Abstract
Pressure and volume overload induce hypertrophic growth of postnatal cardiomyocytes and genetic reprogramming characterized by reactivation of a subset of fetal genes. Despite intense efforts, the nuclear effectors of cardiomyocyte hypertrophy remain incompletely defined. Endothelin-1 (ET-1) plays an important role in cardiomyocyte growth and is involved in mediating the neurohormonal effects of mechanical stress. Here, we show that the phenylephrine-induced complex-1 (PEX1), also known as zinc finger transcription factor ZFP260, is essential for cardiomyocyte response to ET-1 as evidenced in cardiomyocytes with PEX1 knockdown. We found that ET-1 enhances PEX1 transcriptional activity via a PKC-dependent pathway which phosphorylates the protein and further potentiates its synergy with GATA4. Consistent with a role for PEX1 in cardiomyocyte hypertrophy, overexpression of PEX1 is sufficient to induce cardiomyocyte hypertrophy in vitro and in vivo. Importantly, transgenic mice with inducible PEX1 expression in the adult heart develop cardiac hypertrophy with preserved heart function. Together, the results identify a novel nuclear effector of ET-1 signaling and suggest that PEX1 may be a regulator of the early stages of cardiac hypertrophy.
Keywords: Cardiac Hypertrophy, Gene Regulation, Heart, Transcription Factors, Transgenic
Introduction
Cardiac hypertrophy is an adaptive response to various stimuli, physiologic or pathologic, and is characterized by the increased size of terminally differentiated cardiomyocytes and the reexpression of a subset of fetal genes such as atrial natriuretic factor (ANF),4 β-myosin heavy chain (β-MHC), and skeletal muscle α-actin (ACTA1) (1, 2). Persistent hypertrophy induced by pathologic conditions like hypertension or atherosclerosis eventually progresses to heart failure, a major cause of death in industrialized nations (3). The molecular mechanisms underlying the transition from adaptive (compensated) to maladaptive (decompensated) cardiac hypertrophy have been the subject of numerous studies, yet our understanding of this complex process remains incomplete.
Endothelin-1 (ET-1), a 21-amino acid peptide with potent vasoconstrictive properties synthesized primarily by the endothelium, is an important regulator of cardiac contractility and cardiomyocyte growth (4). Cardiac and plasma ET-1 levels are modulated in disease and are elevated in myocardial infarction, hypertension, and heart failure (5–7). ET-1 acts via two G protein-coupled receptors, endothelin receptor type A and B, which are present in the heart and are regulated in pathologic states (8). The hypertrophic effects of ET-1 on cardiac myocytes are mediated by endothelin receptors type A (9, 10), and their antagonists were shown to prevent the development of cardiac hypertrophy (7, 11). Like α1-adrenergic receptors, endothelin receptors type A activate several Gq-coupled signaling cascades, including phospholipase C-PKC, mitogen-activated kinases (MAPKs), and calcium-mediated signaling (12–21). It is generally accepted that these pathways ultimately converge on a set of nuclear effectors, like GATA4 and NFAT, which regulate specific gene programs (14, 22). In particular, the zinc finger protein GATA4, which plays a crucial role in normal heart development, has emerged as a key effector of several hypertrophic stimuli, including α1-adrenergic agonists like phenylephrine (PE) and ET-1 (12, 23). These stimuli alter GATA4 levels or induce post-translational modifications, such as phosphorylation or acetylation, resulting in changes in DNA binding or transcriptional activation capacity (12, 24–27). Enhanced expression of GATA4 in cultured cardiomyocytes is sufficient to induce myocyte hypertrophy, whereas GATA4 knockdown or expression of a dominant negative GATA4 protein inhibits agonist-induced sarcomeric reorganization (12, 28). More generally, GATA4 appears to act as an important component of the heart adaptive response as evidenced by in vivo studies in genetically altered mice (29–31).
Recently, we isolated a novel transcription factor, phenylephrine-induced complex-1 (PEX1), also known as zinc finger protein 260 (ZFP260), involved in α1-adrenergic signaling in the heart (32). PEX1 is a 13-zinc finger-containing protein of the Krüppel family that binds to the conserved phenylephrine response element (PERE) on the ANF promoter (33). The expression profile of PEX1 is remarkably similar to the pattern of ANF expression during embryonic and postnatal development in that PEX1 levels are high during embryonic development and decrease in postnatal ventricles. Moreover, knockdown of PEX1 in cardiomyocytes reduces basal and abrogates PE-induced ANF expression. Thus, PEX1 is one of a handful number of transcription factors that include GATA4 and myocardin (34), a serum response factor cofactor (35, 36) that appears required for nuclear signaling of α1-adrenergic receptors. Interestingly, PEX1 physically and functionally interacts with GATA4 to cooperatively activate transcription of ANF and other hypertrophy-induced genes (32). This raises the intriguing possibility that PEX1 may be a nuclear effector of other growth-promoting stimuli.
