Abstract
Versican is a hyaluronan-binding, extracellular chondroitin sulfate proteoglycan produced by several tumor types, including malignant melanoma, which exists as four different splice variants. The short V3 isoform contains the G1 and G3 terminal domains of versican that may potentially interact directly or indirectly with the hyaluronan receptor CD44 and the EGFR, respectively. We have previously described that overexpression of V3 in MeWo human melanoma cells markedly reduces tumor cell growth in vitro and in vivo. In this study we have investigated the signaling mechanism of V3 by silencing the expression of CD44 in control and V3-expressing melanoma cells. Suppression of CD44 had the same effects on cell proliferation and cell migration than those provoked by V3 expression, suggesting that V3 acts through a CD44-mediated mechanism. Furthermore, CD44-dependent hyaluronan internalization was blocked by V3 expression and CD44 silencing, leading to an accumulation of this glycosaminoglycan in the pericellular matrix and to changes in cell migration on hyaluronan. Furthermore, ERK1/2 and p38 activation after EGF treatment were decreased in V3-expressing cells suggesting that V3 may also interact with the EGFR through its G3 domain. The existence of a EGFR/ErbB2 receptor complex able to interact with CD44 was identified in MeWo melanoma cells. V3 overexpression resulted in a reduced interaction between EGFR/ErbB2 and CD44 in response to EGF treatment. Our results indicate that the V3 isoform of versican interferes with CD44 and the CD44-EGFR/ErbB2 interaction, altering the signaling pathways, such as ERK1/2 and p38 MAPK, that regulate cell proliferation and migration.
Keywords: Cell Adhesion, Hyaluronate, Proteoglycan Structure, Signal Transduction, Tumor, CD44, ErbB Receptor, Melanoma, Versican
Introduction
Versican belongs to the family of the large chondroitin sulfate proteoglycans located within the extracellular matrix (ECM)5 (1, 2). Overproduction of versican is a common feature of several tumor types and it is usually related to poor prognosis (3–10). Versican has been postulated to contribute to the proliferative, adhesive and migratory state of tumor cells, as well as being an important modulator of tumor cell attachment to the interstitial stromal matrix of the tumor. This wide range of functions has been attributed to its ability to interact with other extracellular matrix and cell surface components through its three structural domains: the N-terminal region (G1 domain) that binds hyaluronan (HA), the central G2 domain that carries the glycosaminoglycan chains (GAG-α and GAG-β) and the C-terminal globular region (G3 domain) that interacts with simple carbohydrates, glycosaminoglycans, and other proteins such as tenascin. This latter domain also contains two epidermal growth factor (EGF)-like repeats (11–14). In mammals, versican appears as four possible spliced variants (12, 15). V0 contains G1, G2 (GAG-α and GAG-β), and G3 domains; V1 contains G1, G2 (GAG-β), and G3 domains; V2 contains G1, G2 (GAG-α), and G3 domain; and V3 lacks any GAG subdomain and only contains G1 and G3 domains. Despite the loss of the GAG chains, the V3 isoform maintains the ability to bind hyaluronan through the G1 domain and to interact with EGF receptors (EGFR) through the EGF-like subdomains of G3.
Binding of versican to the hyaluronic acid in the cell matrix results in the formation of versican-hyaluronan tridimensional complexes allowing the cells to regulate cellular responses through the hyaluronan receptors, such as CD44, in coordination with other molecules (16). Indeed, CD44 has been described to interact with many cell components such as integrins, matrix metalloproteinases (MMPs) and hyaluronidase (11). Furthermore, an interaction between CD44 and the ErbB receptor family has been demonstrated in some tumor cell types and shown to regulate tumor cell behavior (17–22).
We previously described for the first time the expression of the large isoforms of versican, V0 and V1, in malignant melanoma, that contribute to the increased cell proliferation rate and decreased cell adhesion of these tumor cells (23, 24). We have also demonstrated that overexpression of the short V3 isoform in MeWo and SK-mel-1.36-1-5 human melanoma cells reversed this phenotype, because transduced cells showed a decrease in cell proliferation and an increase in cell adhesion on hyaluronan (25). The decrease in cell proliferation correlated with a lower activation of the proliferation-related ERK1/2 pathway in response to EGF (26) and a delay in cell cycle progression (25).
These results raised the possibility that V3 isoform could exert its effects through changes in the pericellular coat by competing with the larger chondroitin sulfate-bearing V0 and V1 isoforms of versican. Despite this possible competition mechanism, a direct effect of V3 isoform independent of the presence of the large isoforms should exist, because the diminished proliferation as well as other altered cell functions could be readily observed in cells that lack any versican isoform.
The aim of this work was to elucidate the mechanism by which V3 isoform is acting in cells lacking the large and proliferative V0/V1 isoforms. We examined the potential role of the G1 domain by silencing the expression of CD44 in MeWo cells, as well as the role of the G3 domain by analyzing the activation of signal transduction pathways triggered by EGF. As conclusion, we propose a model in which V3 can interfere with HA-CD44-EGFR/ErbB2 complexes through its G1 and G3 domains, altering signaling pathways that regulate cell proliferation and migration, and modifying HA catabolism.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against EGFR (ErbB1), ErbB2 and ErbB3 used for immunoprecipitation and immunoblotting were obtained from Santa Cruz Biotechnology (Heidelberg, Germany). Anti-human CD44 (clone 156–3C1G2a) was kindly provided by Dr R Vilella (Hospital Clínic de Barcelona, Spain). The antibodies against phospho-ERK5 (BIOSOURCE), and phospho-JNK and JNK (Cell Signaling Technology, Danvers, MA) were a gift from Dr. N. Gómez and Dr. J. M. Lizcano (Dept. Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, Spain). Anti-phospho-p38 MAPK and anti-p38 MAPK antibodies were obtained from Cell Signaling Technology. Anti-actin antibody was obtained from Santa Cruz Biotechnology (Heidelberg, Germany). A biotinylated hyaluronan-binding protein (bHABP) derived from cartilage (Seikagaku Ltd., Tokyo, Japan) was used for detection of hyaluronan.
Cell Culture
MeWo human melanoma cells expressing the V3 isoform of versican (LV3SN) and the empty control vector (LXSN) were obtained as previously described (25). Cells were grown in a humidified atmosphere at 37 °C with 5% CO2 in DMEM medium supplemented with 10% fetal calf serum, 100 IU/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen). For MAPK analysis, subconfluent MeWo cells were cultured in serum-free medium for 24 h and at this time EGF (25 ng/ml, Sigma) was added, and cells were incubated for different times as stated in the figures.
