Abstract
Activation of Kupffer cells (KCs) by gut-derived lipopolysaccharide (LPS) and Toll-Like Receptors 4 (TLR4)-LPS-mediated increase in TNFα production has a central role in the pathogenesis of alcoholic liver disease. Micro-RNA (miR)-125b, miR-146a, and miR-155 can regulate inflammatory responses to LPS. Here we evaluated the involvement of miRs in alcohol-induced macrophage activation. Chronic alcohol treatment in vitro resulted in a time-dependent increase in miR-155 but not miR-125b or miR-146a levels in RAW 264.7 macrophages. Furthermore, alcohol pretreatment augmented LPS-induced miR-155 expression in macrophages. We found a linear correlation between alcohol-induced increase in miR-155 and TNFα induction. In a mouse model of alcoholic liver disease, we found a significant increase in both miR-155 levels and TNFα production in isolated KCs when compared with pair-fed controls. The mechanistic role of miR-155 in TNFα regulation was indicated by decreased TNFα levels in alcohol-treated macrophages after inhibition of miR-155 and by increased TNFα production after miR-155 overexpression, respectively. We found that miR-155 affected TNFα mRNA stability because miR-155 inhibition decreased whereas miR-155 overexpression increased TNFα mRNA half-life. Using the NF-κB inhibitors, MG-132 or Bay11-7082, we demonstrated that NF-κB activation mediated the up-regulation of miR-155 by alcohol in KCs. In conclusion, our novel data demonstrate that chronic alcohol consumption increases miR-155 in macrophages via NF-κB and the increased miR-155 contributes to alcohol-induced elevation in TNFα production via increased mRNA stability.
Keywords: Alcohol, Macrophage, MicroRNA, NF-kB Transcription Factor, Tumor Necrosis Factor (TNF), Ethanol/Administration and Dosage, Kupffer Cells/Immunology, Macrophages/Immunology, TNFalpha/Physiology
Introduction
MicroRNAs (miRs)3 are small non-coding RNA molecules that regulate the expression of target genes involved in a wide range of biological processes (1, 2). Innate immune responses and inflammation are fine-tuned by miR-125b, miR-146a, and miR-155. Upon lipopolysaccharide (LPS) stimulation, both miR-146a and miR-155 are up-regulated in monocytes and macrophages (3, 4), whereas miR-125b is down-regulated (5). Consistent with this, miR-155 exerts a positive regulation on the release of tumor necrosis factor α (TNFα) through enhancing its translation (5). In contrast, miR-146a acts as a negative regulator and decreases the release of inflammatory mediators, such as interleukin (IL)-1β or IL-8 (4), whereas miR-125b acts as a post-transcriptional repressor of TNFα (5).
In alcoholic liver disease (ALD), TNFα production by the resident liver macrophages, Kupffer cells (KCs), plays a central role in disease pathogenesis (6). Chronic alcohol exposure in vitro and in vivo increases inflammatory cell responses, particularly to LPS stimulation (7, 8). Alcohol-induced sensitization of KCs to gut-derived LPS was shown to contribute to the initiation and progression of ALD (9). KC-derived TNFα has been identified as an important mediator of steatosis, inflammation, and hepatocyte damage in ALD (10, 11). Although the involvement of various signaling pathways such as nuclear factor-κB (NF-κB) and Erk and mRNA stability has been studied in KCs from ALD (8, 12, 13), the role of miRs is unknown in resident liver macrophages. A recent report has described the miR expression profile in a murine model of ALD (14), but the functions and physiological activity of specific miRs and their cell-specific role and expression remain to be elucidated.
Considering the potential role of miRs in LPS-induced TNFα production and the importance of macrophage inflammatory activation in ALD, we hypothesized that miR-155, miR-146a, and/or miR-125b could play a role in the development of alcoholic liver injury. Here we report for the first time that chronic alcohol induces miR-155 in macrophages via NF-κB and that elevated miR-155 results in increased TNFα production by increasing TNFα mRNA stability.
EXPERIMENTAL PROCEDURES
Animal Studies and KCs Isolation
All animals received proper care in agreement with animal protocols approved by the Institutional Animal Use and Care Committee of the University of Massachusetts Medical School. Eight-week-old female mice (C57BL/6) were divided into two groups (15–30 mice/group depending on the experiment). The alcohol-fed group received the Lieber-DeCarli diet (Bio-Serv, Frenchtown, NJ) with 5% (v/v) ethanol (32.4% alcohol-derived calories) for 4 weeks; pair-fed control mice received an equal amount of calories as their alcohol-fed counterparts with the alcohol-derived calories substituted with dextrin maltose. Mice were bled by submandibular venipuncture, and serum was separated from whole blood and frozen at −80 °C. For some mice, livers were fixed in formalin and were further paraffin-embedded, sectioned, and stained with hematoxylin and eosin for microscopic analysis. The rest of the mice received anesthesia with ketamine (100 mg/kg), and KCs were isolated as described previously (15). Briefly, the livers were perfused with saline solution for 10 min followed by in vivo digestion with liberase enzyme for 5 min and in vitro digestion for 30 min. The non-hepatocyte content was separated by Percoll gradient and centrifuged for 60 min at 800 × g. The intercushion fraction was washed and adhered to plastic in Dulbecco's modified Eagle's medium with 5% fetal bovine serum. The non-adherent fraction was washed, and the adherent KC population was adjusted to 2 × 106/ml in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Depending on experimental conditions, cells from 5–10 mice were pooled for each experiment given the limited number of KCs available from each animal. For in vitro stimulation, cells were rested overnight, and on the next day, they were stimulated with 25 mm alcohol or 100 ng/ml LPS or both for 6 h; supernatants were collected for TNFα analysis, and total RNA was isolated from cells for miR-155 expression as indicated in Fig. 4, D–E legend.