In this paper, we present evidence supporting a role for PEX1 in ET-1 signaling and cardiac growth both in vivo and in primary myocyte cultures. In vitro, promoter studies and loss-of-function analysis revealed an essential role for PEX1 in mediating ET-1 response, and ET-1 was found to potentiate PEX1 transcriptional activation in a PKC-dependent manner. Gain-of-function experiments confirmed that up-regulation of PEX1 expression is sufficient to induce the morphologic and genetic changes characteristic of cardiac hypertrophy without negatively impacting cardiac function. Thus, PEX1 appears to be a novel component of a transcriptional complex that governs early events in cardiac hypertrophy.
EXPERIMENTAL PROCEDURES
Adenoviral Constructs
A recombinant replication-deficient type 5 adenovirus expressing HA-tagged sense PEX1 (HA-PEX1) was generated using the AdEasy XL Adenoviral Vector system (Stratagene). In summary, a 1296-bp HindIII fragment containing rat HA-PEX1 cDNA was subcloned into a shuttle vector (pshuttle-CMV), and the adenovirus was generated through recombination with the pAdEasy-1 as described previously (32). Transformant bacteria were selected for kanamycin resistance, and recombination was identified via restriction enzyme digestion. Recombinants were then massively produced using the recombination-deficient XL10-Gold strain. Purified recombinant adenoviral plasmid DNA was digested with PacI to expose the terminal repeats and was later transfected to AD-293 cells in which deleted viral assembly genes are complemented in vivo. The viruses were produced in bulk, and titers were determined (37). The cloning and production of adeno-LacZ, adeno-GATA4, and antisense adeno-HA-AS-PEX1 were reported previously (12, 32).
Cell Cultures
Ex vivo experiments were done on primary cultures of rat neonatal cardiomyocytes as described previously (37). Cardiomyocytes were plated and kept overnight in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The next day, cells were extensively washed, and the medium was replaced with serum-free hormone-free medium. Transfections and luciferase assays using ANF reporter plasmids and PEX1 expression vectors were carried out as described previously (32, 37). Cardiomyocytes were infected with different doses of either adeno-LacZ, adeno-PEX1, adeno-GATA4, or an antisense adeno-HA-AS-PEX1 as described in our earlier published work (29, 32, 37). For ET-1 stimulation, cardiomyocytes were treated with 100 nm ET-1 or vehicle in serum-free hormone-free medium for 24 h in the presence or absence of inhibitors: p38 MAPK (SB 203580; 10 μm [SB]), PKC (GF 109203X; 5 μm [GF]), PI3K (LY 294002; 25 μm), ERK1/2 (PD98059; 10 μm) (Calbiochem).
Animal Models
Mice were handled in accordance with institutional guidelines. Experiments were approved by the Institutional Animal Ethics Committee. Mouse PEX1 cDNA was subcloned in the CAG-CAT expression vector (a kind gift from M. Yanagisawa, Howard Hughes Medical Institute, Dallas, TX), in which the expression of PEX1 can be induced via a Cre recombinase-dependent excision of the CAT transgene. Two lines of CAT-PEX1 mice were then crossed with α-MHC/MerCreMer mice expressing a cardiomyocyte-specific, Tamoxifen-inducible Cre recombinase (38). 150-day-old α-MHCMerCreMer (Ctrl) and double-transgenic α-MHCMerCreMer/CAT-PEX1 (TG) mice were treated with Tamoxifen as described previously (39, 40). At 1 and 2 weeks after treatment, groups of mice were anesthetized using 2.0% isoflurane and 80 ml/min 100% O2; their anterior chests were shaved, and two-dimensional guided M-mode echocardiography was performed using the Visual-Sonics VEVO 700 and a 30-MHz linear array transducer as described by Aries et al. (29). The next day, mice were anesthetized with 12–15 μl/g intraperitoneal Avertin (2.5% solution), and either killed for tissue collection or heart-perfused for histologic studies (29). Genotyping was carried out using PCR and quantitative PCR (qPCR) utilizing transgene-specific oligonucleotides.
Histologic and Cytologic Studies
Mouse hearts were perfused with PBS-KCl, fixed with paraformaldehyde, and then paraffin-embedded. Sections were trichrome-stained and were visualized at magnifications of ×1.25 and ×63. Immunohistochemistry and immunofluorescence were performed on tissue sections or cellular preparations as described previously (29, 32), using a rabbit polyclonal rat PEX1 antibody (dilution 1/500) (32), an ANF antibody (dilution 1/1500), sarcomeric α-actinin antibody (1/500) and phalloidin-Alexa Fluor 488 (dilution 1/400).
Western Blotting
Western blots were performed on nuclear extracts from infected cardiac myocytes and adult mouse hearts as described previously (32). PEX1 antibody was used at a dilution of 1/500, GATA4 antibody at 1/2000, and GATA6 antibody at 1/1000. Visualization was done using an anti-rabbit horseradish peroxidase-conjugated antibody (Sigma).
Real-time PCR
Total RNA was isolated from cells or mice tissues with TRIzol (Invitrogen). Transcript levels for the various cardiac markers were determined by real-time PCR carried out as described by Debrus et al. (32). Analysis was done using the ΔΔCT quantitation method, with the ribosomal S-16 serving as the normalizer gene.
