Abstract
Members of the Anoctamin (Ano)/TMEM16A family have recently been identified as essential subunits of the Ca2+-activated chloride channel (CaCC). For example, Ano1 is highly expressed in multiple tissues including airway epithelia, where it acts as an apical conduit for transepithelial Cl− secretion and helps regulate lung liquid homeostasis and mucus clearance. However, little is known about the oligomerization of this protein in the plasma membrane. Thus, utilizing mCherry- and eGFP-tagged Ano1 constructs, we conducted biochemical and Förster resonance energy transfer (FRET)-based experiments to determine the quaternary structure of Ano1. FRET and co-immunoprecipitation studies revealed that tagged Ano1 subunits directly associated before they reached the plasma membrane. This association was not altered by changes in cytosolic Ca2+, suggesting that this is a fixed interaction. To determine the oligomeric structure of Ano1, we performed chemical cross-linking, non-denaturing PAGE, and electromobility shift assays, which revealed that Ano1 exists as a dimer. These data are the first to probe the quaternary structure of Ano1. Understanding the oligomeric nature of Ano1 is an essential step in the development of therapeutic drugs that could be useful in the treatment of cystic fibrosis.
Keywords: Cystic Fibrosis, Fluorescence Resonance Energy Transfer (FRET), Ion Channels, Protein Structure, Protein-Protein Interactions
Introduction
Ca2+-activated Cl− channels (CaCCs)4 play essential roles in many physiological processes including epithelial secretion (1), olfactory transduction (2–4), photoreceptor adaptation (5), regulation of smooth muscle contraction (6), neuronal and cardiac action potential waveform and firing frequency (7), and nociception (8). Because of their physiological significance, CaCCs have attracted attention for decades (9, 10). However, their molecular composition has remained elusive until recently when three laboratories independently identified members of the Anoctamin 1 (Ano1) family, also known as the transmembrane protein 16 (TMEM16) family, as essential subunits of CaCCs (11–13). Ano1/TMEM16A is highly expressed in secretory epithelial tissues including ductal glands, superficial epithelia of the airway, and oviduct, where it has been implicated to play a key role in calcium-dependent chloride secretion (11, 13–16). Ano1 has attracted particular attention in the airways, because it has been suggested that activating this channel might be therapeutically beneficial for treating cystic fibrosis (CF) patients (15). The cystic fibrosis conductance regulator (CFTR) chloride channel is absent or defective in CF epithelia, leaving Ano1 as a potential alternative pathway to effect transepithelial Cl− secretion.
The anoctamin family consists of 10 members, all of which are predicted to have 8 transmembrane domains with cytosolic N and C termini. Ano1 is regulated by both intracellular Ca2+ and by voltage, in a similar fashion to native CaCCs (17), but the mechanisms are not well defined (18). The homology between family members suggests that other Anos may also function as CaCCs (19) and Ano2 has also been shown to mediate the Ca2+-activated Cl− current in olfactory epithelium and photoreceptor synapses (4, 5, 20). With the possible exception of Ano8 and Ano10, anoctamins are also predicted to have a re-entrant loop between transmembrane domains 5 and 6 that may contribute toward Cl− selectivity (21, 22).
A common property of ion channels is their oligomeric nature, which may be homo- or hetero-oligomeric (23). For example, activation of the MthKCa2+-activated K+ channel from Methanobacterium thermoautrophicum involves oligomerization of 4 subunits (24) and it has been suggested that the mammalian Maxi-K K+ channel is activated by a similar mechanism (25). Activation of CFTR Cl− channels involves dimerization of nucleotide binding domains (26), although whether CFTR enters the plasma membrane as a monomer or a dimer remains controversial (27, 28). A first step in understanding how Ano1 is gated and regulated requires knowing whether the channel exists as a monomer or an oligomer and, if it oligomerizes, the number of interacting subunits. Because little is known about how Ano1 forms a Cl− conducting channel in the apical membrane, we have used biochemical techniques and Förster resonance energy transfer (FRET)-based approaches to investigate its subunit assembly.
EXPERIMENTAL PROCEDURES
Cell Culture
Human excess donor lungs and excised recipient lungs were obtained at the time of lung transplantation from portions of the main stem or lumbar bronchi and cells were harvested by enzymatic digestion as previously described under a protocol approved by the University of North Carolina School of Medicine IRB. Human bronchial epithelia cells were transfected with Ano1 constructs in suspension using the Amaxa system (Lonza) as per the manufacturer's instructions and were cultured overnight on 30-mm glass coverslips before imaging. HEK293 cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum and 1× penicillin/streptomycin solution. HEK293 cells were typically used 2–3 days after seeding on 30-mm glass coverslips. Cultures that were ∼75% confluent were transfected for 4–6 h using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions and allowed to incubate in 5% CO2 at 37 °C overnight before use.
Ano1 Constructs and RT-PCR
Mouse Ano1 C-terminal fused with enhanced GFP (Ano1-eGFP) was kindly provided by Dr. Uhtaek Oh (Seoul National University, Korea; variant containing a and c alternative splice sequences, UniProtKB accession number Q8BHY3.2). Ano1-eGFP is in the pEGFP vector (Clontech). The Ano1-mCherry construct was made by replacing eGFP with monomeric Cherry (mCherry, Clontech). In both constructs, the fluorescent tag was separated from the C terminus of Ano1 by a 17-amino acid linker (RILQSTVPRARDPPVAT) to ensure that the fluorescent protein did not affect Ano1 function as previously described (29). The HA-tagged P2Y2 receptor construct was donated by Kendall Harden (Department of Pharmacology, UNC, Chapel Hill, NC) (30). Endogenous expression of all Ano family members (Ano1–10) was determined by RT-PCR using primers as listed in Table 1.
TABLE 1.