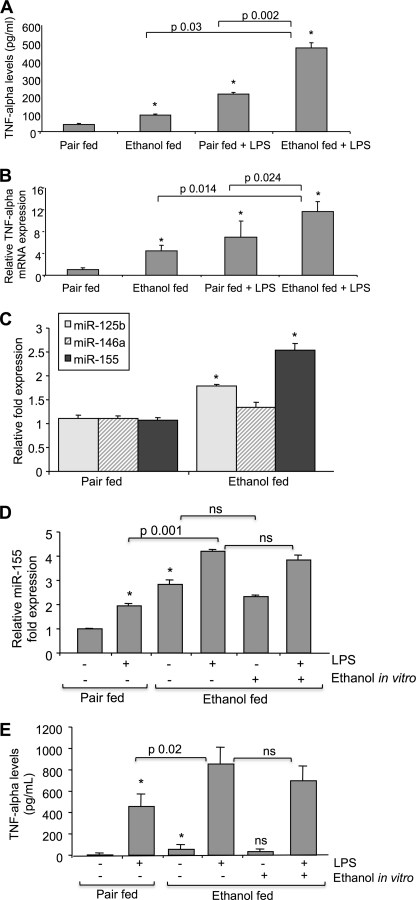
FIGURE 4.
Enhanced miR-155 expression and TNFα in Kupffer cells of chronic alcohol-fed mice. A and B, Kupffer cells isolated from pair-fed or alcohol-fed mice were pooled (n = 5/group) and cultured for 10–12 h followed by stimulation with 0 or 100 ng/ml LPS for 6 h. A, TNFα levels were measured in supernatants by ELISA. Data represent the mean value (S.E. indicated by error bars). B, total RNA was isolated and analyzed for mRNA levels of TNFα using specific primers in real-time PCR. Values of relative TNFα mRNA expression normalized for housekeeping gene 18 S are shown as mean (S.E. indicated by error bars). Statistically significant differences are shown (*, p ≤ 0.05 versus pair-fed control cells). C, Kupffer cells isolated from pair-fed or alcohol-fed mice were pooled (n = 5/group), cultured for 14 h, and harvested. Total RNA was extracted, and expression of miR-125b, miR-146a, and miR-155 was assayed by qPCR. Kupffer cells isolated from pair-fed and alcohol-fed mice were treated or not in vitro with 100 ng/ml LPS, and Kupffer cells isolated from alcohol-fed mice were further stimulated in vitro with 25 mm alcohol or alcohol plus LPS for 6 h. D, total RNA was isolated and analyzed for miR-155 expression. E, supernatants were analyzed for TNFα levels by ELISA. Data were normalized to sno202 control, and the -fold increase in the expression of these miRs in Kupffer cells from alcohol-fed mice versus Kupffer cells from pair-fed mice is shown. Data represent the mean value (S.E. indicated by error bars). Statistically significant differences are shown (*, p ≤ 0.05 versus Kupffer cells from pair-fed mice). ns, non-statistically significant.
Biochemical Assays
Serum alanine aminotransferase activity was determined using a kinetic method (D-Tek LLC, Bensalem, PA), serum endotoxin levels were measured using the limulus amebocyte lysate assay (Lonza, MD), and serum alcohol levels were determined using an alcohol analyzer (Analox Instruments).
Cell Culture and Reagents
RAW 264.7 macrophages were purchased from the American Type Culture Collection and maintained in Dulbecco's modified medium (Invitrogen) containing 10% FBS (HyClone, South Logan, UT) at 37 °C in a 5% CO2 atmosphere. For prolonged alcohol exposure, cells exposed to 50 mm alcohol were placed in a C.B.S. Scientific incubation culture chamber with twice the alcohol concentration in the bottom of the chamber to saturate the chamber and maintain a stable alcohol concentration, as described previously (8). Actinomycin D, MG-132, Bay11-7082, and LPS (Escherichia coli strain 0111:B4) were purchased from Sigma-Aldrich. RAW 264.7 macrophages were stimulated with E. coli-derived LPS (100 ng/ml), 50 mm alcohol, or the combination of LPS and alcohol at the times indicated in Figs. 1, A and B, 2, A–D, 5, A–E, 6, A–D, and 7, A–C legends.
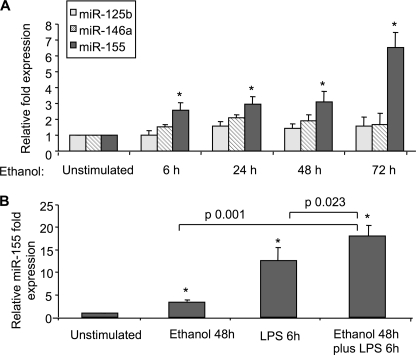
FIGURE 1.
Enhanced miR-155 expression in RAW 264.7 macrophages after LPS and/or alcohol treatment. A, RAW 264.7 macrophages were stimulated with 50 mm alcohol for the indicated time points. B, RAW 264.7 macrophages were stimulated with 50 mm alcohol for 48 h, with LPS for 6 h, or with LPS for 6 h after 48 h of alcohol pretreatment. Expression of miR-125b, miR-146a, and miR-155 was assayed by qPCR, and data were normalized to sno202 control. The -fold increase in the expression of these miRNAs versus non-stimulated cells is shown. Data represent the mean value (S.E. indicated by error bars) of at least three independent experiments. Statistically significant differences are shown (* indicates p ≤ 0.05 versus unstimulated cells).
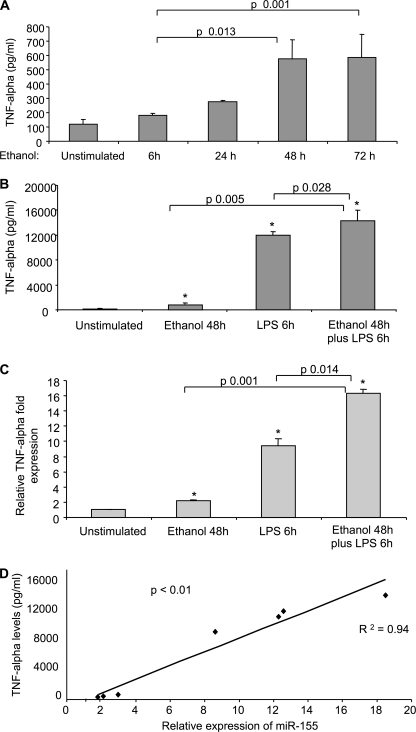
FIGURE 2.