Kinase Assays
The recombinant proteins glutathione S-transferase (GST) and GST-PEX1 were produced as described previously (12). 5 μg of bacterially expressed protein was incubated with 20 ng of the purified catalytic subunit of PKC (Calbiochem) or the AKT kinase (Cell Signaling) in the reaction buffer (25 mm Tris (pH 7.5), 10 mm 6-glycerophosphate, 2 mm DTT, 0.1 mm Na3VO4, 5 μCi ATP) at 30 °C for 30 min. The proteins were resolved by SDS-PAGE (10% w/v gels) and the blots exposed to x-ray films.
Statistical Analysis
Data are means ± S.E., with p < 0.05 by Student's t test being considered as statistically significant.
RESULTS
PEX1 Is a Nuclear Mediator of ET-1 Signaling in Cardiomyocytes
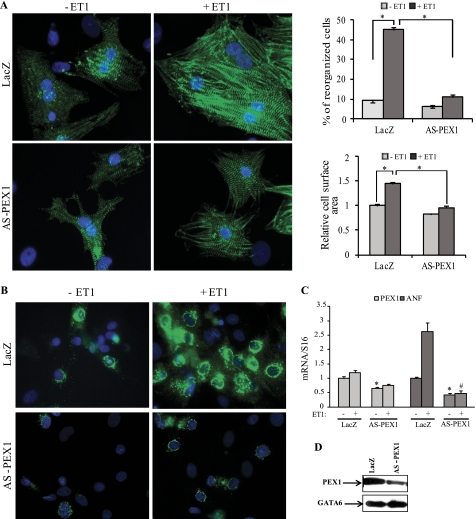
We have shown previously that knocking down PEX1 in postnatal myocytes decreased endogenous ANF expression and interfered with myofibrillar reorganization in response to α1-adrenergic stimulation (32). Given that GATA4 is essential for both α1-adrenergic and ET-1 response (12, 41), we tested whether PEX1 is also involved in ET-1 signaling. To this end, we infected cardiomyocytes with adenoviral vectors expressing either antisense-PEX1 transcripts or LacZ as a control. Cardiomyocytes treated with the antisense PEX1 vector had significantly reduced endogenous PEX1 levels as evidenced by Western blotting (Fig. 1D). Loss of PEX1 blocked ET-1-induced myofibrillar reorganization and cell hypertrophy (Fig. 1A). ANF expression was also inhibited, as assessed at the protein level by immunocytochemistry (Fig. 1B) and at the mRNA level using qPCR (Fig. 1C).
FIGURE 1.
PEX1 is required for ET-1 signaling. A, left, Hoechst (blue) and sarcomeric α-actinin (green) immunofluorescence-labeled slides are shown of the control adeno-LacZ and adeno-HA-AS-PEX1-infected primary cardiomyocytes treated or not with 100 nm ET-1 for 24 h. Costaining was performed using Hoechst to detect cell nuclei and sarcomeric α-actinin to visualize the myofibrils of cardiomyoctes. Note that the loss of PEX1 blocked the myofibrillar reorganization seen in adeno-LacZ-infected cells treated with ET-1. A, right, reorganized cells were counted, and relative cell surface area was measured across 10 fields (×40) in three separate experiments. *, p < 0.05. B, ANF immunofluorescence is shown in cardiomyocytes infected for 3 days with either adeno-LacZ or adeno-HA-AS-PEX1 and treated with vehicle or ET-1. Note how PEX1 down-regulation inhibited ET-1-induced cellular accumulation of ANF. C, ANF mRNA levels in LacZ- or AS-PEX1-transfected cardiomyocytes treated or not with ET-1. *, p < 0.05 versus LacZ; #, p < 0.05 versus LacZ + ET-1. D, PEX1 protein levels in cardiomyocytes infected with the adeno-AS-PEX1 or adeno-LacZ as detected by Western blotting are shown. GATA6 protein was used as an internal control.
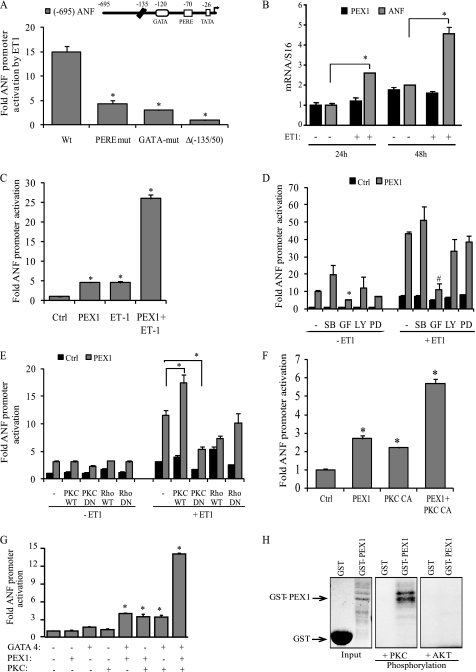
Next, we tested whether the ANF promoter responsiveness to ET-1 requires PERE, which was shown previously to be critical for α1-responsiveness (32). As stated earlier, the GATA elements of the proximal ANF promoter are known to be required for maximal ET-1 response (41). Mutation of the GATA (−120) or PERE (−70) within the ANF promoter significantly reduced its activation by ET-1 (Fig. 2A), indicating that both elements contribute to ET-1 transcriptional response. Together, the results strongly suggest a role for PEX1 as an effector of ET-1 signaling.