Primers used in RT-PCR to detect endogenous anoctamin expression in HEK293 cells
| Gene | Sense | Antisense |
|---|---|---|
| Ano1 | GCGTCCACATCATCAACATC | ATCCTCGTGGTAGTCCATCG |
| Ano2 | TGCCTACCACTACCGGAAAC | ACTTCTTTGCAATGCTGCCT |
| Ano3 | AAACCTGAACCACATCAGCC | TCTTCCCAAAAAGAAAGCGA |
| Ano4 | CATGGGAAGTCCTTGGAAGA | GCCATTGGTAAGCAAACGAT |
| Ano5 | ACACTTCACCAGAATTGGGC | GAAGCTGCTGCTGTTCCTCT |
| Ano6 | CAGTTTGGGTTCGTCACCTT | AGTACGGGTTTCCCTTGCTT |
| Ano7 | CTACTCCTGCCGGTTCAGAG | GTTCCTGCGTGGGTATGTCT |
| Ano8 | ACTTCGCTCTGCTCCTCAAG | CTTCATGACGTTGTTGGGTG |
| Ano9 | TGGAGATCAGCACCTGTGAG | CGAAGTTCACGATTCGGATT |
| Ano10 | GAGGTGCCCAGTTGTTGTTT | CCGAGTGTACCAGGTGTCCT |
Patch Clamping
Whole cell patch clamp recording was performed with an Axopatch 200B amplifier as previously reported (31). The voltage clamp protocol consisted of 750 ms duration steps from a 0-mV holding potential to various potentials between −100 and +100 mV in 20-mV increments. Fluorescent cells were chosen for recording. The pipette solution contained (in mm): 146 CsCl, 2 MgCl2, 5 EGTA, 0 or 5.0143 CaCl2 to make free [Ca2+] of 0 or 1 μm, 10 sucrose, and 8 HEPES, pH 7.3, adjusted with N-methyl-d-glucamine. The standard external solution contained (in mm): 140 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 10 HEPES, pH 7.3.
Acceptor Photobleaching FRET
FRET was performed using a Leica SP5 confocal microscope with a ×63 glycerol immersion objective. The donor (eGFP) was excited at 488 nm and emission collected from 495 to 549 nm, and the acceptor (mCherry) was excited at 561 nm and emission collected from 580 to 654 nm. FRET was measured by the acceptor photobleaching method and analyzed using ImageJ (NIH Freeware). The FRET efficiency (%E) was calculated as: ((donorpostbleach − donorprebleach)/donorpostbleach) × 100.
Measurements of Ca2+i
All cultures were loaded with Fura-2-AM (5 μm at 37 °C for 40 min) and imaged with a 60 × 1.2 NA H2O objective on a Nikon Ti-S microscope. Ca2+i measurements were obtained using an Orca camera (Hamamatsu) and Fura-2 fluorescence was acquired alternately at 340 and 380 nm (emission >450 nm) using Ludl filter wheels with Compix sPCI software. At each excitation wavelength (340 or 380 nm), background light levels were measured by exposing cells to digitonin (15 μm) and MnCl2 (10 mm) and subtracting the emission from the corresponding signal measured in Fura-2-loaded cells before taking the ratio (340/380) (32).
Imaging of the Actin Cytoskeleton
For labeling of actin, HEK293 cells were grown on glass coverslips. Cells were incubated with 1 μm cytochalasin D or vehicle (0.1% DMSO) for 30 min and then fixed in 4% paraformaldehyde for 10 min at room temperature. After washing 3 times in PBS, cells were permeabilized by a 10-min exposure to 1% Triton X-100. After washing 3 times, cells were incubated with Alexa 488-phalloidin (Invitrogen) for 30 min, followed by a deep red HCS cell mask (Invitrogen) for 30 min as per the manufacturer's instructions. Fixed cells were then imaged immediately using the Leica SP5 confocal microscope.
Co-immunoprecipitation and Western Blotting
Co-immunoprecipitation was performed in HEK293 cells transfected with Ano1-mCherry and Ano1-eGFP, adenosine-2B receptor-eGFP (A2BR-eGFP), or eGFP. Cells were lysed in 600 μl of Nonidet P-40 lysis buffer (0.09% Nonidet P-40, 50 mm Tris-HCl, pH 7.4, 10 mm NaMoO4, 150 mm NaCl) supplemented with 1× complete EDTA-free protease inhibitors (Roche). Lysates were then centrifuged at 16,000 × g for 10 min at 4 °C and the supernatants were collected. Protein concentrations were determined by the BCA protein assay kit (Pierce) and equal amounts of protein (800 μg) were diluted to 1.6 μg μl−1 with Nonidet P-40 lysis buffer in spin columns (Pierce) and mixed with anti-mCherry monoclonal antibody (Clontech) and rotated at 4 °C for 2 h. Protein A/G-agarose beads (30 μl, Pierce) were added to lysate and rotated at 4 °C for 4 h. Beads were washed three times with Nonidet P-40 lysis buffer by centrifugation at 1500 × g for 2 min at 4 °C and protein was eluted by boiling in 2× concentrated sample buffer (1% SDS, 5% glycerol, 25 mm Tris-HCl, pH 6.8, 0.01% bromphenol blue, 0.5% 2-mercaptoethanol). Protein was resolved on a 3–8% gradient NuPAGE Tris acetate gel (Invitrogen) and transferred to nitrocellulose using iBlot (Invitrogen). Immunoblots were incubated overnight at 4 °C with either anti-mCherry monoclonal antibody (Clontech, 1:1000) or rabbit polyclonal anti-eGFP antibody (Abcam, 1:1000) in 2% fish gelatin blocking buffer. Proteins were detected using SuperSignal West Pico Chemiluminescent Substrate kit (Pierce) or using goat anit-rabbit IRDye 800CW antibody (1:10,000) in 2% fish gelatin blocking buffer and analyzed with the Odyssey IR imaging system (LI-COR Biosciences).
Chemical Cross-linking
Chemical cross-linking was performed as described (33). Ano1-eGFP-transfected HEK293 cells were lysed in cross-linking buffer (PBS, pH 8). Protein was incubated with varying concentrations of the cross-linker 1,5-difluoro-2,4-dinotrobenzene (Pierce) (water-insoluble, membrane permeable, spacer arm length, 3 Å) at 37 °C for 10 min. The cross-linking reaction was stopped by the addition of 20 mm Tris, pH 7.5, and incubated at room temperature for 20 min. Samples were mixed with 4× NuPAGE LDS sample buffer containing 50 mm dithiothreitol (DTT) and incubated at 70 °C for 10 min. Protein was resolved on a 3–8% NuPAGE Tris acetate gels and analyzed by Western blotting.