TNFα production is increased in RAW 264.7 macrophages after LPS and/or alcohol treatment and correlates with miR-155 expression. A, RAW 264.7 macrophages were stimulated with 50 mm alcohol for the indicated time points, and TNFα levels were measured in supernatants by ELISA. B and C, RAW 264.7 macrophages were stimulated with 50 mm alcohol for 48 h or LPS for 6 h or with LPS for 6 h after 48 h of alcohol pretreatment. TNFα levels were measured in supernatants by ELISA, and TNFα mRNA was quantified using specific primers in real-time PCR. Data represent the mean value (S.E. indicated by error bars) of at least three independent experiments. (* indicates p ≤ 0.05 versus unstimulated cells). Statistically significant differences are shown. D, the correlation between miR-155 expression and TNFα production in RAW 264.7 macrophages under different conditions (50 mm alcohol for 6, 24, and 48 h and 100 ng/ml LPS for 6 h with or without alcohol pretreatment) is shown (R2 = 0.94, p < 0.01). Expression of miR-155 was assayed by qPCR, and data were normalized to sno202 control. TNFα levels were measured in supernatants by ELISA after collection of the medium in the same samples. Each dot represents the average of at least three independent experiments.
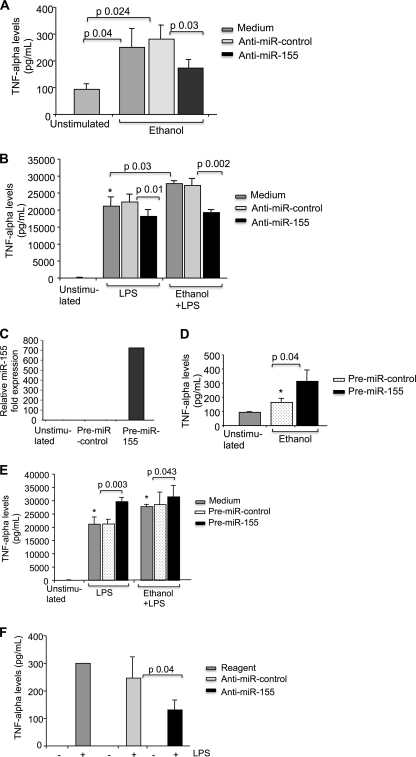
FIGURE 5.
miR-155 regulates TNFα production. A–E, RAW 264.7 macrophages were transfected with anti-miR-control or anti-miR-155 (A and B) with pre-miR-control or pre-miR-155 (C–E), exposed to 50 mm alcohol for 48 h (A, B, D, and E), and further stimulated or not with 100 ng/ml LPS for 6 h as indicated. Culture medium was collected, and supernatants were analyzed for TNFα production by ELISA. Mean values of TNFα (S.E. indicated by error bars) from three independent experiments are shown. C, mature miR-155 expression was assayed by qPCR and normalized to sno202 control after transfection with pre-miR-control or pre-miR-155. Data from two experiments (mean ± S.E.) are shown. F, Kupffer cells from alcohol-fed mice were transfected with anti-miR-control or anti-miR-155 using an Amaxa transfection kit, medium was changed after 4–6 h of transfection and stimulated on the next day or not with 100 ng/ml LPS for 6 h, and culture supernatant was collected and analyzed for TNFα production by ELISA. Mean values of TNFα from three independent experiments are shown. Statistically significant differences are shown (*, p < 0.05).
FIGURE 6.
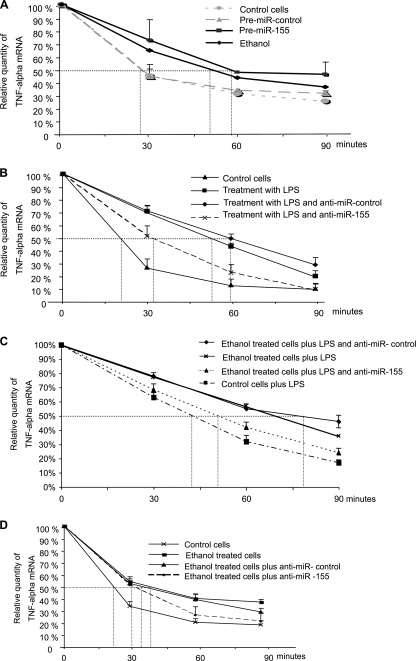
miR-155 increases TNFα secretion by means of affecting TNFα mRNA stability. A–D, RAW 264.7 macrophages were transfected with pre-miR-control or pre-miR-155 and treated or not with alcohol for 48 h (A) or transfected with anti-miR-control or anti-miR-155, treated with LPS or alcohol plus LPS or alcohol alone as indicated (B–D), and further cultured in the presence of 5 μg/ml actinomycin D (A–D). Total RNA was isolated at the times shown, and TNFα mRNA was quantified using specific primers in real-time PCR. Data were normalized with the housekeeping gene 18 S, and half-life is indicated as the percentage of remaining TNFα at different time points showing one experiment out of three with similar results or as absolute numbers (mean ± S.E. (error bars)) from three experiments.
FIGURE 7.
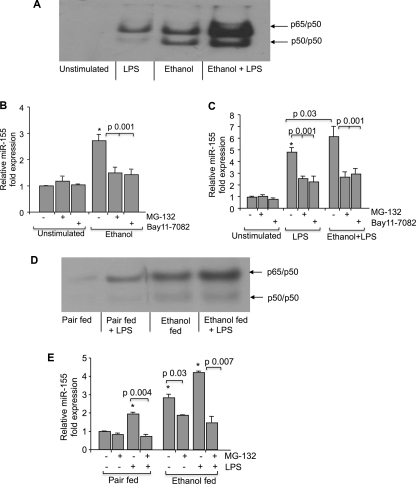
Treatment with NF-κB inhibitors Bay 11-7082 or MG-132 prevented miR-155 increase in response to alcohol. A, RAW 264.7 macrophages were treated with alcohol for 48 h and treated or not with LPS for 30 min, and nuclear proteins were subjected to EMSA. B and C, for NF-κB inhibition, RAW 264.7 macrophages were pretreated with Bay11-7082 (0.1 μm) or MG-132 (0.25 μm) or DMSO as a negative control for 30 min and then exposed or not to alcohol (50 mm) for 48 h and further treated or not with LPS for 6 h. Expression of miR-155 was assayed by qPCR, and data were normalized to sno202 control. The -fold increase in the expression of miR-155 versus non-stimulated cells is shown. Data represent the mean value (S.E. indicated by error bars) of at least three independent experiments. D, Kupffer cells isolated from pair-fed or alcohol-fed animals (n = 8) were pooled and treated or not with 100 ng/ml LPS for 30 min, and 10 μg of whole cell lysate were subjected to EMSA to evaluate NF-κB activation. E, for NF-κB inhibition, Kupffer cells from pair-fed or alcohol-fed animals (n = 10) were pretreated with MG-132 (2.5 μm) or DMSO for 30 min and further stimulated or not with 100 ng/ml LPS for 6 h. Expression of miR-155 was assayed by qPCR, data were normalized to sno202 control, and the -fold increase in the expression of miR-155 in Kupffer cells from alcohol-fed mice versus Kupffer cells from pair-fed mice is shown. Data represent the mean value (S.E. indicated by error bars). Statistically significant differences are shown (*, p < 0.05).