FIGURE 2.
Post-translational regulation of PEX1 by ET-1. A, ANF promoter elements required for ET-1 response in cardiomyocytes. Cells were transfected with wild-type (Wt) and mutated −695ANF or −135ANF promoter luciferase reporter constructs and stimulated for 48 h with 100 nm ET-1. PERE mut corresponds to mutation of the proximal PERE site (position −70 bp); GATA mut corresponds to mutation of the GATA site (position −120 bp). The data are mean of ± S.E. (error bars; n = 4). *, p < 0.05 versus wild type. B, effect of ET-1 on PEX1 mRNA levels. ANF and PEX1 transcript levels were quantified in primary cultured ventricular cardiomyocytes treated or not with 100 nm ET-1 for 24 or 48 h. Data are mean ± S.E. (n = 4). *, p < 0.05 C, ET-1 potentiates PEX1 activation. Cardiomyocytes were cotransfected with the proximal ANF promoter luciferase reporter and 500 ng of PEX1. The cells were then treated with 100 nm ET-1 for 48 h. Data are mean ± S.E. (n = 4). *, p < 0.05 versus Ctrl. D, involvement of PKC but not p38-MAPK or ERK1/2 in ET-1-dependent PEX1 activation of ANF transcription. Cardiomyocytes were cotransfected with the proximal ANF promoter luciferase reporter and 500 ng of PEX1. The cells were then treated with 100 nm ET-1 for 24 h in the presence of inhibitors: p38 MAPK (SB 203580; 10 μm [SB]), PKC (GF 109203X; 5 μm [GF]), PI3K (LY 294002; 25 μm [LY]) ERK1/2 (PD98059; 10 μm [PD]). The data are from one representative experiment carried out in triplicate. *, p < 0.05 versus PEX1 (−ET1); #, p < 0.05 versus PEX1 (+ET1). E and F, cotransfection in cardiomyocytes of the ANF-Luc reporter with the indicated expression vectors treated or not with 100 nm ET-1(PKC CA, PKCβ catalytic domain). Note how cotransfection with PKC-dominant negative (DN) but not Rho-DN abrogates the ET-1 effect on PEX1 activation of the promoter. *, p < 0.05. G, PKCβ synergizes with PEX1 and GATA4 on the ANF promoter. NIH3T3 cells were cotransfected with the ANF-Luc construct 100 ng of PEX1, 10 ng of GATA4, and 200 ng of PKCβ CA. *, p < 0.05 versus Ctrl. H, in vitro PKC phosphorylation of GST-PEX1 fusion protein. In similar kinase assays (such as AKT kinase), PEX1 was not phosphorylated, demonstrating PKC specificity. Coomassie staining was used to show protein loading.
Because PEX1 is already present in the nucleus of unstimulated myocytes, we tested whether ET-1 enhances PEX1 expression. Neither PEX1 mRNA (Fig. 2B) nor protein levels (data not shown) were changed in primary cardiomyocyte cultures stimulated with ET-1 at various time points. However, treatment of cells with ET-1 potentiated the PEX1 transcriptional activation of the ANF promoter (Fig. 2C), suggesting a post-translational regulatory mechanism. Signaling through ETA activates several signal transduction cascades, notably PKCs and MAPKs. Use of pharmacologic inhibitors confirmed the involvement of PKC in ET-1 enhancement of PEX1 transcriptional activity, as evidenced by the ability of PKC inhibitor (GF 109203X) to virtually abrogate promoter activation by PEX1 and ET-1 (Fig. 2D). In contrast, inhibition of p38 MAPK (with SB 203580), ERK1/2 (with PD 98059), or PI3K (with LY 294002) had no significant effect on promoter activation (Fig. 2D). Moreover, ET-1-treated cardiomyocytes cotransfected with the proximal ANF promoter, PEX1 and either PKCβ WT or the constitutively active catalytic domain of PKCβ, showed synergistic activation of the promoter (Fig. 2, E and F). Transfection with dominant negative PKC abrogated the effect of ET-1 and PEX1 on the promoter (Fig. 2E). In NIH3T3 cells, which have low endogenous levels of GATA4 and PEX1, addition of the PKCβ catalytic domain together with either or both transcription factors, greatly enhanced the PEX1/GATA4 synergy over the ANF promoter (Fig. 2G). The PEX1 amino acid sequence contains putative PKC phosphorylation sites, and in vitro kinase assays confirmed that PKCβ, but not other kinases, such as AKT can directly phosphorylate PEX1 (Fig. 2H).