CD44 siRNA Cloning and Cell Transfection
The following small interfering RNA (siRNA) oligonucleotides were used: 5′-GATCCCCGTATGACACATATTGCTTCTTCAAGAGAGAAGCAATATGTGTCATACTTTTTA-3′ and 5′-AGCTTAAAAAGTATGACACATATTGCTTCTCTCTTGAAGAAGCAATATGTGTCATACGGG-3′. The sequences are based on those described in Tzircotis et al. (27). The oligonucleotides (300 μg/ml) were annealed in buffer M (Roche Diagnostics, Mannheim, Germany) by heating at 95 °C for 4 min and slowly cooling down to room temperature. The resulting double-strand sequence was cloned into the pSUPERIOR.puro plasmid (OligoEngine, Seattle, WA) using its BglII/HindIII site. The siRNA construct was stably transfected into MeWo, MeWo LXSN and MeWo LV3SN cell lines using LipofectamineTM reagent (Invitrogen, Paisley, Renfrewshire, UK). All cell lines were also stably transfected with pSUPERIOR.puro plasmid containing the empty vector as a control, giving rise to MeWo pSp, MeWo LXSN pSp, and MeWo LV3SN pSp cell lines. Because parental MeWo cells behavior was indistinguishable from MeWo LXSN cells, and MeWo siCD44 and MeWo LXSN siCD44 also presented identical characteristics, only results from MeWo LXSN-derived cell lines are presented for clarity.
Cell Proliferation, Adhesion, and Migration Assays
For proliferation assays, 6-well plates were seeded with 5 × 104 cells per well. After 3 days in culture, cells were detached by trypsinization and counted in a Neubauer chamber at the indicated times.
For wound healing assays, cells were grown until confluence was reached. At this point, a scratch was done along the cell monolayer using a pipette tip. Cultures were analyzed with a Nikon Eclipse E800 microscope and pictures taken with an integrated camera system.
For adhesion assays, 96-well plates were coated with 5 mg/ml HA (Sigma) or PBS as a negative control for 16 h on a hood. Wells were washed twice with PBS and blocked with 1% BSA in PBS for 60 min at 37 °C. Cells were then plated at a density of 4 × 104 cells per well and allowed to attach to the plate for 2 h. Non-attaching cells were removed and cells adhering to the plate were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Wells were rinsed twice with distilled water and stained with a 0.1% crystal violet solution for 20 min. The extent of adhesion was determined by treating cells with 0.1 m HCl and measuring the absorbance at 620 nm.
For cell migration assays, TranswellTM chambers (6.5-mm diameter; 8-μm pore size polycarbonate membrane) were used. Membranes were coated with 5 mg/ml hyaluronan for 16 h at room temperature, and washed twice with serum-free DMEM. 5 × 104 cells were counted and placed in the upper chamber with DMEM-1% BSA, whereas the lower chamber was loaded with 0.25 ml DMEM medium supplemented with 10% fetal calf serum. Cells were allowed to migrate overnight at 37 °C. The upper and lower sides of the membrane were washed several times with PBS. Cells that stand at the upper chamber were removed with a cotton swab, and cells of the lower side were stained with 0.1% crystal violet for 15 min. Then cells were washed with water and observed at the microscope.
Protein Extraction
For MAPK analyses and immunoprecipitation assays, cell extracts were prepared in 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, 1 mm NaVO4, 10 mm sodium glycerophosphate, 50 mm NaF, 5 mm sodium pyrophosphate, 50 mm saccharose, 50 mm Tris-HCl pH 7.5, 0.5 mm benzamidine, 0.1% β-mercaptoethanol, 25 mm PMSF. The amount of protein applied to the immunoprecipitation assays or gels was normalized after Bradford quantification. For CD44 analyses and hyaluronidase activity assay, membrane extracts were prepared by lysing the cells in 1% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl pH 7.4, and the amount of protein was normalized after Bradford quantification.
Immunoprecipitation and Western Blot Analysis
For immunoprecipitation assays, 2 μg of anti-ErbB or anti-CD44 antibodies were incubated with 100 μl of protein A-Sepharose beads (Pierce). 100 μg of total protein, obtained as described above, were incubated in parallel with 100 μl of protein A-Sepharose for 1 h at 4 °C. After centrifugation at 3000 rpm for 1 min, the clean cell lysate was incubated with the ErbB antibody-protein A-Sepharose complex overnight at 4 °C. The beads were washed three times with 25 mm Tris-HCl, 150 mm NaCl (pH 7.2), and the immunoprecipitated proteins were eluted by boiling the beads in sample buffer (25 μl of lysis buffer and 5 μl of 6× Laemmli buffer).
For Western blot assays, samples (50 μg of protein) were analyzed by SDS-PAGE in a 10% polyacrylamide gel under reducing conditions. After electrophoresis, proteins were transferred onto Immobilon-P membrane (Millipore Corp., Bedford, MA). The blot was placed in a blocking solution consisting of 5% skim milk in TBS-0.05% Tween-20 and incubated for 1 h at room temperature. The membranes were incubated with the primary antibody at 4 °C overnight, washed, incubated with an HRP-labeled secondary antibody, and visualized by chemiluminescence (ECL Plus System, GE Healthcare, Buckinghamshire, UK).
Immunocytochemistry
105 cells were grown in coverslips for 24 h, rinsed with phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde-2% saccharose for 17 min at room temperature. After rinsing with PBS, nonspecific binding sites were blocked with 1% BSA-0.02% goat serum for 20 min. Cells were then incubated at 4 °C overnight with the primary antibody. Cells were washed two times for 5 min and incubated with the secondary antibody labeled with AlexaFluor®488 (Invitrogen, Eugene, OR) or TRITC. Nuclei were visualized by incubating the cover slips with Hoechst 33342 (0.1 mg/ml in PBS) for 3 min. High-power light microscopic images were digitally captured using a Nikon Eclipse E800 epifluorescence microscope with an integrated camera system.
Hyaluronan Staining
Cells were seeded in coverslips at a density of 1 × 105 cells/coverslip for 20 h at 37 °C. Coverslips were rinsed with PBS and fixed with 3% paraformaldehyde-2% saccharose for 17 min at room temperature. After rinsing with PBS, cells were then incubated overnight at 4 °C with 5 μg/ml of biotinylated hyaluronan-binding protein derived from cartilage (b-HABP, Seikagaku Ltd., Tokyo, Japan). Cells were washed two times for 5 min and incubated with a FITC-streptavidin complex for 1 h at room temperature. Samples were then rinsed with PBS, mounted in Vectashield (Vector Laboratories) and analyzed under a Leica TCS SP5 AOBS confocal microscope.
FITC-HA Internalization Assay
Fluorescein isothiocyanate (FITC)-conjugated HA was prepared as previously described (28). Briefly, a stock of FITC-HA was prepared by dissolving 50 mg of high molecular weight HA (Genzyme, Cambridge, MA) in 40 ml of distilled water followed by 20 ml of DMSO (Sigma). In the dark, 25 mg of fluoresceinamine, isotype I (Sigma), in 0.5 ml of DMSO containing 25 μl of acetaldehyde (Sigma) and 25 μl of cyclohexylisocyanide (Sigma) was added to the HA. The pH of the solution was then adjusted and maintained at 4.5 during a 5-h incubation at room temperature. The FITC-HA was brought to 70% ethanol containing 1.3% (weight/volume) potassium acetate and precipitated 2–3 times by overnight incubation at −20 °C. The final FITC-HA pellets were resuspended and diluted to 5 mg/ml in PBS.