Transfection
For inhibition of miR-155, RAW 264.7 macrophages were transfected with anti-miR-155 and anti-miR-negative control 1 (anti-miR-control), and for overexpression, pre-miR-155 and pre-miR-negative control 1 (pre-miR-control) were used to transfect the cells using siPORT NeoFx transfection agent. All these reagents were purchased from Ambion Inc. (Austin, TX). Knockdown efficiency was determined by transfecting the cells with GAPDH siRNA (Ambion), and overexpression efficiency was checked by determining mature miR-155 levels in transfected cells. Transfected cells were treated with and without 50 mm alcohol for 48 h and, where indicated, stimulated with LPS (100 ng/ml) and treated in the presence or absence of actinomycin D according to experimental requirements before the isolation of RNA (RNeasy kit, Qiagen) or supernatant collection. Amaxa mouse macrophage Nucleofector kit was used to transfect KCs according to manufacturer instructions using the Y-001 program (Lonza). Medium was changed after 4–6 h of transfection, and on the next day, KCs were stimulated with LPS as indicated in Fig. 5F legend.
RNA Analysis
Total RNA was isolated using the mirVanaTM miRNA isolation kit (Ambion). The quality of RNA was routinely checked by measurement of optical density (260/280 and 260/230 ratio) and gel electrophoresis. Quantitative RT-PCR analyses for miR-125b, miR-146a, miR-155, and sno202, used as normalizing control, were performed using TaqMan miRNA assays with reagents, primers, and probes obtained from Ambion. In brief, a stem loop primer was used for reverse transcription (30 min, 16 °C; 30 min, 42 °C; 5 min 85 °C) followed by qPCR employing TaqMan probes and primers in an Eppendorf Realplex Mastercycler (Eppendorf, Westbury, NY). For TNFα and 18 S mRNA expression, RNA was cDNA transcribed with the reverse transcription system (Promega Corp., Madison, WI). Real-time quantitative polymerase chain reaction was performed using the iCycler (Bio-Rad Laboratories), as described (16). The primer sequences were as follows: 18 S, forward, 5′-GTA ACC CGT TGA ACC CCA TT-3′, and reverse, 5′-CCA TCC AAT CGG TAG TAG CG-3′; TNFα, forward, 5′-CAC CAC CAT CAA GGA CTC AA-3′, and reverse, 5′-AGG CAA CCT GAC CAC TCT CC-3′. Relative expression was calculated using the comparative threshold cycle (Ct) method.
Electrophoretic Mobility Shift Assay (EMSA)
To examine NF-κB activation, RAW 264.7 macrophages were exposed to 50 mm alcohol for 48 h and treated or not with 100 ng/ml LPS for 30 min, and nuclear proteins were isolated. KCs from pair-fed or alcohol-fed mice were isolated, rested overnight, and treated or not with LPS (100 ng/ml) for 30 min, and whole cell lysate was isolated given the limited number of cells. Five μg of nuclear protein from RAW 264.7 macrophages or 10 μg of whole cell lysate from KCs were labeled with NF-κB probe as described (17).
NF-κB Inhibition
To inhibit NF-κB activity, RAW 264.7 macrophages were first treated with Bay11-7082 or MG-132 for 30 min prior to alcohol or LPS stimulation, and then cells were exposed or not to 50 mm alcohol for 48 h as indicated in the Fig. 7, B–C legend. For KCs, cells were rested overnight and then treated with MG-132 for 30 min prior to any in vitro stimulation as described in Fig. 7E legend. Total RNA was extracted with the mirVana miRNA isolation kit and analyzed for miR-155 expression as described earlier.
ELISA
The amount of TNFα in cell-free supernatants was determined by ELISA according to the manufacturer's instructions (BD Biosciences).
Statistical Analysis
Data are presented as mean ± S.E., and groups were compared by means of Student's t test or Mann-Whitney U test according to data distribution. Correlation was assessed by means of Spearman's rho test. p < 0.05 was regarded as significant.
RESULTS
Increase of miR-155 Expression Correlates with Increased TNFα Production in Macrophages after Alcohol and/or LPS Stimulation
Prolonged alcohol exposure leads to increased inflammatory cell responses, particularly up-regulation of LPS-induced TNFα production in macrophages and KCs (8, 18). Recent studies demonstrated that TNFα is regulated by miR-125b (5), miR-146a (19), and miR-155 (5). Thus, we hypothesized that alcohol may affect TNFα production via regulation of miRs. First, we studied RAW 264.7 cells that represent a surrogate model of KCs with respect to alcohol-induced TNFα production (20). Alcohol treatment at a 50 mm dose, which corresponds to a 0.2 g/dl blood alcohol level found in chronic alcoholics, resulted in significant up-regulation of miR-155 (Fig. 1A). Alcohol increased miR-155 within 6–72 h with the highest induction after prolonged alcohol exposure for 72 h (Fig. 1A). We identified that the alcohol-induced increase was specific to miR-155 as there were no significant changes in miR-146a or miR-125b levels (Fig. 1A). This observation led us to further study miR-155 in alcohol-treated macrophages.
The Toll-like receptor 4 (TLR4) ligand, LPS, has been implicated in the pathogenesis of ALD (15), and it has been proposed that alcohol sensitizes liver macrophages to LPS-induced TNFα production. Although alcohol or LPS treatment alone resulted in a significant increase in miR-155 expression in RAW macrophages, alcohol pretreatment augmented the LPS-induced increase in miR-155 levels (Fig. 1B). These data suggest that miR-155 could be involved in alcohol-induced changes in macrophage activation.