PEX1 Overexpression Is Sufficient to Induce Cardiomyocyte Hypertrophy
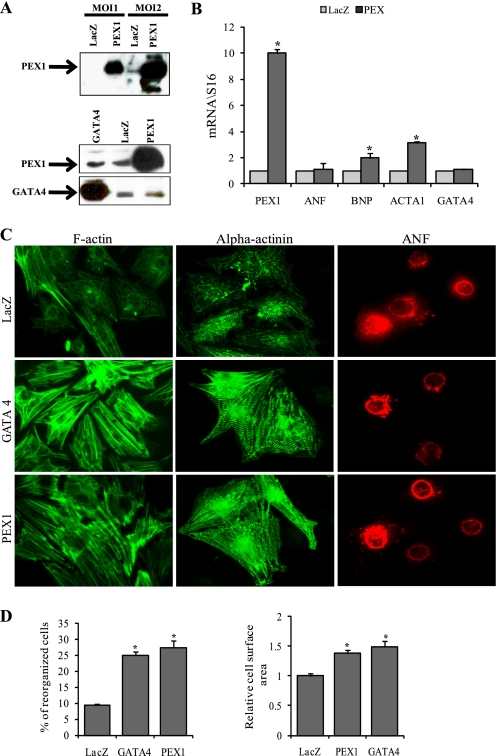
To test the causal relationship between PEX1 and myocyte hypertrophy, we generated an adenovirus expressing HA-tagged PEX1 and examined the consequences of up-regulating PEX1 levels in neonatal cardiomyocytes. Infecting the myocytes with this construct led to a significant increase in PEX1 mRNA and protein levels, as assessed by qPCR and Western blotting (Fig. 3, A and B). Levels of ACTA1 and BNP transcripts were also increased (Fig. 3B). Elevated PEX1 levels induced myofibrillar reorganization and cell hypertrophy and mimicked the effects of GATA4 overexpression both on the cytoskeleton and on ANF (Fig. 3, C and D). Interestingly, PEX1 overexpression affected neither the mRNA nor the protein levels of GATA4. In addition, PEX1 protein levels were unchanged in cardiomyocytes overexpressing GATA4, suggesting that these proteins act in two different, yet convergent pathways (Fig. 3A).
FIGURE 3.
Overexpression of PEX1 is sufficient to induce myocyte hypertrophy. A, upper, representative Western blot confirming overexpression of PEX1 in adeno-HA-PEX1-infected cardiomyocytes. A, lower, Western blot showing no change in PEX1 levels in cardiomyocytes overexpressing GATA4 and no change in GATA4 levels in myocytes overexpressing PEX1. B, histograms showing mRNA fold change of PEX1, ANF, BNP, ACTA1, and GATA4 in adeno-LacZ- or adeno-HA-PEX1-infected cardiomyocytes. The data are expressed as fold change relative to LacZ with S16 as the normalizer gene. Overexpression of PEX1 caused an up-regulation of BNP and ACTA1 mRNA. *, p < 0.05 versus LacZ. C, F-actin detected with the phalloidin-Alexa Fluor 488 (green in left panel), Sarcomeric α-actinin (green in middle panel) and ANF (red in right panel) immunofluoresence-labeled adeno-LacZ-, adeno-GATA4-, or adeno-HA-PEX1-infected cardiomyocytes. Note how induction of either GATA4 or PEX1 induces myofibrillar reorganization (as shown in both the phalloidin and the α-actinin staining) but not ANF accumulation. D, reorganized cells were counted and relative cell surface area measured across 10 fields (×40) in three separate experiments. *, p < 0.05 versus LacZ.
Conditional Overexpression of PEX1 in the Myocardium Induces Cardiac Hypertrophy
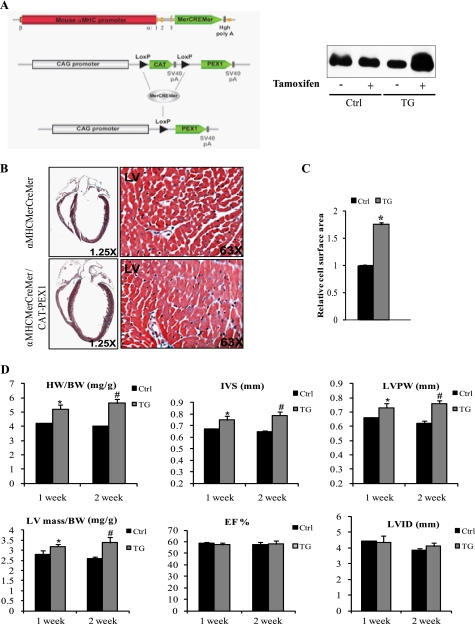
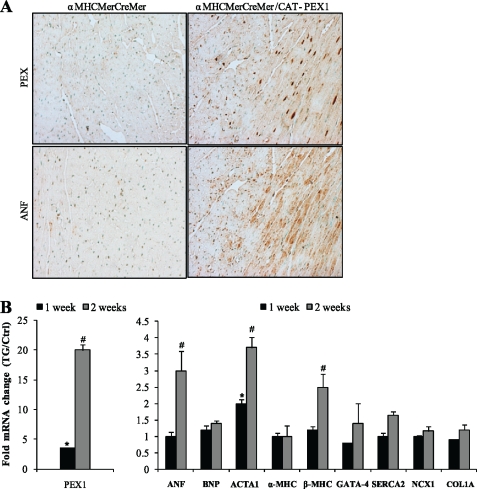
We set out to generate transgenic mice overexpressing PEX1 specifically in the heart under the control of the α-MHC promoter. Unfortunately, overexpression of PEX1 in the embryo interfered with normal development; expressing embryos were lost between E9.5 and E12.5 with evidence of cardiac enlargement and failure. To avoid interfering with normal development and to specifically manipulate PEX1 expression in postnatal hearts, we generated two transgenic lines with conditional cardiac-specific overexpression of PEX1 by crossing CAT-PEX1 and α-MHCMerCreMer mice (38) (Fig. 4A). Adult (150-day-old) a-MHCMerCreMer (controls) and α-MHCMerCreMer/CAT-PEX1 mice (referred to as PEX1 mice) were treated with Tamoxifen (0.5 mg/kg/day, intraperitoneally). At 1 and 2 weeks after treatment, groups of mice were analyzed by echocardiography and then killed to harvest hearts for immunohistochemistry, protein, or RNA analysis. Following Tamoxifen treatment, both transgenic lines had increased PEX1 expression compared with