For the internalization assay, the method described by Embry and Knudson (2003) was followed. Briefly, 5 × 105 cells were seeded in cover slips. Sixteen hours after seeding, they were incubated with 2 units/ml of Streptomyces hyalurolyticus hyaluronidase (Sigma) overnight at 37 °C in DMEM containing 10% FCS to remove cell surface-bound HA and to expose all CD44 binding sites. Next, cells were washed with PBS and 100 μg/ml of FITC-HA was added. To visualize the cell surface-bound probe, cells were labeled with the probe for 1 h at 4 °C. In parallel, FITC-HA was added to another set of coverslips for 6 h at 37 °C. A CellMaskTM staining was used to visualize cell membrane (Invitrogen). Cells were washed with PBS, fixed with 1% paraformaldehyde in PBS and visualized using a Leica TCS SP5 AOBS confocal microscope (Servei Microscòpia, UAB, Barcelona). Images were processed with IMARIS software.
Hyaluronan Quantification and Hyaluronidase Activity Assays
To determine the hyaluronan concentration in the culture medium an ELISA-like assay was performed by the method described by Underhill et al. (28) with some modifications. 105 cells were seeded in 9-cm2 plates, and after 24 h the medium was replaced with serum-free DMEM. After 16 h, conditioned media were harvested and digested overnight at 37 °C with a 0.1 mg/ml protease solution (Sigma). After boiling, 130 μl of each media were incubated with 130 μl of a biotinylated hyaluronan-binding protein (b-HABP, Seikagaku, Japan) solution, 5 μg/ml in PBS + 10% calf serum, 5 h at 4 °C. At the same time, a 96-well plate was coated with an HA-BSA solution (200 μg/ml HA + 1% BSA) for 2 h at 4 °C and blocked with a 10% calf serum (CS) solution for 3 h at 4 °C. After discarding the blocking solution, 80 μl of each media were added to the wells in triplicate, and the plate was incubated overnight at 4 °C. Wells were then rinsed with PBS and 80 μl of a 2 μg/ml solution of streptavidin-peroxidase (Sigma) + 10% CS were added to each well. The plate was incubated for 1.5 h at 4 °C. Finally, wells were rinsed with PBS, and 70 μl of a 0.05% AEBT (Sigma) solution in 0.1 m sodium citrate (pH 4.2) + 0.001% hydrogen peroxide were added to the wells. After 20 min of incubation, absorbance of the samples was determined at 405 nm.
For hyaluronidase activity assays, a modification of the ELISA-like assay developed by Stern and Stern (29) was used. 20 μg of protein in hyaluronidase buffer (0.1 sodium formate, 0.15 NaCl, 0.02% BSA, pH 4.0) were added to the HA-BSA-coated 96 wells plate and incubated for 16 h at 37 °C. After this incubation, wells were washed, blocked with a 10% CS solution, and 40 μl of a 1 μg/ml b-HABP solution + 10% CS were added to the wells. The plate was incubated for 16 h at 4 °C. Wells were rinsed with PBS and incubated with the streptavidin-peroxidase and AEBT solutions as previously described.
Statistical Analysis
Statistical significance was determined using the Student's t test. Values of p ≤ 0.05 were considered to be significant.
RESULTS
CD44 Silencing in MeWo LXSN and MeWo LV3SN Human Melanoma Cells
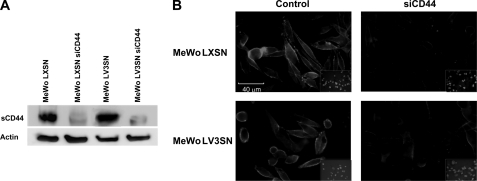
Because the V3 isoform maintains the ability of the other members of the versican family for binding to HA through its G1 domain, and, under the hypothesis that some of the effects of V3 in MeWo melanoma cells could be mediated by CD44, we silenced the expression of CD44 in MeWo LV3SN cells to analyze whether the effects of V3 are maintained when CD44 is knocked down. MeWo LV3SN cells as well as parental MeWo cells and MeWo LXSN cells were transfected with a siRNA sequence against CD44 giving rise to MeWo siCD44, MeWo LXSN siCD44 and MeWo LV3SN siCD44 cell lines. Several clones were isolated after transfection of the cell lines. Efficiency of CD44 inhibition was checked by Western blot and densitometry, and clones with at least 80% silencing were selected in MeWo LXSN as well as in MeWo LV3SN cell lines. Immunocytochemistry confirmed its reduced expression, as illustrated for MeWo siCD44 clone 4 and MeWo LV3SN siCD44 clone 7 (Fig. 1). Expression of V3 did not alter the levels of CD44, as we had previously described (25). The standard form of CD44 (sCD44) is the only CD44 isoform expressed by these cell lines. At least five clones were tested for each cell line. Results presented in this work are those corresponding to clones 4 and 7.
FIGURE 1.
siCD44 transfection critically diminishes CD44 expression in human melanoma MeWo cells. A, immunoblotting of cell membrane extracts from control (MeWo LXSN and MeWo LV3SN) and siRNA-transfected (MeWo LXSN siCD44 and MeWo LV3SN siCD44) cell lines. Membrane extracts were resolved in a 10% SDS-PAGE and blotted with an antibody against CD44 (1:1000). The standard form of CD44 (sCD44) is the only CD44 isoform expressed by these cell lines. B, immunocytochemistry analysis of CD44 in MeWo melanoma cells. 105 cells were seeded onto coverslips, fixed, and incubated with an antibody against sCD44. The signal is given by the incubation of the samples with AlexaFluor®488.
Role of CD44 in V3-induced Effects on MeWo Melanoma Cells
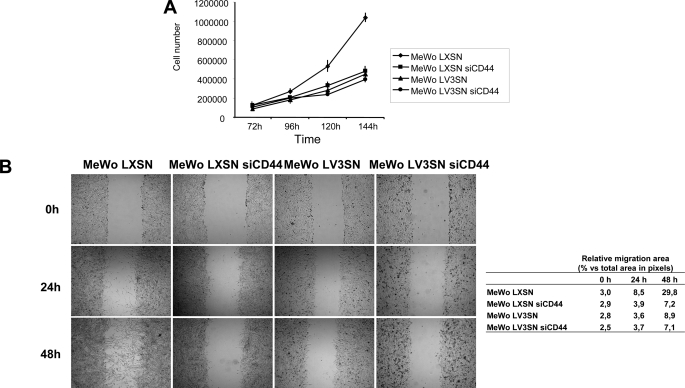
One of the main effects of V3 expression in MeWo cells is a marked reduction in cell proliferation (25). To check whether this effect is mediated through CD44, a proliferation assay was performed in LXSN siCD44 and LV3SN siCD44 cell lines, as well as in their control counterparts. As shown in Fig. 2A, MeWo LV3SN cells presented a delay in cell proliferation in comparison to LXSN cells, as expected. Silencing of CD44 in LXSN cells caused a similar decrease in cell proliferation. Finally, CD44 inhibition in MeWo LV3SN cell line did not cause a further decrease in cell growth, indicating that V3 versican effect on cell proliferation is probably mainly mediated through CD44.
FIGURE 2.