Because miR-155 is involved in LPS-induced TNFα regulation (5) and TNFα is increased in ALD (18), next we confirmed enhanced TNFα production in chronic alcohol-treated macrophages. Prolonged alcohol treatment alone resulted in a time-dependent induction of TNFα in RAW cells (Fig. 2A), and LPS alone also stimulated TNFα production (Fig. 2, B and C). More important, prolonged alcohol pretreatment augmented LPS-induced TNFα protein production (Fig. 2B) and mRNA expression (Fig. 2C). We identified that changes in TNFα production and miR-155 expression were parallel in macrophages. Indeed, a significant positive correlation (R2 = 0.94) existed between TNFα levels and miR-155 expression after alcohol and/or LPS stimulation (Fig. 2D).
miR-155 Is Up-regulated in Vivo in KCs after Chronic Alcohol Feeding
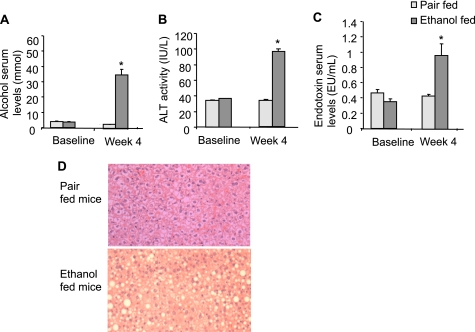
KCs, the resident macrophages in the liver, are the main source of TNFα in this organ (21), and activation of KCs plays a key role in ALD (18). Therefore, we next evaluated whether KCs in vivo would have increased miR-155 levels in ALD. Chronic alcohol feeding in mice resulted in increased serum alcohol levels (Fig. 3A) and significant liver damage indicated by increased serum alanine aminotransferase levels when compared with isocaloric diet feeding (Fig. 3B). We found increased serum endotoxin levels in alcohol-fed mice, suggesting a role for LPS (Fig. 3C). Evaluation of liver histology revealed features of ALD including steatosis and inflammatory cell infiltrates in alcohol-fed but not in pair-fed mice (Fig. 3D).
FIGURE 3.
Chronic alcohol feeding induced liver steatohepatitis in mice as well as increased alcohol, alanine aminotransferase, and endotoxin serum levels. A–C, mice (15/group) received the Lieber-DeCarli diet for 4 weeks as described under “Experimental Procedures.” Blood was collected after 4 weeks of feeding, and serum was separated and analyzed for alcohol (A), alanine aminotransferase (ALT) (B), and endotoxin levels (C). Mean values with S.E. (error bars) are shown (n = 10). (* indicates p < 0.05 when compared with pair-fed mice.) EU, endotoxin units. D, representative sections of formalin-fixed, paraffin-embedded livers stained with hematoxylin and eosin of each group are shown.
Given the features of ALD in vivo, next we tested the hypothesis that miR-155 and TNFα were increased in vivo in KCs in ALD. We determined that KCs isolated from alcohol-fed mice showed increased TNFα production and TNFα mRNA expression when compared with pair-fed controls even without ex vivo stimulation (Fig. 4, A and B). LPS stimulation in KCs from alcohol-fed mice resulted in significantly higher TNFα production at both protein and mRNA levels when compared with KCs from LPS-stimulated pair-fed mice as well as from LPS-naive KCs of alcohol fed mice (Fig. 4, A and B). In addition, we found significantly higher expression of miR-155 in KCs of mice with alcohol feeding when compared with mice with isocaloric control diet (Fig. 4C). Interestingly, the level of miR-125b but not miR-146a was also increased after alcohol feeding in KCs (Fig. 4C). In vitro LPS stimulation amplified alcohol-induced miR-155 expression in KCs isolated from alcohol-fed mice when compared with pair-fed mice (Fig. 4D). In contrast, in vitro alcohol exposure showed no significant effect on miR-155 level in KCs from alcohol-fed mice with or without in vitro LPS stimulation (Fig. 4D). A similar pattern was seen on TNFα level because in vitro alcohol exposure showed no significant effect on TNFα production in KCs from alcohol-fed mice treated or not with LPS in vitro (Fig. 4E). These data suggested that miR-155 up-regulation occurs in vivo in KCs after chronic alcohol intake in the liver.
miR-155 Regulates TNFα Production
The correlation between increased miR-155 expression and TNFα levels after alcohol treatment prompted us to evaluate whether a causative relationship existed between the alcohol-induced increases in miR-155 and TNFα levels. First, transfection with a miR-155-specific inhibitor reduced alcohol-induced TNFα production (Fig. 5A). More important, inhibition of miR-155 not only reduced LPS-induced TNFα production but also decreased TNFα production in response to alcohol plus LPS (Fig. 5B). These results revealed that up-regulation of TNFα could be prevented by the anti-miR-155 but not by the anti-miR-control in RAW 264.7 macrophages treated with alcohol, LPS, or their combination (Fig. 5, A and B).
Next, we sought to evaluate whether miR-155 overexpression could augment LPS-induced TNFα production because this approach could be expected to mimic up-regulation of TNFα in alcohol-treated cells. Overexpression of miR-155 by transfection of pre-miR-155, but not the pre-miR-control, resulted in increased mature miR-155 levels as measured by real-time PCR for mature miR-155 expression (Fig. 5C). Overexpression of miR-155 increased TNFα production in alcohol-treated cells when compared with cells transfected with pre-miR-control (Fig. 5D). Likewise, increased TNFα production after LPS stimulation was found in cells transfected with pre-miR-155 in both alcohol-naive and alcohol-pretreated cells (Fig. 5E), suggesting that miR-155 could augment the effect of LPS on TNFα production. However, we found no significant increase in TNFα production in cells transfected with pre-mir-155 and treated with alcohol plus LPS when compared with LPS alone-stimulated cells. These data suggest that cells may reach a threshold in miR-155 effect on TNFα after LPS stimulation, where the miR-155 effect is saturated and cannot result in further increase in TNFα production (Fig. 5E).