vehicle-treated transgenics or to Tamoxifen-treated α-MHCMerCreMer littermates which were used as controls in the study. First, echocardiography revealed a significant increase in left ventricular mass in PEX1 mice as evidenced by the increase in interventricular septum and left ventricular posterior wall thickness (Fig. 4D). On the other hand, neither ejection fraction nor left ventricular internal dimensions differed between transgenics and controls, indicating that cardiac function was preserved. At death, heart weight was significantly increased in the transgenic mice (Fig. 4D). Histologic analysis clearly showed that PEX1-overexpressing hearts were proportionately larger; trichrome staining did not reveal evidence of fibrosis, but myocyte size was increased in PEX1 mice relative to controls (Fig. 4, B and C). Because pathologic hypertrophy is often accompanied by apoptosis, we measured myocyte apoptosis using TUNEL assays; hearts overexpressing PEX1 did not have increased TUNEL-positive myocytes, rather they showed a consistent reduction in the percentage of apoptotic nuclei relative to control (0.1% versus 0.3% for Ctrl; p < 0.05). Immunohistochemistry and qPCR analysis showed up-regulated ANF levels in PEX1-expressing ventricles (Fig. 5, A and B). Other markers of cardiac hypertrophy were also increased in these tissues (ACTA1 and β-MHC). GATA4 mRNA levels, however, remained unchanged. Importantly, no significant change was observed in genes which are dysregulated in pathologic hypertrophy (42) such as SERCA2, NCX1, α-MHC, and COL1A1 (Fig. 5B). In summary, cardiac-specific overexpression of PEX1 in adult hearts induced cardiac hypertrophy with maintained systolic function, in the absence of histologic and genetic features of pathologic remodeling even with significant and sustained up-regulation of PEX1.
FIGURE 4.
Conditional overexpression of PEX1 in adult mice hearts leads to cardiac hypertrophy. A, strategy used for conditional expression of PEX1 using the Cre/LoxP system. α-MHCMerCreMer mice were crossed with CAT-PEX1 transgenic mice to obtain a conditional transgenic line that overexpresses PEX1 strictly in cardiomyocytes once Tamoxifen is administered. Right, transgenic mice receiving tamoxifen showing increased PEX1 protein levels, whereas those injected with peanut oil and controls receiving tamoxifen showing no change. B, image magnified ×1.25 of trichrome-stained heart sections from 150-day-old α-MHCMerCreMer or α-MHCMerCreMer/CAT-PEX1 mice treated with Tamoxifen. Notice the hypertrophied heart when PEX1 expression is induced. Magnification ×63 shows increased myocyte size. C, relative cell surface area was measured across 10 fields (×40) in transgenic (TG) and control (Ctrl) mice (2 of each). *, p < 0.05 versus control. D, echocardiographic analysis of mice hearts 1 week and 2 weeks after Tamoxifen administration. Heart weight-to-body weight ratios (HW/BW) are also shown. IVS, interventricular septum thickness; LVPW, left ventricular posterior wall thickness; LVID, left ventricular internal dimension; LV mass, left ventricular mass; EF, ejection fraction. Notice how ejection fraction was not changed despite the increase in left ventricular mass/body weight ratio suggestive of preserved heart function. The data are means ± S.E. (error bars; n = 4–5 for each group). *, p < 0.05 versus control (Ctrl; 1 week); #, p < 0.05 versus control (2 weeks).
FIGURE 5.