Effect of CD44 silencing in cell proliferation and migration of MeWo melanoma cells expressing or not the V3 isoform of versican. A, CD44 silencing causes a decrease in cell proliferation in MeWo LXSN cells but does not cause an additional decrease in MeWo LV3SN cells. 6-well plates were seeded with 5 × 104 cells per well. After 3 days in culture, cells were detached by trypsinization and counted in a Neubauer chamber at the indicated times. A representative experiment out of four performed is shown. B, CD44 silencing causes a decrease in wound healing ability in MeWo LXSN cells but does not cause an additional decrease in MeWo LV3SN cells. Cells were grown until confluence was reached. At this point, a scratch was done along the cell monolayer using a pipette tip. Cultures were analyzed with a Nikon Eclipse E800 microscope and pictures taken with an integrated camera system. Values are mean ± S.D.
When cell migration was checked in wound healing assays, we observed that MeWo LV3SN cells migrated slower than MeWo LXSN control cells. CD44 silencing also provoked a decrease in cell migration rate (Fig. 2B). Similar to cell proliferation assays, MeWo LV3SN siCD44 cells did not show an additional decrease in cell migration, indicating that the inhibition caused by V3 versican expression is mediated through an interaction with CD44.
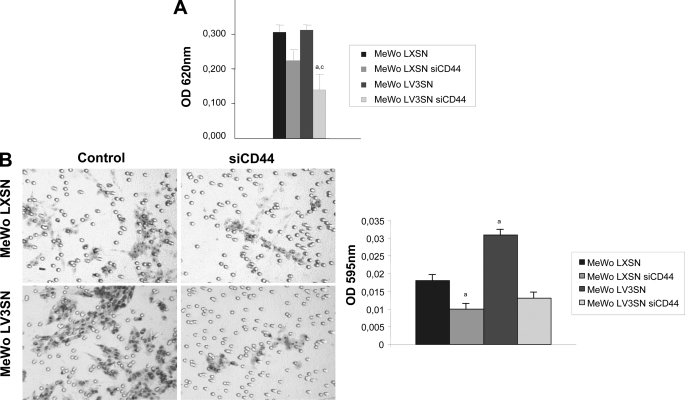
Because CD44 is the main cell membrane receptor for HA, the effect of V3 expression and CD44 silencing on cell adhesion on HA-coated plates and on cell migration on HA-coated Transwells was analyzed. As expected, lack of CD44 decreased the ability of MeWo melanoma cells to adhere to HA and to migrate onto HA-coated surfaces. In contrast, expression of V3 moderately increased the ability of cells to migrate in these surfaces (Fig. 3), in consonance with our previous results (25). No significant differences were found between LXSN siCD44 and MeWo LV3SN siCD44 cells (Fig. 3).
FIGURE 3.
Effect of CD44 silencing in cell adhesion and migration on hyaluronan of MeWo melanoma cells expressing or not the V3 isoform of versican. A, CD44 silencing causes a decrease in cell adhesion onto HA in MeWo LXSN and MeWo LV3SN cells. 96-well plates were coated with 5 mg/ml HA or PBS as a negative control. Cells were plated at a density of 4 × 104 cells per well and allowed to attach to the plate for 16 h. Wells were stained with a 0.1% crystal violet solution for 20 min. The extent of adhesion was determined by treating cells with 0.1 m HCl and measuring the absorbance at 620 nm. A representative experiment out of four performed is shown. Small letters indicate p < 0.05 (a: versus MeWo LXSN; c: versus MeWo LV3SN). B, CD44 silencing causes a decrease in cell migration through HA in MeWo LXSN and MeWo LV3SN cells. Transwell membranes were coated with hyaluronan (5 mg/ml), cells were allowed to migrate overnight at 37 °C, and the lower side of the membranes was stained with crystal violet. A representative experiment out of three performed is shown. Small letters indicate p < 0.05 (a: versus MeWo LXSN). Values are mean ± S.D.
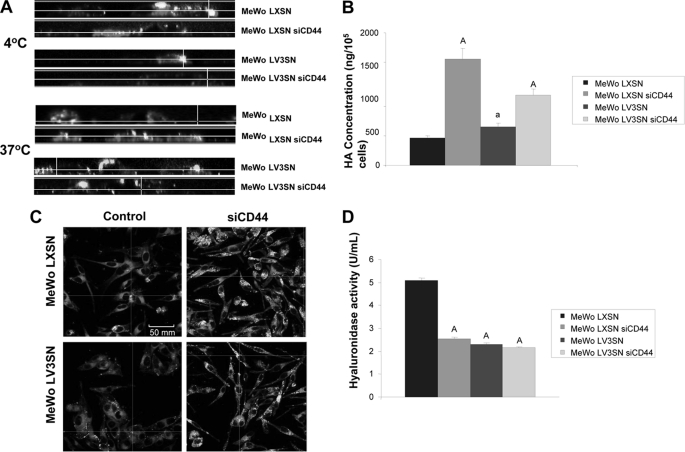
Because HA internalization is widely recognized as a CD44-dependent process (30), we checked the ability of V3 to alter the internalization of an exogenous HA-FITC complex. Cells were seeded on coverslips and treated with hyaluronidase to remove endogenous HA. Then FITC-labeled HA was added to the medium, and cells were incubated for 1h at 4 °C to check the binding to the membrane, or 5h at 37 °C to allow the cells to internalize it. As shown in a cross-section obtained by scanning confocal microscopy, MeWo LXSN siCD44 and MeWo LV3SN cells presented lower amounts of FITC-HA bound to the membrane when compared with MeWo LXSN control cell line, while no binding was detected in MeWo LV3SN siCD44 cells (Fig. 4A). When cells were treated for 5h at 37 °C, FITC-HA was internalized by MeWo LXSN cells, but when V3 versican was expressed and/or CD44 was silenced, the complex remained at the cell surface, indicating that HA internalization had been blocked in these cell lines (Fig. 4A). Furthermore, CD44 silencing causes a clear increase of HA levels in conditioned media from MeWo LXSN siCD44 cells and MeWo LV3SN siCD44 cell line, and less potently, in MeWo LV3SN cells (Fig. 4B). These results were confirmed when cells were incubated with a biotinylated-HABP that allows endogenous HA staining, and analyzed by confocal microscopy (Fig. 4C). CD44 is also required for the full activity of HYAL2 hyaluronidase (31); thus, the effect of V3 expression and/or CD44 silencing on cell membrane hyaluronidase activity was analyzed. In MeWo LXSN siCD44 cells, LV3SN cells and the double transfectants, hyaluronidase activity was reduced by a 50–60% compared with control LXSN cells. (Fig. 4D). These results indicate that hyaluronidase is mainly depending on CD44 in MeWo melanoma cells and suggest that the negative effect of V3 versican on enzyme activity is probably mediated through an interaction of CD44 with HYAL2.
FIGURE 4.