Next, we transfected KCs from alcohol-fed mice with anti-miR-155 or anti-miR-control using the Amaxa mouse macrophage Nucleofector kit. Inhibition of miR-155 resulted in decreased TNFα production when compared with cells transfected with anti-miR-control (Fig. 5F). These results suggest that miR-155 regulates TNFα production in KCs in response to alcohol.
miR-155 Increases TNFα mRNA Stability
Previous reports indicated that prolonged alcohol treatment increases LPS-induced TNFα mRNA stability both in RAW 264.7 macrophages and in primary rat KCs (22). Thus, we evaluated the hypothesis that miR-155 could affect TNFα production in part by regulating mRNA stability in alcohol-treated cells. First, we found that alcohol treatment alone increased TNFα half-life (Fig. 6A). Second, transfection with pre-miR-155, which is a miR-155 precursor, increased TNFα half-life in alcohol-naive cells when compared with cells treated with pre-miR-control or unstimulated cells (Fig. 6A). Together these results suggested that alcohol might increase TNFα RNA half-life in part via miR-155. We next evaluated TNFα RNA half-life in cells transfected with anti-miR-155 or anti-miR-control and further stimulated with LPS. LPS treatment alone increased TNFα mRNA half-life when compared with unstimulated cells, and half-life was decreased in cells transfected with anti-miR-155 (Fig. 6B). We also found that alcohol pretreatment increased TNFα mRNA stability when compared with LPS stimulation alone (Fig. 6C). However, administration of a miR-155 inhibitor reduced LPS-induced TNFα mRNA half-life (∼55 min) when compared with anti-miR-control in alcohol-pretreated cells (∼80 min) (Fig. 6C), suggesting that alcohol may exert its effects on TNFα via miR-155. The increase in TNFα mRNA half-life induced by alcohol alone was slightly decreased in cells transfected with anti-miR-155 (Fig. 6D).
NF-κB Mediates miR-155 Increase after Alcohol Exposure
Previous studies from our laboratory demonstrated that both chronic alcohol exposure and LPS stimulation activate NF-κB (8, 15), and it was also shown that miR-155 expression is regulated by NF-κB activation (23). In search of mechanisms by which alcohol may regulate miR-155, we tested the hypothesis that NF-κB was involved. Similar to our previous observation, we found that prolonged alcohol treatment resulted in an increase in NF-κB nuclear binding of both p65/p50 and p50/p50 dimers and augmented LPS-induced NF-κB activation (Fig. 7A). Here we show that inhibition of NF-κB by MG-132, a proteasome inhibitor (24), or by Bay 11-7082, an IκB kinase inhibitor (25), decreased miR-155 levels in alcohol, LPS, or alcohol plus LPS-treated macrophages (Fig. 7, B and C), suggesting that alcohol-induced miR-155 expression was mediated through the NF-κB pathway. To verify the in vivo relevance of this finding, we next evaluated the role of NF-κB in KCs of pair-fed and alcohol-fed mice. First, we found increased NF-κB activation, specifically p65/p50 heterodimer binding, in KCs of alcohol-fed mice; this was further enhanced upon in vitro LPS stimulation (Fig. 7D). Moreover, inhibition of NF-κB with MG-132 decreased miR-155 expression in KCs from alcohol- or pair-fed mice treated or not with LPS (Fig. 7E). All these findings in KCs were similar to those observed in RAW 264.7 macrophages (Fig. 7, A–C). Collectively, these data suggested a functional role for NF-κB activation in miR-155 up-regulation by alcohol in KCs.
DISCUSSION
The discovery of miRs has revolutionized our understanding of many biological processes, and these molecules are now recognized to have key roles in health and disease. Among them, miR-155 has been identified as a central mediator of macrophage inflammatory response (3, 26). Here we report for the first time that chronic alcohol treatment increases miR-155 levels in macrophages both in vitro and in vivo. miR-155 levels were increased upon LPS stimulation, and it has been hypothesized that its enhanced expression further augments TNFα secretion (5, 27). Our novel observation demonstrates that chronic alcohol exposure augments TNFα production via miR-155 in liver macrophages. Our results also indicate that miR-155 stabilizes TNFα mRNA after alcohol exposure, thus increasing the release of this inflammatory mediator. We report for the first time that miR-155 overexpression itself increases TNFα mRNA stability in macrophages even in the absence of alcohol. In accordance with our observation, more TNFα transcripts were observed in wild type mice when compared with miR-155−/− mice (28).
TNFα is a major cytokine involved in the pathogenesis of ALD (6, 29), and our data indicate that alcohol regulates its production via targeting miR-155. The expression of TNFα is tightly regulated by both transcriptional and post-transcriptional modifications. Post-transcriptional regulation of TNFα includes mRNA stability, polyadenylation, and translational initiation (30, 31) via targeting AU-rich elements in its 3′-untranslated region (UTR) (32). Proteins such as HuR, TIA-1, etc. that bind to 3′-UTR of TNFα regulate TNFα mRNA stability (33, 34). Given the fact that miRs regulate multiple targets, it is likely that miR-155 may affect one of the proteins that bind to 3′-UTR of TNFα and hence may be able to enhance its production. In fact, chronic alcohol is known to increase TNFα mRNA stability (12) via HuR protein (36) and thus increase production of TNFα. Based on our study showing that miR-155 may enhance TNFα mRNA stability, we propose that this mechanism may explain, at least in part, the effect of miR-155 on alcohol-induced TNFα production. We have to consider the fact, however, that a single miR can target multiple genes within the same pathway and that several genes can be directly or indirectly regulated by the same miR. Therefore, it may also be possible that the phenotype described in our mechanistic study could be the result of cumulative effects of miR-155 on multiple genes involved in TNFα regulation.
In addition to miR-155, which augments TNFα production, miR-146a acts as a negative regulator of TNFα and has a role in endotoxin tolerance (37). We found no change in the expression of miR-146a in alcohol-treated cells; however, there was a slight increase in miR-125b after in vivo alcohol administration in KCs. Given the potential of miR-125b as a post-transcriptional repressor, the minimal up-regulation of miR-125b after alcohol treatment may represent a suboptimal counter-regulatory mechanism in response to the pro-inflammatory effects of significant induction of miR-155 and TNFα (5).
TNFα has been recognized as a central mediator of pathomechanism as well as clinical symptoms in ALD (38). Indeed, TNFα has been a target of therapeutic interventions in ALD based on the clinical observations that serum TNFα levels correlated with the severity of acute alcoholic steatohepatitis and experimental data demonstrating attenuation of alcohol-induced liver damage in mice deficient in the expression of TNFR1 or after administration of antibodies to TNFα (10, 39, 40). However, early human clinical trials with anti-TNFα- or TNF receptor-blocking antibodies failed in acute alcoholic hepatitis due to side effects related to increased infections (41–43). Perhaps the most promising currently used clinical intervention is pentoxyphyllin, a non-selective phosphodiesterase inhibitor that attenuates, but does not fully block, TNFα production (44). Our experiments indicate that miR-155 inhibition achieved attenuation but not full inhibition of TNFα production induced by prolonged alcohol feeding and LPS in KCs. Thus, TNFα inhibition remains to be an attractive therapeutic target, and the applicability of our novel findings on alcohol-induced regulation of miR-155 and TNFα production merits further evaluation.