Genetic changes in PEX1-expressing hearts. A, histologic sections stained with either the PEX1 antibody or ANF antibody and counterstained with methyl green. Note the increased nuclear distribution of PEX1 and the concomitant up-regulation of ANF in the Tamoxifen-treated transgenics versus Tamoxifen-treated controls. B, -fold change in transcript levels relative to controls for PEX1, ANF, BNP, ACTA1, α-MHC, β-MHC, GATA4, SERCA2, NCX1, and COL1A1 in hearts of transgenic mice after 1 and 2 weeks of Tamoxifen treatment (ratio of transgenic (TG) to controls (Ctrl)). The controls of 1 and 2 weeks of Tamoxifen treatment were set at 1.The data are means ± S.E. (error bars; n = 4–5 for each group). S16 was used as the normalizer gene. *, p < 0.05 versus control (1 week); #, p < 0.05 versus control (2 weeks).
DISCUSSION
Essential Role of PEX1 in Endothelin Signaling
In this paper, we provide evidence that the zinc finger transcription factor PEX1 is required for cellular and genetic response of cardiomyocytes to ET-1. Previously we showed that cardiomyocyte response to the α1-adrenergic agonist PE also required PEX1 (32). Cardiomyocyte signaling through α1-adrenergic and ETA G protein-coupled receptors involves the activation of several signaling cascades and transcription factors, including proto-oncogenes, STAT proteins, NFAT, NF-κB, serum response factor, and GATA4 (22, 43). However, few transcription factors have been shown to be required for nuclear signaling by the G protein-coupled receptors. Several lines of evidence support our conclusion that PEX1 is an obligatory effector of ET-1 signaling in cardiomyocytes. First, knocking down PEX1 abrogates ET-1-dependent myofibrillar reorganization and ANF up-regulation (Fig. 1A). Second, mutation of the PEX1 binding site on the ANF promoter reduced ET-1 activation by >70% (Fig. 2A). These effects are reminiscent of those observed when the GATA binding site of the ANF promoter is mutated (Fig. 2A) and when GATA4 levels are similarly down-regulated in cardiomyocytes (12). Interestingly, ET-1 stimulation enhances PEX1 transcriptional activity through a PKC-dependent pathway (Fig. 2, C–F), whereas ET-1 activation of GATA4 involves MAPK (12). PEX1 contains two putative PKC phosphorylation sites that are evolutionary conserved. We found that PKC-β indeed phosphorylates PEX1 and potentiates its transcriptional activity and its synergy with GATA4 (Fig. 2, G and H). No significant effect of ET-1 or PKC on PEX1 nuclear localization was evident. These results raise the intriguing possibility that a multiprotein complex containing GATA4 and PEX1 may transduce nuclear signaling by ET-1 and that the different downstream kinases target specific components therein. Molecular characterization of GATA4 or PEX1 protein complexes will provide valuable insight into ET-1 nuclear signaling pathways.
Other than its role in cardiomyocyte growth, ET-1 is essential for normal embryonic development, and disruption of the ET-1 or ETA genes in mice results in craniofacial abnormalities and great vessel defects (44, 45). Postnatally, ET-1 exerts profound effects on vascular smooth muscle cells, and its dysregulation is linked to numerous vascular diseases (46). In addition to cardiomyocytes, PEX1 is expressed in smooth muscle cells as well as in other ET-1 target tissues including lung and kidney (32). These cells also express members of the GATA family, most notably GATA6 which plays a critical role in vascular smooth muscle cell growth and differentiation as well as in lung and great vessel development (47–51). In the future, it will be interesting to test whether cooperative interaction between PEX and GATA proteins may represent a general mechanism for nuclear signaling by endothelin through its ETA receptor.
PEX1, a Rate-limiting Regulator of Cardiomyocyte Hypertrophy
Previously, we showed that in genetic models of progressive hypertrophy PEX1 mRNA and protein levels are induced at the onset of cardiac hypertrophy, as observed in spontaneously hypertensive rats or in the angiotension receptor transgenic mice (32). Additionally, we found that two hypertrophic stimuli, ET-1 (this study) and α1-adrenergic agonists (32) enhance PEX1 activity or level. Given that PEX1 levels decrease in post-mitotic cardiomyocytes (32), we tested whether up-regulation of PEX1 in postnatal cardiomyocytes participates in the development of hypertrophy. This was achieved by adenovirus-mediated overexpression of PEX1 in cultured postnatal cardiomyocyte and by an inducible cardiac-specific transgenic mouse model.
Both in vitro and in vivo, PEX1 induced genetic and phenotypic features of cardiac hypertrophy. In vitro, PEX1 overexpression was sufficient to induce myofibrillar reorganization and up-regulate ACTA1 mRNAs levels. These effects mimic those obtained when GATA4 is similarly overexpressed (12), but neither GATA4 transcripts nor protein levels were induced by PEX1 (Fig. 3, A and B). Likewise, overexpression of GATA4 did not result in increased PEX1 levels (Fig. 3A). Previously we reported that knockdown of PEX1 had no effect on GATA4 levels and vice versa. These results suggest that PEX1 and GATA4 are not hierarchically related; rather, they are consistent with a role for PEX1 and GATA4 as mutual coactivators of a genetic program underlying cardiac hypertrophy.
