, TLR4, and TLR7 ligand-induced levels of activation, already at low ligand concentrations, and TLR5 and IL-1R were only sensitive to S34Y at high ligand concentrations (cf. supplemental Fig. S3B). In contrast, the analysis of the mutant R98C revealed distinct dominant negative effects between different receptor pathways; R98C attenuated IL-1R, TLR2, and TLR5 signals significantly but only entirely blocked TLR4 and TLR7 ligand-induced effects (cf. Fig. 2B). These results might indicate differences in MyD88-dependent receptor-proximal downstream events. Given that Mal-MyD88 interactions were comparable with WT in S34Y and R98C (cf. supplemental Fig. S4), upstream coupling to TLR2 and 4 via Mal and, thus, TIR-mediated TLR2/4-Mal-MyD88 associations are presumably intact. Consequently, the dominant-negative effect would manifest itself at the level of the DD assembly. Targeting different interfaces of DD assembly (S34Y, type 3; R98C, type 2) in a dominant-negative fashion appears to affect specific TLR-induced Myddosomes differently. These assemblies may, therefore, show greater receptor-specific differences than previously thought, and a model of signal transduction in which all MyD88-dependent pathways are converted to the same downstream signal at the level of MyD88 is likely to be oversimplified. A certain degree of specificity is potentially maintained from incoming signals via different TLRs at and beyond the level of post-receptor complexes. Reconstitution experiments into MyD88-deficient cells endogenously expressing TLRs may be ways to confirm this hypothesis. Additionally, studies involving LUMIER assays between different TLR TIR domains and MyD88 mutants or proteomics approaches involving mass spectrometry may help to unravel the structural framework for the specificity of different MyD88 TLR interactions.
In conclusion, we present here a detailed functional and biochemical analysis of two rare MYD88 loss of function variants whose significance for human disease waits to be explored more fully. Our study provides valuable insights into the assembly of MyD88-IRAK4 Myddosomes. Our data point to the fact that this process is regulated by non-DD regions in both MyD88 and IRAK4 and presumably also IRAK1 or -2. The precise contribution of these domains will need to be clarified functionally, biochemically, and cell biologically to understand and therapeutically interfere with, the function of the Myddosome as a post-receptor TLR signaling complex.
Supplementary Material
Acknowledgments
We gratefully acknowledge J. Meckler, M. Scheuermann, T. Schmidt, M. Kögl, and the other members of the German Cancer Research Centre Proteomics core facility for technical assistance. We thank M. Frank and T. Holz for support regarding molecular dynamics and computers. J. C. Crook, K. Knox, and the Oxford Pneumococcal Surveillance Group, A. Berkley, K. Marsh, N. Peshu, A. G. Scott, T. Williams (Kilifi, Kenya), S. Roy, S. K. Hazra, and B. Saha (Kolkata, India), were involved in the collection of samples of the different study populations investigated. We thank R. Jerala for critical reading of this manuscript and helpful comments.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- TLR
- Toll-like receptor
- AP
- activating protein
- DD
- death domain
- ID
- intermediate domain
- IL-1R
- IL-1 receptor
- IRAK
- IL-1 receptor-associated kinase
- Mal
- MyD88-adaptor like
- MyD88
- myeloid differentiation primary response gene 88
- ns
- non-synonymous
- SNP
- single nucleotide polymorphism
- TIR
- Toll/interleukin 1 receptor
- d(H)
- hydrodynamic diameter
- FL
- full-length.
REFERENCES
- 1. Kawai T., Akira S. (2008) Ann. N.Y. Acad. Sci. 1143, 1–20 [DOI] [PubMed] [Google Scholar]
- 2. Rock F. L., Hardiman G., Timans J. C., Kastelein R. A., Bazan J. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwasaki A., Medzhitov R. (2004) Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 4. Watters T. M., Kenny E. F., O'Neill L. A. (2007) Immunol. Cell Biol. 85, 411–419 [DOI] [PubMed] [Google Scholar]
- 5. O'Neill L. A. (2008) Immunol. Rev. 226, 10–18 [DOI] [PubMed] [Google Scholar]
- 6. Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. (2003) J. Exp. Med. 197, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weber C. H., Vincenz C. (2001) Trends Biochem. Sci. 26, 475–481 [DOI] [PubMed] [Google Scholar]
- 8. Scott F. L., Stec B., Pop C., Dobaczewska M. K., Lee J. J., Monosov E., Robinson H., Salvesen G. S., Schwarzenbacher R., Riedl S. J. (2009) Nature 457, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park H. H., Wu H. (2007) Acta Crystallogr. Sect F Struct. Biol. Cryst. Commun. 63, 229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 11. Brikos C., Wait R., Begum S., O'Neill L. A., Saklatvala J. (2007) Mol. Cell. Proteomics 6, 1551–1559 [DOI] [PubMed] [Google Scholar]
- 12. Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., Saitoh T., Kawai T., Takeuchi O., Akira S. (2008) Nat. Immunol. 9, 684–691 [DOI] [PubMed] [Google Scholar]