Native PAGE
Cells were sonicated in 600 μl of native PAGE 4× sample buffer containing 2% n-dodecyl β-d-maltoside (Invitrogen). Lysate was centrifuged at 16,000 × g for 10 min at 4 °C. Supernatant was collected and 10 μg of protein was treated with varying concentrations of SDS and treated with 0.5% G-250 (Invitrogen). Protein was resolved on a Native PAGE Novex 4–16% BisTris gel and detected by Western blotting.
Electromobility Shift Assay
Whole cell HEK293 lysate expressing Ano1-eGFP and Ano1-mCherry was obtained for electromobility shift assay (EMSA) analysis. Whole cell lysate containing 7.5 μg of protein was prebound with anti-mCherry at 4 °C for 1 h. Prebound lysate was then subjected to non-denaturing PAGE. Protein shift was resolved by Western blotting with anti-GFP.
Statistical Methods
All data are presented as the mean ± S.E. for n number of cells. Each transfection was repeated on at least three separate days. Differences between means were tested for statistical significance using paired or unpaired t tests or their non-parametric equivalent as appropriate to the experiment. From such comparisons, differences yielding p ≤ 0.05 were judged to be significant.
RESULTS
Ano Expression and Verification of Ano1 Construct Function
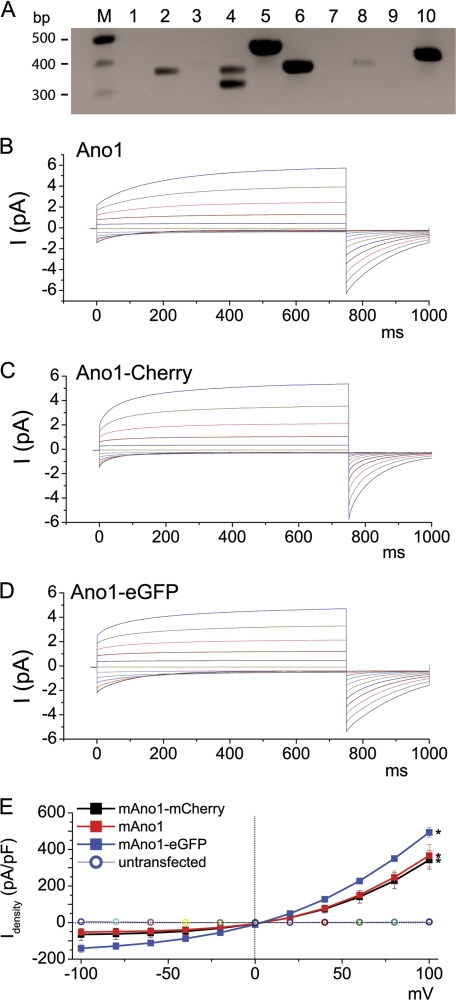
We have previously shown that HEK293 cells do not endogenously express a Ca2+-activated Cl− current (34). However, to determine that these cells did not endogenously express Ano1, we performed RT-PCR (Fig. 1A). Consistent with the lack of an endogenous CaCC (see also Fig. 1E), these cells did not express Ano1. However, Ano2, -4, -5, -6, -8, and -10 were endogenously expressed, suggesting that these additional Ano family members are not capable of forming a CaCC in these cells.
FIGURE 1.
Ca2+-activated Cl− currents induced by fluorescent protein-tagged mouse Ano1 constructs are identical to wild type Ano1. HEK293 cells were transiently transfected and subjected 1 day later to whole cell patch clamp recording. A, total RNA was collected from untransfected HEK293 cells and RT-PCR was performed to determine which Ano proteins are endogenously expressed. Ano5, -6, and -10 were strongly expressed, whereas Ano2 and -4 were mildly expressed. Ano1, -3, -7, -8, and -9 were not detectable. B, wild type mouse Ano1 (variant containing a and c alternative splice sequences, accession Q8BHY3.2). C, Ano1 tagged with eGFP on the C terminus. D, Ano1 tagged with mCherry on the C terminus. E, steady-state current-voltage relationships (red square, wild type; black square, mCherry-tagged Ano1; blue square, eGFP-tagged Ano1; circle, untransfected). All n = 10. N.B., in some cases, error bars were obscured by the symbols. * denotes p < 0.05 different to basal currents.
To probe the quaternary structure of Ano1, we fused eGFP and mCherry to the C termini of Ano1. We then verified that these FRET constructs could still be activated by intracellular Ca2+ to conduct Cl−. With nominally zero intracellular Ca2+, both tagged constructs and the untagged wild type Ano1 failed to display significant currents (<0.05 nA; Fig. 1E). However, in the presence of 1 μm free intracellular Ca2+, whole cell currents were typically >5 nA for all Ano1s (Fig. 1, B–E). Importantly, the currents from both constructs exhibited outward rectification and time dependence that was typical of CaCC currents at this Ca2+ concentration and was identical to untagged Ano1 (Fig. 1, B–E) (35). These results demonstrate that attaching eGFP or mCherry had no obvious effect on Ano1 function.
Ano1-eGFP and Ano1-mCherry Undergo FRET in the Plasma Membrane of HEK293 Cells and Airway Epithelia
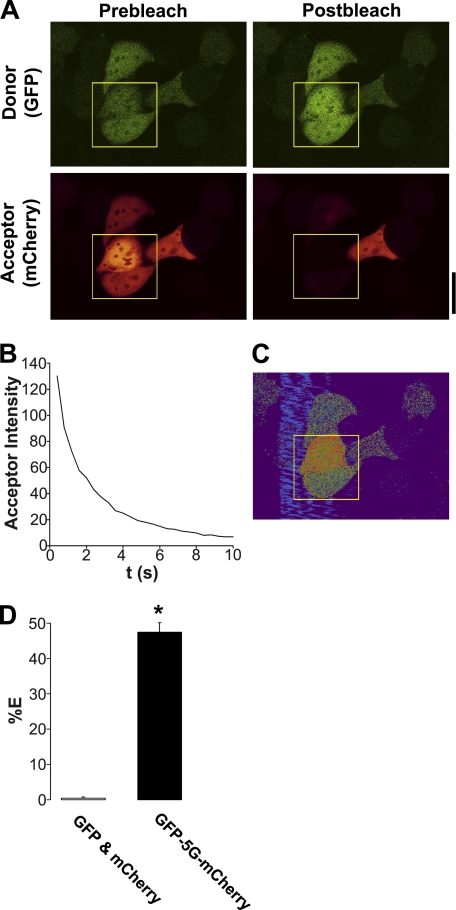
We first used FRET to search for Ano1-Ano1 interactions because this method allows for live cell quantification. As a positive FRET control, we directly linked eGFP to mCherry by five glycines (in the absence of Ano1) and measured the % FRET with this construct in HEK293 cells using the acceptor photobleaching approach (36). Typical acceptor photobleaching of this FRET pair is shown in Fig. 2, A and B. We were able to measure 48 ± 1% FRET with this construct (Fig. 2, C and D), which is close to the maximal FRET that is measurable between eGFP and mCherry (50%) (37), suggesting that we can accurately measure FRET with our system. In contrast, unlinked eGFP and mCherry did not undergo FRET when co-expressed in HEK cells (Fig. 2D).