Effect of CD44 silencing on hyaluronan catabolism of MeWo melanoma cells expressing or not the V3 isoform of versican. A, CD44 silencing causes a decrease in cell-surface binding and internalization of a FITC-HA complex in MeWo LXSN and MeWo LV3SN cells. Subconfluent cultures were treated overnight with hyaluronidase and incubated with FITC-HA for 1 h at 4 °C or 6 h at 37 °C. A CellMaskTM staining was used to visualize cell membrane. Cells were fixed and visualized using a Leica TCS SP5 AOBS confocal microscope. Images were processed using IMARIS software. B, CD44 silencing increases HA content in the culture media of MeWo LXSN and MeWo LV3SN cells. Conditioned media from cells cultured in serum-free medium for 24 h were collected and digested with protease solution. HA was determined with an ELISA-like assay. A representative experiment out of three performed is shown. Small letters indicate p < 0.05 and capital letters indicate p < 0.01 (versus MeWo LXSN). C, CD44 silencing increases endogenous expression of hyaluronan in MeWo LXSN and MeWo LV3SN melanoma cells. Subconfluent cultures from LXSN and LV3SN MeWo melanoma cells were fixed and incubated with a hyaluronan-binding protein (HABP). Cells were incubated with a FITC-streptavidin complex, mounted in Vectashield and analyzed under a Leica TCS SP5 AOBS confocal microscope. D, CD44 silencing causes a decrease in MeWo LXSN hyaluronidase activity but does not cause an additional decrease in MeWo LV3SN melanoma cells. Cell protein extracts were incubated in plates coated with BSA-conjugated hyaluronan. The remaining HA was detected with b-HABP as described under “Experimental Procedures.” A representative experiment out of three performed is shown. Capital letters indicate p < 0.01 (versus MeWo LXSN). Values are mean ± S.D.
ErbB Receptors in MeWo Human Melanoma Cells and Their Association with CD44 Interference by V3
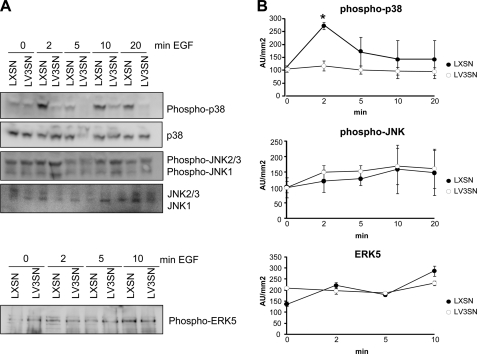
Because V3 can also potentially interact with the EGFR through the EGF-like repeats in its G3 domain (32), we determined whether the activation of signal transduction pathways by EGF is hampered in MeWo LV3SN cells. Parental MeWo LXSN and MeWo LV3SN cells were used since the primary goal of the present work was to find out the mechanism by which V3 is acting in these cells. We have previously described that V3 expression in MeWo melanoma cells interferes with activation of the ERK1/2 pathway by EGF and suggested that the growth inhibition observed in LV3SN MeWo melanoma cells may be related to the partial blocking of this mitogenic pathway (26). In the present work, we have analyzed the activation state of other MAPK pathways. To this end, MeWo LXSN and LV3SN cells were treated with 25 ng/ml of EGF for 2 min, and the phosphorylation level of p38 MAPK, JNK, and ERK5 were determined by immunoblotting. As shown in Fig. 5, MeWo LV3SN cells showed a markedly reduced activation of p38 MAPK in response to EGF compared with control LXSN cells. No significant differences were observed between LXSN and LV3SN MeWo cells in JNK and ERK5 signaling pathways. No effects were observed either in the Akt/PKB pathway (not shown). Both delayed activation of ERK1/2 and blocking of p38 MAPK pathways indicate that signaling mediated through the EGFR pathway is hampered in V3-expressing MeWo melanoma cells and may contribute to the cell growth inhibition observed in this cell line.
FIGURE 5.
Versican V3 overexpression decreases p38 MAPK activation and does not alter JNK or ERK5 activation. A, subconfluent cells were treated with 25 ng/ml EGF in serum-free medium for the indicated times. After the treatment, cell extracts were prepared as described under “Experimental Procedures” followed by immunoblotting with anti-phospho-p38MAPK, anti-phospho-JNK1/2/3, and anti-phospho-ERK5 antibodies or anti-non-phosphorylated forms antibodies (as a loading control). B, densitometric analyses of the ratio of the area of the phosphorylated versus total kinases from LXSN and LV3SN MeWo cells. In the case of JNK, both bands corresponding to JNK1 and JNK2/3 were considered. A representative experiment out of four performed is shown. Values are mean ± S.D. *, p < 0.05 (versus t = 0 min).
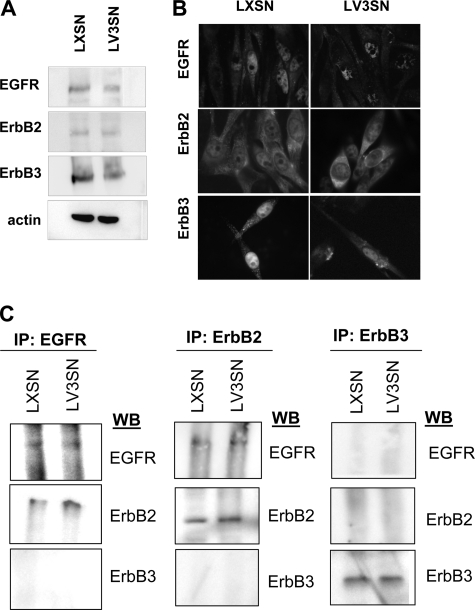
Because MAPK activation after EGF treatment is lower in V3-expressing melanoma cells and it has been described that CD44 is able to interact with members of the ErbB family in other cell types (21, 22), we determined whether CD44-ErbB complexes formed in MeWo melanoma cells. Firstly, we analyzed which members of the ErbB family were expressed by immunoblotting analysis using a panel of anti-ErbB family antibodies. Our results indicated that EGFR, ErbB2, and ErbB3 were expressed at different levels in MeWo cells, consistent with other melanoma cell lines (33). No significant differences between MeWo LXSN and LV3SN melanoma cells were observed (Fig. 6A). Similar results were obtained when the expression of these receptors was examined by immunocytochemistry (Fig. 6B), where it is shown that these molecules can be found in the cytoplasm as well as in the cell membrane. ErbB receptors form homo- and/or heterodimers after EGF binding resulting in the phosphorylation of tyrosine residues in the C-terminal tail of the receptor and their consequent activation (34). To address the question of which ErbB family members were able to interact with each other in MeWo LXSN and LV3SN melanoma cells following EGF stimulation, we carried out a pull-down experiment using anti-ErbB antibodies to immunoprecipitate the receptor followed by immunoblotting. The results shown in Fig. 6C indicated that only the heterodimer between EGFR and ErbB2 was formed in MeWo melanoma cells, whereas ErbB3 did not appear to be able to form complexes with the other members of the family. No differences were observed between MeWo LXSN and LV3SN cells.
FIGURE 6.
EGFR, ErbB2, and ErbB3 are expressed at similar levels and EGFR/ErbB2 heterodimers are formed in MeWo LXSN and MeWo LV3SN melanoma cells. A, immunoblotting of cell membrane extracts from LXSN and LV3SN MeWo melanoma cells. Membrane extracts were resolved in a 10% SDS-PAGE and blotted with an antibody against EGFR, ErbB2, ErbB3, or with anti-actin antibody (as a loading control). B, ErbB expression in LXSN and LV3SN MeWo melanoma cells detected by immunocytochemistry. 105 cells were seeded on coverslips, allowed to attach, and fixed by 2% paraformaldehyde. Samples were then stained for EGFR, ErbB2, and ErbB3, and with nuclear staining Hoechst 33342. Magnification: ×400. C, EGFR-ErbB2 is the only ErbB heterodimer formed in MeWo melanoma cells and is not altered by V3 overexpression. Cell lysates obtained from MeWo melanoma cells (LXSN) and V3-expressing MeWo melanoma cells (LV3SN) treated with 25 ng/ml EGF for 2 min were immunoprecipitated (IP) with anti-EGFR, anti-ErbB2, anti-ErbB3 antibodies followed by immunoblotting.