Chronic alcohol exposure in vitro sensitizes human and murine macrophages to LPS (8, 18) and, likewise, chronic alcohol intake sensitizes KCs to activation by intestinal LPS, amplifying TNFα production and resulting in liver inflammation, a hallmark of ALD (45). The intracellular signaling mechanisms of alcohol-induced macrophage sensitization, however, are still under investigation. Our present work indicates for the first time that miR-155 has a causative role in the pro-inflammatory state induced by alcohol in macrophage and KCs after chronic alcohol feeding. We found a linear correlation between increased miR-155 expression and TNFα production in KCs after chronic alcohol feeding, and given the current role of TNFα in ALD, our data both directly and indirectly suggest an involvement of miR-155 in alcoholic liver injury in vivo. The positive correlation between miR-155 and TNFα is supported by previous reports where mice overexpressing miR-155 in B cells had higher TNFα production after an LPS challenge and mice lacking miR-155 showed decreased production of this inflammatory mediator (28). Tili et al. (5) also found increased production of TNFα in Eμ-miR-155 transgenic mice when challenged with LPS. Another report supporting the concept that miR-155 positively regulates TNFα came from a model of infection with Francisella novicida, where it was shown that miR-155 positively regulates TNFα and IL-6 production in response to Francisella infection (46).
Apart from this, miR-155 promotes T helper 1 versus type 2 differentiation (28, 47), which is consistent with the increase in TNFα and other pro-inflammatory cytokine serum levels found in patients with excessive alcohol intake (48). Liver expression of miR-155 was also up-regulated in a murine model of non-alcoholic steatohepatitis, highlighting its potential role in inflammation-related liver diseases (49).
We also showed that alcohol-induced miR-155 increase involves NF-κB activation. NF-κB plays a crucial role in TNFα production, and its induction in chronic alcohol-treated cells has been shown previously both in liver KCs as well as in isolated blood monocytes from patients with alcoholic hepatitis or after prolonged in vitro treatment of blood monocytes with alcohol (50, 51). We previously reported that chronic alcohol increased LPS-induced NF-κB activation via augmentation of LPS-induced IRAK-1 and IκB kinase activity and degradation of IκBα (8). Although NF-κB directly binds to the TNFα promoter in chronic alcohol-exposed monocytes and macrophages to enhance LPS-induced promoter activity (8), it is likely that NF-κB regulates miR-155 transcription via binding to its promoter. Our data clearly show that NF-κB inhibition diminishes chronic alcohol-induced up-regulation of miR-155, confirming a regulatory role for NF-κB in macrophages. Regulation of miR-155 via an NF-κB-dependent mechanism is documented in several studies (35, 46, 49, 52).
In summary, we have identified miR-155 as a novel regulator of alcohol-induced macrophage/KC activation and inflammatory cytokine induction. Our results support the concept that miRs contribute to the development of alcoholic liver injury and indicate that miR-155 could be a potential therapeutic target in ALD.
Acknowledgments
We thank Jan Petrasek and Odette Taha for help in animal work. Core resources supported by the Diabetes Endocrinology Research Center Grant DK32520 were used for this study.
This work was supported, in whole or in part, by National Institutes of Health Grants AA011576 and AA008577 (to G. S.). This work was also supported by a scholarship from the Alfonso Martin Escudero Foundation, Spain (to M. M.).
- miR
- microRNA
- ALD
- alcoholic liver disease
- KC
- Kupffer cell
- DMSO
- dimethyl sulfoxide
- qPCR
- quantitative PCR.
REFERENCES
- 1. Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. (2008) Nat. Immunol. 9, 839–845 [DOI] [PubMed] [Google Scholar]
- 2. Bi Y., Liu G., Yang R. (2009) J. Cell. Physiol. 218, 467–472 [DOI] [PubMed] [Google Scholar]
- 3. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc Natl. Acad. Sci. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perry M. M., Moschos S. A., Williams A. E., Shepherd N. J., Larner-Svensson H. M., Lindsay M. A. (2008) J. Immunol. 180, 5689–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 6. Thurman R. G. (1998) Am. J. Physiol. 275, G605–G611 [DOI] [PubMed] [Google Scholar]
- 7. Romics L., Jr., Mandrekar P., Kodys K., Velayudham A., Drechsler Y., Dolganiuc A., Szabo G. (2005) Alcohol Clin. Exp. Res. 29, 1018–1026 [DOI] [PubMed] [Google Scholar]
- 8. Mandrekar P., Bala S., Catalano D., Kodys K., Szabo G. (2009) J. Immunol. 183, 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagy L. E. (2003) Exp. Biol. Med. 228, 882–890 [DOI] [PubMed] [Google Scholar]
- 10. Iimuro Y., Gallucci R. M., Luster M. I., Kono H., Thurman R. G. (1997) Hepatology 26, 1530–1537 [DOI] [PubMed] [Google Scholar]
- 11. Uesugi T., Froh M., Arteel G. E., Bradford B. U., Wheeler M. D., Gäbele E., Isayama F., Thurman R. G. (2002) J. Immunol. 168, 2963–2969 [DOI] [PubMed] [Google Scholar]
- 12. Kishore R., McMullen M. R., Nagy L. E. (2001) J. Biol. Chem. 276, 41930–41937 [DOI] [PubMed] [Google Scholar]
- 13. Kishore R., Hill J. R., McMullen M. R., Frenkel J., Nagy L. E. (2002) Am. J. Physiol. Gastrointest. Liver Physiol. 282, G6–G15 [DOI] [PubMed] [Google Scholar]