FIGURE 2.
eGFP linked to mCherry with 5 glycines (eGFP-5G-mCherry) undergoes near-maximal FRET. A, typical confocal micrographs of eGFP-5G-mCherry (green, donor; red, acceptor) before and after photobleaching of the acceptor fluorophore. B, graph showing typical acceptor photobleaching over 10 s. C, resultant FRET image from A. D, mean FRET efficiency (%E) of unlinked eGFP and mCherry cotransfected in the same cells (n = 10) and from eGFP-5G-mCherry (n = 14). * denotes p < 0.05 different between linked and unlinked eGFP and mCherry. Scale bar represents 25 μm.
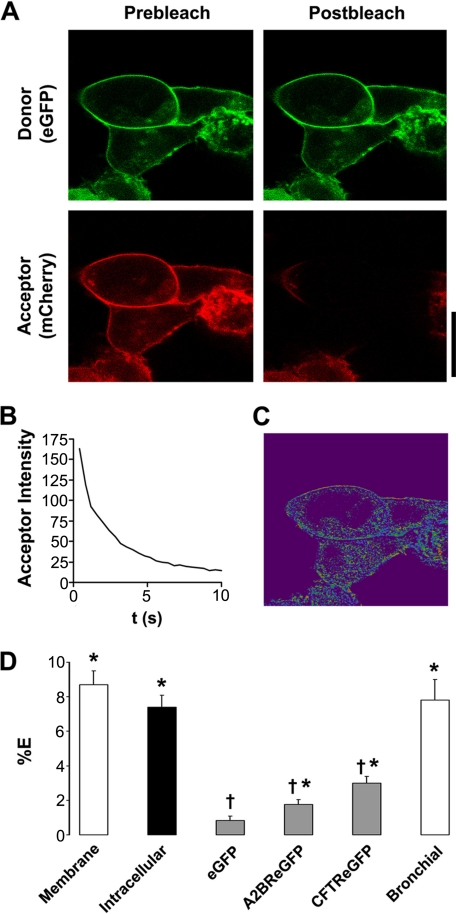
24 h after transfection in HEK293 cells, Ano1 was predominantly expressed in the plasma membrane (Fig. 3A) and after acceptor photobleaching (Fig. 3B) we detected a significant 8% of FRET (Fig. 3, C and D), suggesting that Ano1 subunits were close enough to interact. To determine whether Ano1 channels associate before they reach the plasma membrane, we measured Ano1 FRET ∼15 h post-transfection when more Ano1 is located intracellularly. Intracellular Ano1-eGFP − Ano1-mCherry FRET was also ∼8% (Fig. 3D), suggesting that Ano1 oligomers assemble before reaching the plasma membrane.
FIGURE 3.
Ano1-eGFP and Ano1-mCherry undergo specific FRET in the plasma membrane. A, typical confocal micrographs of Ano1-eGFP (green, donor) and Ano1-mCherry (red, acceptor) before and after photobleaching of Ano1-mCherry. B, graph showing typical acceptor photobleaching over 10 s. C, resultant FRET image calculated from A. D, mean FRET efficiency (%E). Open bars, plasma membrane FRET in HEK293 cells (n = 56) and human bronchial epithelial cells (n = 13). Closed bar, intracellular FRET in HEK293 cells (n = 36). Gray bars, FRET between Ano1-mCherry and the denoted acceptor molecule. eGFP, Ano1-mCherry (n = 19); A2BR-eGFP, Ano1-mCherry (n = 19); CFTR-eGFP, Ano1-mCherry (n = 22). * denotes FRET that is p < 0.05 different from 0. † denotes FRET that is p < 0.05 different from plasma membrane Ano1 FRET in HEK293 cells. Scale bar represents 25 μm.
We failed to detect any FRET between eGFP alone and Ano1-mCherry, suggesting that the Ano1 FRET was due to the specific assembly of Ano1 subunits. To test for the possibility that the plasma membrane of a HEK293 cell may become crowded following transient transfections, forcing an intimateness that would not normally occur, we co-expressed Ano1-mCherry and eGFP-tagged A2B adenosine receptor (A2BR-eGFP), a G-protein-coupled receptor that resides in the plasma membrane but has not previously been shown to interact with Ano1. In this case, there was a small, but significant, amount of FRET (2%), which was significantly less than the 8% FRET seen between Ano1 subunits (Fig. 3D). Coexpressed Ano1-mCherry and eGFP-tagged CFTR also returned moderate FRET (∼3.5%) that was also significantly less than the 8% FRET seen between Ano1 subunits (Fig. 3D). Together, these low levels of FRET are perhaps indicative of the restricted two-dimensional diffusion that limits distances between two proteins that do not physically interact in the plasma membrane.
To demonstrate that Ano1-Ano1 FRET was not influenced by the expression system used, we co-expressed the Ano1 constructs in single primary human bronchial epithelial cells. We again observed close to 8% FRET in these cells, suggesting that the Ano1 interaction is an intrinsic property of Ano1, rather than a discreteness forced by the HEK293 expression system (Fig. 3D).