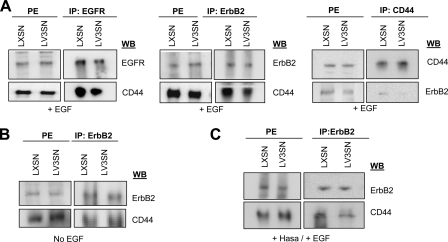
Secondly, we observed that EGFR-ErbB2 heterodimer was able to interact with CD44 in LXSN and LV3SN cells in co-immunoprecipitation experiments when cell extracts were pulled down with EGFR/ErbB2 antibodies and blotted with the antibody against CD44. More importantly, a lower amount of CD44 was found in immunoprecipitates from LV3SN cells, indicating that the interaction between CD44 and EGFR/ErbB2 was weaker in cells overexpressing the V3 isoform of versican. Similar results were obtained when cell extracts were pulled down with CD44 antibody and blotted with the antibody against ErbB2 (Fig. 7A). The amount of CD44 in protein extracts is similar in both cell types. These findings indicate that CD44 and EGFR/ErbB2 are closely associated with each other and that there is a decrease in their EGF-induced association in LV3SN cells compared with LXSN cells. The complex CD44-ErbB2 is also formed in the absence of EGF treatment but, in this case, the interaction between both molecules is not significantly affected by V3, indicating that EGF should be present in the extracellular medium for V3 to exert its effect on complex formation. Formation of the complex by V3 is also impaired when cells were previously treated with hyaluronidase (Fig. 7B).
FIGURE 7.
Effect of versican V3 overexpression on ErbB heterodimers/CD44 interaction. A, versican V3 overexpression impairs EGFR and ErbB2-CD44 interaction in MeWo melanoma cells. Cell lysates obtained from LXSN and LV3SN MeWo melanoma cells treated with 25 ng/ml EGF for 2 min were immunoprecipitated (IP) with anti-EGFR or anti-ErbB2 or anti-CD44 followed by immunoblotting. B, ErbB/CD44 interaction is not impaired in LV3SN cells in the absence of EGF. Cell lysates obtained from LXSN and LV3SN MeWo melanoma cells were immunoprecipitated (IP) with anti-ErbB2 followed by immunoblotting with anti-CD44 or reblotting with anti-ErbB2. C, ErbB/CD44 interaction is also impaired by V3 in hyaluronidase-treated cells. Cells were incubated with 2 units/ml hyaluronidase for 2 h at 37 °C before treatment with 25 ng/ml EGF for 2 min. Cell lysates obtained from LXSN and LV3SN MeWo melanoma cells were immunoprecipitated (IP) with anti-ErbB2 followed by immunoblotting with anti-CD44 or reblotting with anti-ErbB2. PE: protein extracts.
DISCUSSION
Some years ago, we described for the first time a high expression of the large isoforms of versican V0 and V1 by malignant melanoma cells (23). The exogenous expression of the short isoform V3 had the opposite effects than V0 and V1, since it decreased cell proliferation and migration of melanoma cells (25). Because the V3 isoform of versican only contains the N- and C-terminal G1 and G3 globular domains and it lacks chondroitin sulfate chains, the action of V3 should be attributed to the binding of G1 and/or G3 to their partners: G1 domain mediates the association between versican and hyaluronan, and G3 mediates versican-EGFR interaction through its EGF-like repeats (11). The initial working hypothesis proposed a competition model between V3 and V0/V1 versican isoforms (25, 35). Nevertheless, because the expression of V3 also decreased the cell proliferation rate in V0/V1-lacking human MeWo melanoma cells, we propose another model for V3 signaling mechanism.
V3 Interferes with CD44 Action
The G1 domain is able to interact with HA (11) and, indirectly, with CD44, which is a main signaling mechanism because it is able to interact with transduction pathway components and the cytoskeleton (16). To seek out the mechanism for V3, we silenced CD44 expression in control and V3-expressing MeWo cells. Our results demonstrate that CD44 has a main role in the control of melanoma cell proliferation and cell migration in wound healing assays since these abilities are greatly impaired in siCD44 MeWo cells. These results are in accordance with other tumor cell types where the effect of CD44 silencing has been investigated (36–38). The inhibitory effect of siCD44 on these cellular functions is mimicked by V3-expressing cells and by the double transfectants, supporting the hypothesis that interfering with CD44 action is one of the main mechanisms for V3 versican isoform. Because CD44 is a main surface receptor for HA, we also analyzed the behavior of V3-expressing cells, siCD44-cells and the double transfectants on HA-coated surfaces. As expected, CD44 silencing provoked a decrease on cell adhesion and cell migration on this substrate. In contrast, V3 expression had a positive effect on cell migration on HA. Although the potency of the effect is moderate, these differences between V3 expressing and siCD44 cells indicate that there is an uncoupling of the migration and proliferation pathways in cells with V3 versican.
If V3 interferes with CD44 action, other CD44-dependent processes should be also affected in V3-expressing cells, as HA uptake and catabolism. The current model states that HA fragments are initially generated by HYAL2 hyaluronidase near the cell surface, internalized via CD44 and delivered ultimately to lysosomes (30, 31). Furthermore, CD44 and HYAL2 are able to directly interact with each other leading to a regulation of HYAL2 activity by CD44 (30, 31, 39). According to this model, siCD44 and V3-expressing cells should have lower hyaluronidase activity and lower ability to internalize HA, leading to an accumulation of HA. Our results indicate that this is indeed the case: accumulation of HA is more pronounced in siCD44 cells, because these cells should have a very low capacity for HA uptake whereas V3-expressing cells should retain some of this ability since CD44 is still present at the cell surface. The existence of a thicker HA surface in V3-expressing cells, together with the presence of CD44, may explain why LV3SN cells are able to adhere and migrate better onto HA-coated surfaces than their control LXSN cells. The expression of HAS isoforms was not altered in any of the cell lines (not shown). Taking all these results into account, we propose that V3, through its G1 domain, interferes with CD44-dependent processes as cell proliferation and migration, as well as hyaluronan uptake and catabolism.
V3 can potentially interact with the EGFR through the EGF-like repeats of its G3 domain, and this mechanism has been already proposed by others in cells overexpressing the G3 domain of versican (32, 40). We have suggested that this may be also the case in MeWo melanoma cells, where V3 expression causes a decrease in ERK1/2 signaling triggered by EGF (25). To gain further insight into the possible role of the EGFR in V3 mechanism, we have analyzed the activation state of other signal transduction pathways in LXSN and LV3SN cells. We observed a blocked p38 MAPK phosphorylation by EGF in V3 expressing cells. p38 appears to be downstream of the EGF pathway, because several studies indicate that EGF induces the phosphorylation of p38 (41–43). In melanoma, both ERK and p38 pathways are required for cell migration and growth in vivo (44). Furthermore, several works describe that Ras may regulate p38 MAPK pathway (45, 46) suggesting that Ras could exert as a hinge molecule that regulates ERK1/2 as well as p38 MAPK activation.