- 13. Wan Y., Xiao H., Affolter J., Kim T. W., Bulek K., Chaudhuri S., Carlson D., Hamilton T., Mazumder B., Stark G. R., Thomas J., Li X. (2009) J. Biol. Chem. 284, 10367–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conner J. R., Smirnova, Poltorak A. (2009) J. Exp. Med. 206, 1615–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. D. (1997) Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 16. Motshwene P. G., Moncrieffe M. C., Grossmann J. G., Kao C., Ayaluru M., Sandercock A. M., Robinson C. V., Latz E., Gay N. J. (2009) J. Biol. Chem. 284, 25404–25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin S. C., Lo Y. C., Wu H. (2010) Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C. L., Chrabieh M., Mustapha I. B., Ghandil P., Camcioglu Y., Vasconcelos J., Sirvent N., Guedes M., Vitor A. B., Herrero-Mata M. J., Aróstegui J. I., Rodrigo C., Alsina L., Ruiz-Ortiz E., Juan M., Fortuny C., Yagüe J., Antón J., Pascal M., Chang H. H., Janniere L., Rose Y., Garty B. Z., Chapel H., Issekutz A., Maródi L., Rodriguez-Gallego C., Banchereau J., Abel L., Li X., Chaussabel D., Puel A., Casanova J. L. (2008) Science 321, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Misch E. A., Hawn T. R. (2008) Clin. Sci. 114, 347–360 [DOI] [PubMed] [Google Scholar]
- 20. Li X., Commane M., Burns C., Vithalani K., Cao Z., Stark G. R. (1999) Mol. Cell. Biol. 19, 4643–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. George J., Kubarenko A. V., Rautanen A., Mills T. C., Colak E., Kempf T., Hill A. V., Nieters A., Weber A. N. (2010) J. Immunol. 184, 3025–3032 [DOI] [PubMed] [Google Scholar]
- 22. Barrios-Rodiles M., Brown K. R., Ozdamar B., Bose R., Liu Z., Donovan R. S., Shinjo F., Liu Y., Dembowy J., Taylor I. W., Luga V., Przulj N., Robinson M., Suzuki H., Hayashizaki Y., Jurisica I., Wrana J. L. (2005) Science 307, 1621–1625 [DOI] [PubMed] [Google Scholar]
- 23. Chiu R., Boyle W. J., Meek J., Smeal T., Hunter T., Karin M. (1988) Cell 54, 541–552 [DOI] [PubMed] [Google Scholar]
- 24. Sali A., Overington J. P. (1994) Protein Sci. 3, 1582–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubarenko A., Frank M., Weber A. N. (2007) Biochem. Soc. Trans. 35, 1515–1518 [DOI] [PubMed] [Google Scholar]
- 26. Kubarenko A. V., Ranjan S., Colak E., George J., Frank M., Weber A. N. (2010) Protein Sci. 19, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. (2003) Nature 426, 789–796 [DOI] [PubMed] [Google Scholar]
- 28. Ramensky V., Bork P., Sunyaev S. (2002) Nucleic Acids Res. 30, 3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) J. Biol. Chem. 273, 12203–12209 [DOI] [PubMed] [Google Scholar]
- 30. Li S., Strelow A., Fontana E. J., Wesche H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5567–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suzuki H., Suzuki Y., Narita I., Aizawa M., Kihara M., Yamanaka T., Kanou T., Tsukaguchi H., Novak J., Horikoshi S., Tomino Y. (2008) J. Am. Soc. Nephrol. 19, 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S. O., Goode J., Lin P., Mann N., Mudd S., Crozat K., Sovath S., Han J., Beutler B. (2003) Nature 424, 743–748 [DOI] [PubMed] [Google Scholar]
- 33. Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 34. Khor C. C., Chapman S. J., Vannberg F. O., Dunne A., Murphy C., Ling E. Y., Frodsham A. J., Walley A. J., Kyrieleis O., Khan A., Aucan C., Segal S., Moore C. E., Knox K., Campbell S. J., Lienhardt C., Scott A., Aaby P., Sow O. Y., Grignani R. T., Sillah J., Sirugo G., Peshu N., Williams T. N., Maitland K., Davies R. J., Kwiatkowski D. P., Day N. P., Yala D., Crook D. W., Marsh K., Berkley J. A., O'Neill L. A., Hill A. V. (2007) Nat. Genet. 39, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Picard C., Puel A., Bonnet M., Ku C. L., Bustamante J., Yang K., Soudais C., Dupuis S., Feinberg J., Fieschi C., Elbim C., Hitchcock R., Lammas D., Davies G., Al-Ghonaium A., Al-Rayes H., Al-Jumaah S., Al-Hajjar S., Al-Mohsen I. Z., Frayha H. H., Rucker R., Hawn T. R., Aderem A., Tufenkeji H., Haraguchi S., Day N. K., Good R. A., Gougerot-Pocidalo M. A., Ozinsky A., Casanova J. L. (2003) Science 299, 2076–2079 [DOI] [PubMed] [Google Scholar]
- 36. Zhang S. Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., Picard C., Chapgier A., Plancoulaine S., Titeux M., Cognet C., von Bernuth H., Ku C. L., Casrouge A., Zhang X. X., Barreiro L., Leonard J., Hamilton C., Lebon P., Héron B., Vallée L., Quintana-Murci L., Hovnanian A., Rozenberg F., Vivier E., Geissmann F., Tardieu M., Abel L., Casanova J. L. (2007) Science 317, 1522–1527 [DOI] [PubMed] [Google Scholar]
- 37. Pothlichet J., Burtey A., Kubarenko A. V., Caignard G., Solhonne B., Tangy F., Ben-Ali M., Quintana-Murci L., Heinzmann A., Chiche J. D., Vidalain P. O., Weber A. N., Chignard M., Si-Tahar M. (2009) PLoS One 4, e7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schork N. J., Murray S. S., Frazer K. A., Topol E. J. (2009) Curr. Opin. Genet. Dev. 19, 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Janssens S., Burns K., Tschopp J., Beyaert R. (2002) Curr. Biol. 12, 467–471 [DOI] [PubMed] [Google Scholar]
- 40. Loiarro M., Gallo G., Fantò N., De Santis R., Carminati P., Ruggiero V., Sette C. (2009) J. Biol. Chem. 284, 28093–28103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.