Ano1 FRET Is Due to a Protein-Protein Interaction
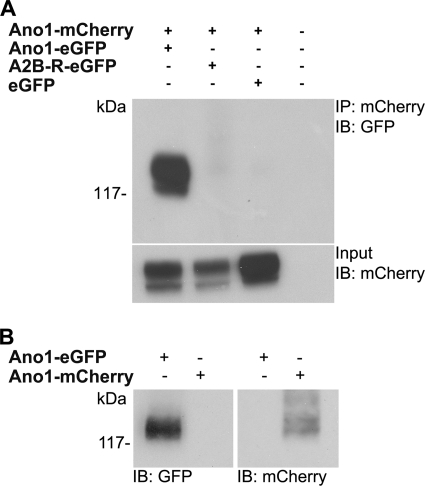
To confirm that FRET between Ano1-eGFP and Ano1-mCherry was due to a physical interaction, we co-immunoprecipitated Ano1-mCherry and Ano1-eGFP using antibodies specifically raised against eGFP and mCherry (Fig. 4A). As a negative control, we immunoprecipitated Ano1-mCherry from HEK293 cells co-expressing A2BR-eGFP or eGFP and probed with an eGFP antibody (Fig. 4A). No bands were detected after blotting for eGFP in control lanes, indicating that these proteins do not interact with Ano1. Western blotting confirmed that both antibodies are specific for eGFP/mCherry, respectively, and neither detected the other fluorescent protein (Fig. 4B).
FIGURE 4.
Ano1-eGFP and Ano1-mCherry co-immunoprecipitate. A, HEK293 cells were transfected with Lipofectamine alone, or with Ano1-mCherry and Ano1-eGFP, A2B-R-eGFP, and eGFP. Input represents 5% of total protein used in co-immunoprecipitation. B, Western blot indicating specificity of eGFP and mCherry antibodies. HEK293 cells were transfected with either Ano1-eGFP or Ano1-mCherry. The eGFP antibody only detected Ano1-eGFP (left), whereas mCherry antibody only detected Ano1-mCherry (right). The blot is representative of three separate experiments. IB, immunoblot.
Ano1 FRET Is Not Altered by Changes in Intracellular Ca2+ or Actin Cytoskeleton Disruption
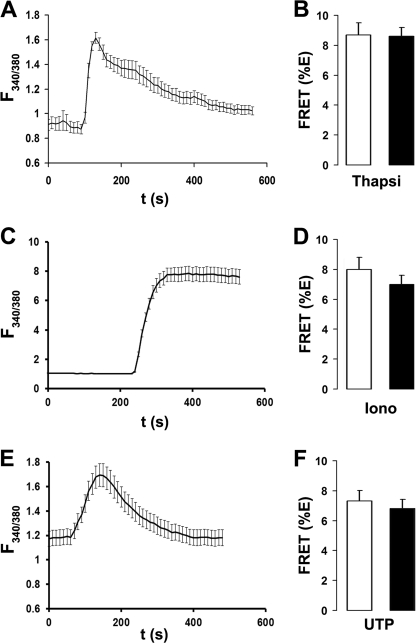
We next tested whether changes in intracellular Ca2+ could alter Ano1 FRET. Intracellular Ca2+ was raised with 2 μm thapsigargin (17), 100 μm UTP (32), or 10 μm ionomycin (38). Cells treated with UTP were also transfected with an HA-tagged P2Y2 receptor construct (P2Y2R-HA) to ensure a consistent UTP response. HEK293 cells displayed a robust increase in the Fura-2 emission ratio after all three treatments (Fig. 5, A, C, and E). However, these maneuvers failed to affect Ano1 FRET (Fig. 5, B, D, and F), suggesting that Ano1-Ano1 interactions are not influenced by changes in intracellular Ca2+.
FIGURE 5.
Ano1-eGFP and Ano1-mCherry FRET are not affected by changes in intracellular Ca2+. A, typical Fura-2 emission ratio observed following exposure to 2 μm thapsigargin (n = 10). B, mean FRET (%E) from Ano1 in the plasma membrane of HEK293 cells after vehicle (open bars, n = 25) and following a 5-min exposure to 2 μm thapsigargin (closed bars, n = 27). C, typical Fura-2 emission ratio observed following exposure to 1 μm ionomycin (n = 10). D, mean FRET (%E) from Ano1 in the plasma membrane of HEK293 cells after vehicle (open bars, n = 20) and following a 5-min exposure to 1 μm ionomycin (closed bars, n = 21). E, typical Fura-2 emission ratio observed following exposure to 10 μm UTP (n = 10). F, mean FRET (%E) from Ano1 in the plasma membrane of HEK293 cells after vehicle (open bars, n = 20) and following a 5-min exposure to 10 μm UTP (closed bars, n = 20).
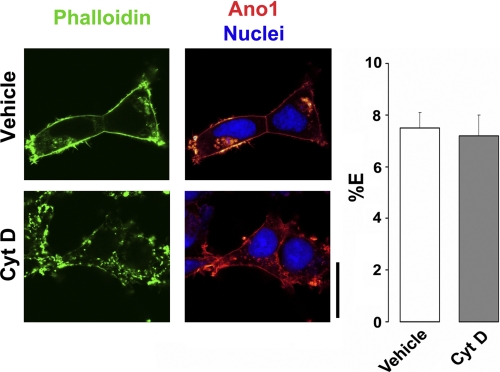
Epithelial ion channels have been shown to be linked to the cytoskeletal infrastructure by postsynaptic density 95 discs largezonula occludens-1 (PDZ) binding motifs and these interactions can be ablated by disrupting F-actin polymerization with cytochalasin D (39). However, a 30-min treatment with 1 μm cytochalasin D had no effect on plasma membrane Ano1 FRET, suggesting that oligomerization of Ano1 is not contingent on binding to the actin cytoskeleton (Fig. 6).
FIGURE 6.
Ano1-eGFP and Ano1-mCherry FRET are not dependent on an intact actin cytoskeleton. Left, confocal micrographs of Ano1-mCherry in HEK293 cells (red) before and after disruption of actin cytoskeleton (phalloidin/green) with 1 μm cytochalasin D for 30 min. Right, mean % E in HEK293 cells after vehicle (open bars, n = 36) and after 30 min cytochalasin D (Cyt D) exposure (gray bars, n = 49). Scale bar represents 25 μm.