- 14. Dolganiuc A., Petrasek J., Kodys K., Catalano D., Mandrekar P., Velayudham A., Szabo G. (2009) Alcohol Clin. Exp. Res. 33, 1704–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hritz I., Mandrekar P., Velayudham A., Catalano D., Dolganiuc A., Kodys K., Kurt-Jones E., Szabo G. (2008) Hepatology 48, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romics L., Jr., Kodys K., Dolganiuc A., Graham L., Velayudham A., Mandrekar P., Szabo G. (2004) Hepatology 40, 376–385 [DOI] [PubMed] [Google Scholar]
- 17. Mandrekar P., Jeliazkova V., Catalano D., Szabo G. (2007) J. Immunol. 178, 7686–7693 [DOI] [PubMed] [Google Scholar]
- 18. Mandrekar P., Szabo G. (2009) J. Hepatol. 50, 1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc Natl. Acad. Sci. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szabo G., Mandrekar P. (2009) Alcohol Clin. Exp. Res. 33, 220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong Z., Wei H., Sun R., Tian Z. (2007) Cell. Mol. Immunol 4, 241–252 [PubMed] [Google Scholar]
- 22. Nagy L. E. (2004) Alcohol 33, 229–233 [DOI] [PubMed] [Google Scholar]
- 23. Rai D., Karanti S., Jung I., Dahia P. L., Aguiar R. C. (2008) Cancer Genet. Cytogenet. 181, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qureshi N., Perera P. Y., Shen J., Zhang G., Lenschat A., Splitter G., Morrison D. C., Vogel S. N. (2003) J. Immunol. 171, 1515–1525 [DOI] [PubMed] [Google Scholar]
- 25. Pierce J. W., Schoenleber R., Jesmok G., Best J., Moore S. A., Collins T., Gerritsen M. E. (1997) J. Biol. Chem. 272, 21096–21103 [DOI] [PubMed] [Google Scholar]
- 26. Ruggiero T., Trabucchi M., De Santa F., Zupo S., Harfe B. D., McManus M. T., Rosenfeld M. G., Briata P., Gherzi R. (2009) FASEB J. 23, 2898–2908 [DOI] [PubMed] [Google Scholar]
- 27. Pedersen I. M., Otero D., Kao E., Miletic A. V., Hother C., Ralfkiaer E., Rickert R. C., Gronbaek K., David M. (2009) EMBO Mol. Med 1, 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Science 316, 604–608 [DOI] [PubMed] [Google Scholar]
- 29. Tilg H., Diehl A. M. (2000) N. Engl. J. Med. 343, 1467–1476 [DOI] [PubMed] [Google Scholar]
- 30. Han J., Brown T., Beutler B. (1990) J. Exp. Med. 171, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang E., Ma W. J., Aghajanian C., Spriggs D. R. (1997) Cancer Res. 57, 5426–5433 [PubMed] [Google Scholar]
- 32. Han J., Beutler B. (1990) Eur. Cytokine Netw. 1, 71–75 [PubMed] [Google Scholar]
- 33. Dean J. L., Wait R., Mahtani K. R., Sully G., Clark A. R., Saklatvala J. (2001) Mol. Cell. Biol. 21, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piecyk M., Wax S., Beck A. R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Anderson P. (2000) EMBO J. 19, 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gatto G., Rossi A., Rossi D., Kroening S., Bonatti S., Mallardo M. (2008) Nucleic Acids Res. 36, 6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McMullen M. R., Cocuzzi E., Hatzoglou M., Nagy L. E. (2003) J. Biol. Chem. 278, 38333–38341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nahid M. A., Pauley K. M., Satoh M., Chan E. K. L. (2009) J. Biol. Chem. 284, 34590–34599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McClain C. J., Cohen D. A. (1989) Hepatology 9, 349–351 [DOI] [PubMed] [Google Scholar]
- 39. Gaetke L. M., Oz H. S., Frederich R. C., McClain C. J. (2003) J. Am. Coll. Nutr. 22, 415–420 [DOI] [PubMed] [Google Scholar]
- 40. Yin M., Wheeler M. D., Kono H., Bradford B. U., Gallucci R. M., Luster M. I., Thurman R. G. (1999) Gastroenterology 117, 942–952 [DOI] [PubMed] [Google Scholar]
- 41. Tilg H., Jalan R., Kaser A., Davies N. A., Offner F. A., Hodges S. J., Ludwiczek O., Shawcross D., Zoller H., Alisa A., Mookerjee R. P., Graziadei I., Datz C., Trauner M., Schuppan D., Obrist P., Vogel W., Williams R. (2003) J. Hepatol. 38, 419–425 [DOI] [PubMed] [Google Scholar]
- 42. Menon K. V., Stadheim L., Kamath P. S., Wiesner R. H., Gores G. J., Peine C. J., Shah V. (2004) Am. J. Gastroenterol. 99, 255–260 [DOI] [PubMed] [Google Scholar]
- 43. Naveau S., Chollet-Martin S., Dharancy S., Mathurin P., Jouet P., Piquet M. A., Davion T., Oberti F., Broët P., Emilie D. (2004) Hepatology 39, 1390–1397 [DOI] [PubMed] [Google Scholar]
- 44. Akriviadis E., Botla R., Briggs W., Han S., Reynolds T., Shakil O. (2000) Gastroenterology 119, 1637–1648 [DOI] [PubMed] [Google Scholar]
- 45. Thakur V., McMullen M. R., Pritchard M. T., Nagy L. E. (2007) J. Gastroenterol. Hepatol. 22, Suppl. 1, S53–S56 [DOI] [PubMed] [Google Scholar]
- 46. Cremer T. J., Ravneberg D. H., Clay C. D., Piper-Hunter M. G., Marsh C. B., Elton T. S., Gunn J. S., Amer A., Kanneganti T. D., Schlesinger L. S., Butchar J. P., Tridandapani S. (2009) PLoS One 4, e8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McClain C. J., Barve S., Deaciuc I., Kugelmas M., Hill D. (1999) Semin. Liver Dis. 19, 205–219 [DOI] [PubMed] [Google Scholar]
- 49. Wang B., Majumder S., Nuovo G., Kutay H., Volinia S., Patel T., Schmittgen T. D., Croce C., Ghoshal K., Jacob S. T. (2009) Hepatology 50, 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hill D. B., Barve S., Joshi-Barve S., McClain C. (2000) J. Lab. Clin. Med. 135, 387–395 [DOI] [PubMed] [Google Scholar]
- 51. Hill D. B., Devalaraja R., Joshi-Barve S., Barve S., McClain C. J. (1999) Clin. Biochem. 32, 563–570 [DOI] [PubMed] [Google Scholar]
- 52. Kluiver J., van den Berg A., de Jong D., Blokzijl T., Harms G., Bouwman E., Jacobs S., Poppema S., Kroesen B. J. (2007) Oncogene 26, 3769–3776 [DOI] [PubMed] [Google Scholar]