Our results support then a model in which V3 expression in MeWo melanoma cells is able to interact directly or indirectly with CD44 and the EGFR, interfering with these signaling pathways and altering the growth and migratory abilities of MeWo melanoma cells. This conclusion lead us to analyze whether these two receptors, CD44 and members of the EGFR family, interact in MeWo melanoma cells as it has been proposed in other tumor cell types to regulate signaling crosstalk and tumor cell progression (21, 22).
We have shown that MeWo cells expressed EGFR, ErbB2 and ErbB3 at different levels, consistent with other melanoma cell lines (33) and with the important role of ErbB receptors in this tumor type (47). Our results show also that the main heterodimer for the ErbB family present on MeWo melanoma cells arises from the combination of EGFR-ErbB2. It should be taken into account that ErbB2 does not bind extracellular ligands but is poised to associate with ligand-activated forms of the other family members, such as the EGFR (48).
Finally, the interaction between CD44 and the ErbB receptor family has been examined in LXSN and LV3SN cells under the hypothesis that V3, being a molecule with dual ability to bind both receptors through its G1 and G3 versican subdomains, may interfere with this binding and subsequent signaling mechanisms. Our results showed that V3 expression causes indeed a significant decrease in the amount of CD44 recruited into the EGFR-ErbB2 complex. This decrease is only observed when cells are treated with EGF, suggesting that a competition between both molecules exist. In contrast, the effect is independent of the presence of HA. Thus, our observations strongly suggest that V3 directly or indirectly interferes with the interaction between EGFR-ErbB2 and CD44 leading to a decreased signaling.
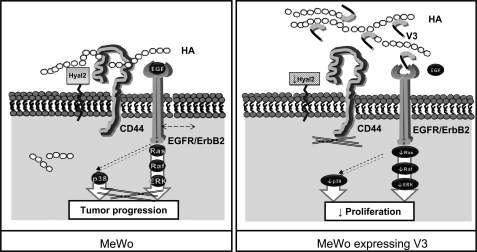
In summary, our current model (Fig. 8) proposes that V3 isoform interferes with HA-CD44 and competes with EGF to bind the EGFR, possibly through its G1 domain and the EGF-like repeats in the G3 domain, respectively. In consequence, the interaction between EGFR-ErbB2 and CD44 is altered in the presence of V3. In one hand, EGF-dependent activation of ERK1/2 and p38 MAPK pathways is down-regulated because of the direct interference of V3 and decreased ability of CD44 to bind the EGFR-ErbB2 complex. In other hand, V3 isoform causes a decrease in CD44-mediated signaling to proliferative pathways and the cytoskeleton, affecting cell growth and migration. Finally, V3 provokes an accumulation of HA on the cell surface because of hyaluronidase down-regulation and blockage of HA uptake into the cell, which are both CD44-dependent mechanisms. Altogether, we propose a complex mechanism to explain the functional properties of V3-expressing cells. This may be a good example of how cell behavior may be modified by manipulation of the extracellular matrix in cancer cells.
FIGURE 8.
Proposed model for V3-mediated effects in MeWo melanoma cells. Left, CD44 and the ErbB2/EGFR complex are main signaling pathways in MeWo melanoma cells, leading to tumor cell growth and migration. CD44 also regulates HA catabolism by regulating HYAL2 activity and allowing HA internalization. Right, in V3-expressing cells, V3 interferes with HA-CD44-EGFR/ErbB2 interaction through its G1 and G3 domains, leading to a delayed activation of EGFR-mediated signaling pathways, such as ERK1/2 and p38 MAPK. CD44-dependent mechanisms, such as hyaluronan internalization and HYAL2 activity, are inhibited. Cell proliferation and migration on plastic are decreased due to impairment of both transduction pathways. Nevertheless, adhesion and migration on HA-coated surfaces are enhanced because of the accumulation of HA and the presence of CD44.
Acknowledgments
We thank Dr. R. Vilella (Hospital Clinic, Barcelona, Spain) for providing the anti-CD44 antibodies, Dr. F. X. Real (I. M. I. M., Barcelona, Spain) for providing the melanoma cell lines, Dr. N. Gómez and Dr. J. M. Lizcano (Departament de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, Spain) for the antibodies against ERK5 and JNK, and Anna Vilalta for excellent technical assistance.
This work was supported in part by Grant AGL2006-02365 from the Ministerio de Ciencia y Tecnologia, Grant 2009 SGR-1091 from the Generalitat de Catalunya (to A. B.), and the FEDER program from the European Union.
- ECM
- extracellular matrix
- HA
- hyaluronan
- GAG
- glycosaminoglycan
- HABP
- hyaluronan-binding protein.
REFERENCES
- 1. Wight T. N. (2002) Curr. Opin. Cell Biol. 14, 617–623 [DOI] [PubMed] [Google Scholar]
- 2. Ricciardelli C., Sakko A. J., Ween M. P., Russell D. L., Horsfall D. J. (2009) Cancer Metastasis Rev. 28, 233–245 [DOI] [PubMed] [Google Scholar]
- 3. Ricciardelli C., Mayne K., Sykes P. J., Raymond W. A., McCaul K., Marshall V. R., Horsfall D. J. (1998) Clin. Cancer Res. 4, 963–971 [PubMed] [Google Scholar]
- 4. Ricciardelli C., Brooks J. H., Suwiwat S., Sakko A. J., Mayne K., Raymond W. A., Seshadri R., LeBaron R. G., Horsfall D. J. (2002) Clin. Cancer Res. 8, 1054–1060 [PubMed] [Google Scholar]
- 5. Suwiwat S., Ricciardelli C., Tammi R., Tammi M., Auvinen P., Kosma V. M., LeBaron R. G., Raymond W. A., Tilley W. D., Horsfall D. J. (2004) Clin. Cancer Res. 10, 2491–2498 [DOI] [PubMed] [Google Scholar]
- 6. Voutilainen K., Anttila M., Sillanpää S., Tammi R., Tammi M., Saarikoski S., Kosma V. M. (2003) Int. J. Cancer 107, 359–364 [DOI] [PubMed] [Google Scholar]