Biochemical Evidence That Ano1 Is a Dimer
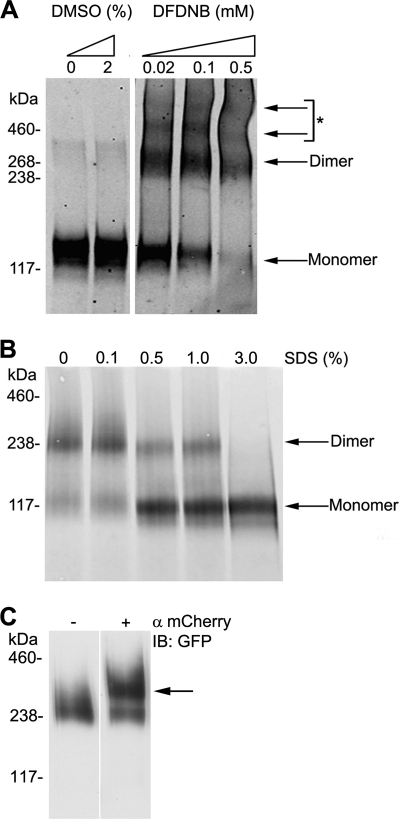
To investigate the stoichiometry of Ano1, we performed chemical cross-linking experiments on cell lysates obtained from HEK293 cells expressing Ano1-eGFP (Fig. 7A). Under control (i.e. reducing) conditions, Ano1-eGFP was seen predominantly as a ∼130 kDa monomer. However, after treating whole cell lysate with the cross-linker 1,5-difluoro-2,4-dinotrobenzene, an additional band was observed that corresponded to an Ano1-eGFP dimer (∼260 kDa). Two faint higher order bands were also visible. However, the molecular weight of these bands was harder to ascertain and may represent nonspecific cross-linking of Ano1 to other membrane proteins.
FIGURE 7.
Determination of Ano1 subunit stoichiometry by chemical cross-linking and non-denaturing gel electrophoresis. A, determination of possible oligomeric structures of Ano1-eGFP using chemical cross-linking. Cell lysates were treated with increasing concentrations of 1,5-difluoro-2,4-dinotrobenzene, membrane permeable (DFDNB). *, denotes possible higher order complexes or adducts. B, determination of oligomeric structure of Ano1-eGFP under non-denaturing (native) PAGE conditions. Cell lysates were incubated with 0–3% of SDS to denature protein complexes and probed. Under non-denaturing conditions, Ano1 is found mainly as a dimer. C, EMSA analysis from HEK293 cell lysate expressing Ano1-eGFP and Ano1-mCherry, showing a mobility shift elicited by anti-mCherry (arrow). IB, immunoblot.
We next performed non-denaturing PAGE on lysate obtained from HEK293 cells to validate the cross-linking experiments (Fig. 7B). Cells were lysed by 2% n-dodecyl β-d-maltoside and sonicated in the absence of SDS. The majority of Ano1 appeared as a dimer with no presence of higher order bands. Inclusion of SDS induced the emergence of a ∼117 kDa band indicating that Ano1 had been fully denatured and that we were observing the monomeric protein. This band was slightly smaller than the predicted size of Ano1-eGFP (∼130 kDa), but is consistent with an apparent reduction in protein size seen under non-reducing conditions (40). To ensure that we were observing Ano1-eGFP dimers and not Ano1-eGFP in association with an unidentified endogenous protein, we performed an EMSA by exposing lysate from HEK293 cells expressing Ano1-eGFP and Ano1-mCherry to an mCherry antibody before non-denaturing PAGE (Fig. 7C). We then blotted for eGFP to test whether Ano1-eGFP was shifted to a higher molecular weight by antibody pre-binding. A portion of Ano1-eGFP remained at 230 kDa, which likely represented a homodimer population of Ano1-eGFP alone. Importantly, we observed a distinct shift of Ano1-eGFP to a higher molecular weight, which corresponded to a complex containing Ano1-eGFP, Ano1-mCherry, and the mCherry antibody.
DISCUSSION
We examined the stoichiometry of Ano1 using biophysical and biochemical approaches. We used HEK293 cells because they do not endogenously express Ano1 (Fig. 1A) and do not have a spontaneously active CaCC, even with 1 μm Ca2+ in the patch pipette (Fig. 1E). For our studies, we C-terminally labeled murine Ano1 with either eGFP or mCherry. To ensure that these fluorescent proteins did not interfere with Ano1 function, we included a 17-amino acid linker between Ano1 and eGFP or mCherry. Importantly, these fluorescent proteins had no effect on Ano1-mediated Cl− currents, suggesting that their use in this endeavor was valid (Fig. 1).
We chose eGFP/mCherry over the more commonly used CFP/YFP FRET pair because CFP photobleaches easily, which reduces its ability to donate photons during FRET, whereas eGFP is significantly more photostable (41). This approach was confirmed using a FRET-positive construct where the acceptor was linked to the donor by 5 glycines (Fig. 2). The theoretical maximum FRET that can occur between two fluorescent proteins (including CFP, eGFP, mCherry, and YFP) is 50%, which is limited by the size of the fluorescent proteins themselves (37). Using CFP linked to YFP, we measured 38 ± 3% FRET (n = 6). In contrast, we measured 48 ± 1% FRET (n = 14) with eGFP linked to mCherry, confirming previous reports that the latter FRET pair is significantly more suited for confocal microscopy (Fig. 2) (41).
Using acceptor-photobleaching FRET, we found a remarkably constant 8% FRET between Ano1-eGFP − Ano1-mCherry (Fig. 3), which suggested that Ano1 can oligomerize. Co-immunoprecipitation experiments revealed that Ano1-eGFP and Ano1-mCherry are associated. In contrast, Ano1-mCherry did not co-immunoprecipitate with A2BR-eGFP, suggesting that the observed 2% FRET between these proteins was due to the moderate proximity in the plasma membrane, and not a direct physical interaction. Many other apical membrane ion channels are known to oligomerize. For example, ENaC is a heteromultimer that returns between 5 and 15% FRET between different ENaC subunits (42). ENaC is also known to interact with CFTR (43) and FRET was 7% between CFTR and all three ENaC subunits (43). A study by Kerschensteiner et al. (44) used acceptor-photobleaching FRET to determine the stoichiometry of voltage-gated K+ (Kv) channels that are known to be heteromeric oligomers. They obtained ∼25% FRET efficiency when Kv2.1-CFP and Kv9.3-YFP were expressed compared with a ∼10% FRET efficiency when Kv2.1-YFP and Kv9.3-CFP were expressed. Thus, the 8% FRET that we record between Ano1 subunits is comparable with both FRET seen between ion channel subunits and between distinct ion channels. Perhaps, surprisingly, whereas CFTR has previously been shown to negatively regulate CaCC (45, 46), we detected only minimal FRET between CFTR and Ano1 (Fig. 3), suggesting that regulation of Ano1 by CFTR is not due to a direct interaction. Because we found Ano1-Ano1 FRET to be 8% when measured intracellularly (Fig. 3), it is likely that Ano1 subunits come together en route to the plasma membrane. It should be noted that we did not differentiate between intracellular compartments during our analysis and subunits may oligomerize as early as the ER or as late as endosomes.
CFTR indirectly binds to the actin cytoskeleton via its C-terminal PDZ-binding motif and disruption of F-actin polymerization with cytochalasin D affects CFTR activity (47, 48). Performing a computational search with the Eukaryotic Linear Motif resource, we found that murine Ano1 also has a predicted C-terminal PDZ-binding motif (GDAL), which may serve to link Ano1 to the actin cytoskeleton through intermediary scaffolding proteins. However, the ability of Ano1 to undergo FRET was not affected by cytochalasin D exposure, suggesting that Ano1 subunit interactions are not cytoskeleton-dependent (Fig. 6). However, the effect of cytochalasin D on trafficking remains to be determined. We also found Ano1 FRET to be insensitive to intracellular Ca2+ levels (Fig. 5), suggesting that Ano1 oligomerization is fixed and independent of the gating ability of Ano1, which is Ca2+-dependent (11–13).
Chemical cross-linking and non-denaturing PAGE assays indicated that Ano1 is a dimer (Fig. 7). A distinct dimer band was observed following chemical cross-linking (Fig. 7A), which was not observed under control conditions. Additional higher order complexes were also observed after cross-linking. However, these may be proteins that are non-specifically cross-linked to Ano1-eGFP. Following non-denaturing PAGE a strong dimer band was also observed (Fig. 7B). This dimerization was only disrupted with 3% SDS (Fig. 7B). Higher order oligomers were not detected by non-denaturing PAGE, suggesting those observed following cross-linking are not true oligomers. We next performed EMSA to determine whether the dimer band consists of two Ano1 proteins or Ano1 with an unidentified endogenous protein (Fig. 7C). This approach revealed a shift of Ano1-eGFP to a higher molecular weight when prebound with an anti-mCherry antibody, confirming that Ano1-eGFP was present as a dimer with Ano1-mCherry and not with any other protein. Because only Ano1 is present under non-denaturing conditions, it is likely that the co-immunoprecipitation is indicative of a direct interaction between Ano1 subunits (Fig. 4). A portion of Ano1-eGFP did not shift to a higher molecular weight. This is likely due to the fact that HEK293 cells transfected with Ano1-eGFP and Ano1-mCherry should yield three distinct populations of Ano1 dimers, consisting of Ano1-eGFP/Ano1-eGFP, Ano1-eGFP/Ano1-mCherry, and Ano1-mCherry/Ano1-mCherry.
Although Ano1 has been identified as an essential subunit of CaCC (11–13), transiently expressed Ano1 could form a heteromultimer with other anoctamin family members because many of these proteins are ubiquitously expressed (Fig. 1A). However, our data suggests that this is not the case and that Ano1 specifically forms a homodimer and in conclusion, we have shown that Ano1-eGFP and Ano1-mCherry are functional proteins that form a dimer. Determining the oligomeric structure of CaCC is an important step toward a better understanding of how this recently identified channel functions. Further work is needed to determine how Ano1 subunits come together, whether other proteins facilitate the process, and how this affects activation of CaCC and conductance of Cl−.
Acknowledgments
The mouse Ano1 clone used in these studies was kindly provided by Dr. Uhtaek Oh (Seoul National University, Korea). The technical support of Michael Watson and Nancy Quiney is gratefully acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grants HL084934, HL034322, EY0148852, and GM60448, the Cystic Fibrosis Foundation, and the Children's Healthcare of Atlanta Center for Cystic Fibrosis Research.