- 7. Pirinen R., Leinonen T., Böhm J., Johansson R., Ropponen K., Kumpulainen E., Kosma V. M. (2005) Hum. Pathol. 36, 44–50 [DOI] [PubMed] [Google Scholar]
- 8. Pukkila M., Kosunen A., Ropponen K., Virtaniemi J., Kellokoski J., Kumpulainen E., Pirinen R., Nuutinen J., Johansson R., Kosma V. M. (2007) J. Clin. Pathol. 60, 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nikitovic D., Zafiropoulos A., Katonis P., Tsatsakis A., Theocharis A. D., Karamanos N. K., Tzanakakis G. N. (2006) IUBMB Life 58, 47–53 [DOI] [PubMed] [Google Scholar]
- 10. Isogai Z., Shinomura T., Yamakawa N., Takeuchi J., Tsuji T., Heinegård D., Kimata K. (1996) Cancer Res. 56, 3902–3908 [PubMed] [Google Scholar]
- 11. Wu Y. J., La Pierre D. P., Wu J., Yee A. J., Yang B. B. (2005) Cell Res. 15, 483–494 [DOI] [PubMed] [Google Scholar]
- 12. Dours-Zimmermann M. T., Zimmermann D. R. (1994) J. Biol. Chem. 269, 32992–32998 [PubMed] [Google Scholar]
- 13. Iozzo R. V. (1998) Annu. Rev. Biochem. 67, 609–652 [DOI] [PubMed] [Google Scholar]
- 14. Matsumoto K., Shionyu M., Go M., Shimizu K., Shinomura T., Kimata K., Watanabe H. (2003) J. Biol. Chem. 278, 41205–41212 [DOI] [PubMed] [Google Scholar]
- 15. Ito K., Shinomura T., Zako M., Ujita M., Kimata K. (1995) J. Biol. Chem. 270, 958–965 [DOI] [PubMed] [Google Scholar]
- 16. Marhaba R., Zöller M. (2004) J. Mol. Histol. 35, 211–231 [DOI] [PubMed] [Google Scholar]
- 17. Bourguignon L. Y., Zhu H., Zhou B., Diedrich F., Singleton P. A., Hung M. C. (2001) J. Biol. Chem. 276, 48679–48692 [DOI] [PubMed] [Google Scholar]
- 18. Wang S. J., Bourguignon L. Y. (2006) Arch. Otolaryngol. Head Neck Surg. 132, 771–778 [DOI] [PubMed] [Google Scholar]
- 19. Bourguignon L. Y., Gilad E., Peyrollier K. (2007) J. Biol. Chem. 282, 19426–19441 [DOI] [PubMed] [Google Scholar]
- 20. Misra S., Hascall V. C., Berger F. G., Markwald R. R., Ghatak S. (2008) Connect Tissue Res. 49, 219–224 [DOI] [PubMed] [Google Scholar]
- 21. Bourguignon L. Y., Zhu H., Chu A., Iida N., Zhang L., Hung M. C. (1997) J. Biol. Chem. 272, 27913–27918 [DOI] [PubMed] [Google Scholar]
- 22. Ghatak S., Misra S., Toole B. P. (2005) J. Biol. Chem. 280, 8875–8883 [DOI] [PubMed] [Google Scholar]
- 23. Touab M., Villena J., Barranco C., Arumí-Uria M., Bassols A. (2002) Am. J. Pathol. 160, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Touab M., Arumi-Uría M., Barranco C., Bassols A. (2003) Am. J. Clin. Pathol. 119, 587–593 [DOI] [PubMed] [Google Scholar]
- 25. Serra M., Miquel L., Domenzain C., Docampo M. J., Fabra A., Wight T. N., Bassols A. (2005) Int. J. Cancer 114, 879–886 [DOI] [PubMed] [Google Scholar]
- 26. Miquel-Serra L., Serra M., Hernández D., Domenzain C., Docampo M. J., Rabanal R. M., de Torres I., Wight T. N., Fabra A., Bassols A. (2006) Lab. Invest. 86, 889–901 [DOI] [PubMed] [Google Scholar]
- 27. Tzircotis G., Thorne R. F., Isacke C. M. (2005) J. Cell Sci. 118, 5119–5128 [DOI] [PubMed] [Google Scholar]
- 28. Hua Q., Knudson C. B., Knudson W. (1993) J. Cell Sci. 106, 365–375 [DOI] [PubMed] [Google Scholar]
- 29. Stern M., Stern R. (1992) Matrix 12, 397–403 [DOI] [PubMed] [Google Scholar]
- 30. Knudson W., Chow G., Knudson C. B. (2002) Matrix Biol. 21, 15–23 [DOI] [PubMed] [Google Scholar]
- 31. Harada H., Takahashi M. (2007) J. Biol. Chem. 282, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Cao L., Yang B. L., Yang B. B. (1998) J. Biol. Chem. 273, 21342–21351 [DOI] [PubMed] [Google Scholar]
- 33. Stove C., Stove V., Derycke L., Van Marck V., Mareel M., Bracke M. (2003) J. Invest. Dermatol. 121, 802–812 [DOI] [PubMed] [Google Scholar]
- 34. Tao R. H., Maruyama I. N. (2008) J. Cell Sci. 121, 3207–3217 [DOI] [PubMed] [Google Scholar]
- 35. Lemire J. M., Merrilees M. J., Braun K. R., Wight T. N. (2002) J. Cell Physiol. 190, 38–45 [DOI] [PubMed] [Google Scholar]
- 36. Shi Y., Tian Y., Zhou Y. Q., Ju J. Y., Qu L., Chen S. L., Xiang Z. G., Liu Y., Zhu L. P. (2007) Oncol. Rep. 18, 397–403 [PubMed] [Google Scholar]
- 37. Subramaniam V., Vincent I. R., Gilakjan M., Jothy S. (2007) Exp. Mol. Pathol. 83, 332–340 [DOI] [PubMed] [Google Scholar]
- 38. Li C. Z., Liu B., Wen Z. Q., Li H. Y. (2008) Folia Biol. 54, 180–186 [PubMed] [Google Scholar]
- 39. Bourguignon L. Y., Singleton P. A., Diedrich F., Stern R., Gilad E. (2004) J. Biol. Chem. 279, 26991–27007 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y., Cao L., Kiani C., Yang B. L., Hu W., Yang B. B. (1999) J. Cell Biochem. 73, 445–457 [PubMed] [Google Scholar]
- 41. Harris V. K., Coticchia C. M., Kagan B. L., Ahmad S., Wellstein A., Riegel A. T. (2000) J. Biol. Chem. 275, 10802–10811 [DOI] [PubMed] [Google Scholar]
- 42. New L., Li Y., Ge B., Zhong H., Mansbridge J., Liu K., Han J. (2001) J. Cell Biochem. 83, 585–596 [DOI] [PubMed] [Google Scholar]
- 43. Ostrander J. H., Daniel A. R., Lofgren K., Kleer C. G., Lange C. A. (2007) Cancer Res. 67, 4199–4209 [DOI] [PubMed] [Google Scholar]
- 44. Estrada Y., Dong J., Ossowski L. (2009) Pigment Cell Melanoma Res. 22, 66–76 [DOI] [PubMed] [Google Scholar]
- 45. Palsson E. M., Popoff M., Thelestam M., O'Neill L. A. (2000) J. Biol. Chem. 275, 7818–7825 [DOI] [PubMed] [Google Scholar]
- 46. McDermott E. P., O'Neill L. A. (2002) J. Biol. Chem. 277, 7808–7815 [DOI] [PubMed] [Google Scholar]
- 47. Gordon-Thomson C., Jones J., Mason R. S., Moore G. P. (2005) Melanoma Res. 15, 21–28 [DOI] [PubMed] [Google Scholar]
- 48. Burgess A. W. (2008) Growth Factors 26, 263–274 [DOI] [PubMed] [Google Scholar]