- CaCC
- Ca2+-activated Cl− channel
- Ano1
- anoctamin 1
- TMEM16
- transmembrane protein 16
- CF
- cystic fibrosis
- mCherry
- monomeric cherry
- A2BR
- adenosine 2b receptor
- CFTR
- cystic fibrosis transmembrance conductance regulator
- ENaC
- epithelial sodium channel
- eGFP
- enhanced GFP
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Eggermont J. (2004) Proc. Am. Thorac. Soc. 1, 22–27 [DOI] [PubMed] [Google Scholar]
- 2. Hengl T., Kaneko H., Dauner K., Vocke K., Frings S., Möhrlen F. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6052–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rasche S., Toetter B., Adler J., Tschapek A., Doerner J. F., Kurtenbach S., Hatt H., Meyer H., Warscheid B., Neuhaus E. M. (2010) Chem. Senses 35, 239–245 [DOI] [PubMed] [Google Scholar]
- 4. Stephan A. B., Shum E. Y., Hirsh S., Cygnar K. D., Reisert J., Zhao H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11776–11781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stöhr H., Heisig J. B., Benz P. M., Schöberl S., Milenkovic V. M., Strauss O., Aartsen W. M., Wijnholds J., Weber B. H., Schulz H. L. (2009) J. Neurosci. 29, 6809–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Large W. A., Wang Q. (1996) Am. J. Physiol. Cell Physiol. 271, C435–454 [DOI] [PubMed] [Google Scholar]
- 7. Duan D. (2009) J. Physiol. 587, 2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu B., Linley J. E., Du X., Zhang X., Ooi L., Zhang H., Gamper N. (2010) J. Clin. Invest. 120, 1240–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barish M. E. (1983) J. Physiol. 342, 309–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miledi R. (1982) Proc. R. Soc. Lond. B. Biol. Sci. 215, 491–497 [DOI] [PubMed] [Google Scholar]
- 11. Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L. J. (2008) Science 322, 590–594 [DOI] [PubMed] [Google Scholar]
- 12. Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y. (2008) Cell 134, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Y. D., Cho H., Koo J. Y., Tak M. H., Cho Y., Shim W. S., Park S. P., Lee J., Lee B., Kim B. M., Raouf R., Shin Y. K., Oh U. (2008) Nature 455, 1210–1215 [DOI] [PubMed] [Google Scholar]
- 14. Huang F., Rock J. R., Harfe B. D., Cheng T., Huang X., Jan Y. N., Jan L. Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21413–21418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rock J. R., O'Neal W. K., Gabriel S. E., Randell S. H., Harfe B. D., Boucher R. C., Grubb B. R. (2009) J. Biol. Chem. 284, 14875–14880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romanenko V. G., Catalán M. A., Brown D. A., Putzier I., Hartzell H. C., Marmorstein A. D., Gonzalez-Begne M., Rock J. R., Harfe B. D., Melvin J. E. (2010) J. Biol. Chem. 285, 12990–13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lytton J., Westlin M., Hanley M. R. (1991) J. Biol. Chem. 266, 17067–17071 [PubMed] [Google Scholar]
- 18. Zhu M. H., Kim T. W., Ro S., Yan W., Ward S. M., Koh S. D., Sanders K. M. (2009) J. Physiol. 587, 4905–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schreiber R., Uliyakina I., Kongsuphol P., Warth R., Mirza M., Martins J. R., Kunzelmann K. (2010) J. Biol. Chem. 285, 7838–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pifferi S., Dibattista M., Menini A. (2009) Pflugers Arch. 458, 1023–1038 [DOI] [PubMed] [Google Scholar]
- 21. Das S., Hahn Y., Walker D. A., Nagata S., Willingham M. C., Peehl D. M., Bera T. K., Lee B., Pastan I. (2008) Cancer Res. 68, 6306–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duran C., Thompson C. H., Xiao Q., Hartzell H. C. (2010) Annu. Rev. Physiol. 72, 95–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwappach B. (2008) Mol. Membr. Biol. 25, 270–278 [DOI] [PubMed] [Google Scholar]
- 24. Kuo M. M., Baker K. A., Wong L., Choe S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2151–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cui J., Yang H., Lee U. S. (2009) Cell Mol. Life Sci. 66, 852–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hwang T. C., Sheppard D. N. (2009) J. Physiol. 587, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J. H., Chang X. B., Aleksandrov A. A., Riordan J. R. (2002) J. Membr. Biol. 188, 55–71 [DOI] [PubMed] [Google Scholar]
- 28. Ramjeesingh M., Kidd J. F., Huan L. J., Wang Y., Bear C. E. (2003) Biochem. J. 374, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weatherman R. V., Chang C. Y., Clegg N. J., Carroll D. C., Day R. N., Baxter J. D., McDonnell D. P., Scanlan T. S., Schaufele F. (2002) Mol. Endocrinol. 16, 487–496 [DOI] [PubMed] [Google Scholar]
- 30. Sromek S. M., Harden T. K. (1998) Mol. Pharmacol. 54, 485–494 [DOI] [PubMed] [Google Scholar]
- 31. Xiao Q., Yu K., Cui Y. Y., Hartzell H. C. (2009) J. Physiol. 587, 4379–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tarran R., Loewen M. E., Paradiso A. M., Olsen J. C., Gray M. A., Argent B. E., Boucher R. C., Gabriel S. E. (2002) J. Gen. Physiol. 120, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Penna A., Demuro A., Yeromin A. V., Zhang S. L., Safrina O., Parker I., Cahalan M. D. (2008) Nature 456, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fischmeister R., Hartzell H. C. (2005) J. Physiol. 562, 477–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuruma A., Hartzell H. C. (2000) J. Gen. Physiol. 115, 59–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kenworthy A. K. (2001) Methods 24, 289–296 [DOI] [PubMed] [Google Scholar]
- 37. Albertazzi L., Arosio D., Marchetti L., Ricci F., Beltram F. (2009) Photochem. Photobiol. 85, 287–297 [DOI] [PubMed] [Google Scholar]
- 38. Ribeiro C. M., McKay R. R., Hosoki E., Bird G. S., Putney J. W., Jr. (2000) Cell Calcium 27, 175–185 [DOI] [PubMed] [Google Scholar]
- 39. Lamprecht G., Seidler U. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G766–777 [DOI] [PubMed] [Google Scholar]
- 40. Wittig I., Schägger H. (2008) Proteomics 8, 3974–3990 [DOI] [PubMed] [Google Scholar]
- 41. Tramier M., Zahid M., Mevel J. C., Masse M. J., Coppey-Moisan M. (2006) Microsc. Res. Tech. 69, 933–939 [DOI] [PubMed] [Google Scholar]
- 42. Staruschenko A., Medina J. L., Patel P., Shapiro M. S., Booth R. E., Stockand J. D. (2004) J. Biol. Chem. 279, 27729–27734 [DOI] [PubMed] [Google Scholar]
- 43. Berdiev B. K., Cormet-Boyaka E., Tousson A., Qadri Y. J., Oosterveld-Hut H. M., Hong J. S., Gonzales P. A., Fuller C. M., Sorscher E. J., Lukacs G. L., Benos D. J. (2007) J. Biol. Chem. 282, 36481–36488 [DOI] [PubMed] [Google Scholar]
- 44. Kerschensteiner D., Soto F., Stocker M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6160–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson L. G., Boyles S. E., Wilson J., Boucher R. C. (1995) J. Clin. Invest. 95, 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei L., Vankeerberghen A., Cuppens H., Cassiman J. J., Droogmans G., Nilius B. (2001) Pflugers Arch. 442, 280–285 [DOI] [PubMed] [Google Scholar]
- 47. Fischer H., Illek B., Machen T. E. (1995) J. Physiol. 489, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haggie P. M., Stanton B. A., Verkman A. S. (2004) J. Biol. Chem. 279, 5494–5500 [DOI] [PubMed] [Google Scholar]