Interestingly, induction of PEX1 expression in adult hearts resulted in cardiac hypertrophy and activation of ANF. The reason why ANF was up-regulated by PEX1 in adult hearts but not in neonatal cardiomyocytes is not evident at this stage but may suggest that endogenous PEX1 levels in neonate cardiomyocytes, where they are much higher than in adult ventricles, are sufficient for maximal ANF gene transcription. Alternatively, other neurohormonal signals present in vivo but not in vitro may be needed to recruit other PEX1 collaborators and induce ANF. Heart failure is generally accompanied by morphologic and molecular changes that include disruption of sarcomeric calcium homeostasis evidenced by a decrease in SERCA2 and a reciprocal increase in sodium-calcium exchanger III (42). Increased myocyte apoptosis and collagen deposition (associated with enhanced COL1A1 and COL3A1 levels) are characteristic features of maladaptive hypertrophy leading to cardiac remodeling. Although mice hearts overexpressing PEX1 were significantly hypertrophied, they neither showed sign of fibrosis, nor abnormal remodeling. Changes in their genetic profile did not resemble those found in failing hearts, and importantly, cardiac function was not altered by 20-fold increase in PEX1 levels. In this respect, a difference in the phenotype of mice with inducible PEX1 or GATA4 in adult hearts using the same approach was noted. Indeed, whereas ANF levels were similarly increased in both cases, increased GATA4 levels were not accompanied by detectable evidence of cardiac hypertrophy, a result consistent with the reported phenotype of GATA4 overexpression in transgenic mice using a different inducible system (52). Thus, PEX1 may be an obligatory cofactor of GATA4 required for hypertrophic growth.
Although a handful of transcription factors such as GATA4, myocardin (34), and serum response factor (53) have been shown to be required for hypertrophic signaling in vitro, the consequences of their in vivo expression suggest that they are either insufficient to induce cardiac hypertrophy of the adult heart (as for GATA4) or that their overexpression results in lethal cardiomyopathies, severe cardiac dysfunction, and premature death as reported for serum response factor (54). In contrast, enhanced PEX1 expression in adult hearts induces left ventricular hypertrophy with preserved cardiac function, raising the likelihood that PEX1 may be a critical effector of neurohormonal signaling activated during early stages of hypertrophy.
Acknowledgments
We thank Chantal Lefèbvre, Michel Robillard, and Annie Lavallée for technical assistance; Hélène Touchette for secretarial support; and Dr. Jae-Won Soh for the PKCβ WT and DN.
This work was supported by grants from the Canadian Institutes of Health Research.
- ANF
- atrial natriuretic factor
- ACTA1
- skeletal muscle α-actin
- ET-1
- endothelin-1
- BNP
- brain natriuretic peptide
- MHC
- myosin heavy chain
- PE
- phenylephrine
- PERE
- phenylephrine response element
- PEX1
- phenylephrine-induced complex 1
- qPCR
- quantitative PCR
- ZFP
- zinc finger protein.
REFERENCES
- 1. MacLellan W. R., Schneider M. D. (2000) Annu. Rev. Physiol. 62, 289–319 [DOI] [PubMed] [Google Scholar]
- 2. Olson E. N., Schneider M. D. (2003) Genes Dev. 17, 1937–1956 [DOI] [PubMed] [Google Scholar]
- 3. Frey N., Olson E. N. (2003) Annu. Rev. Physiol. 65, 45–79 [DOI] [PubMed] [Google Scholar]
- 4. Ito H., Hirata Y., Adachi S., Tanaka M., Tsujino M., Koike A., Nogami A., Murumo F., Hiroe M. (1993) J. Clin. Invest. 92, 398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwanaga Y., Kihara Y., Hasegawa K., Inagaki K., Yoneda T., Kaburagi S., Araki M., Sasayama S. (1998) Circulation 98, 2065–2073 [DOI] [PubMed] [Google Scholar]
- 6. Wei C. M., Lerman A., Rodeheffer R. J., McGregor C. G., Brandt R. R., Wright S., Heublein D. M., Kao P. C., Edwards W. D., Burnett J. C., Jr. (1994) Circulation 89, 1580–1586 [DOI] [PubMed] [Google Scholar]
- 7. Sakai S., Miyauchi T., Kobayashi M., Yamaguchi I., Goto K., Sugishita Y. (1996) Nature 384, 353–355 [DOI] [PubMed] [Google Scholar]
- 8. Miyauchi T., Masaki T. (1999) Annu. Rev. Physiol. 61, 391–415 [DOI] [PubMed] [Google Scholar]
- 9. Ammarguellat F. Z., Gannon P. O., Amiri F., Schiffrin E. L. (2002) Hypertension 39, 679–684 [DOI] [PubMed] [Google Scholar]
- 10. Spieker L. E., Noll G., Ruschitzka F. T., Lüscher T. F. (2001) Heart Fail. Rev. 6, 301–315 [DOI] [PubMed] [Google Scholar]
- 11. Sakai S., Miyauchi T., Sakurai T., Kasuya Y., Ihara M., Yamaguchi I., Goto K., Sugishita Y. (1996) Circulation 93, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 12. Charron F., Tsimiklis G., Arcand M., Robitaille L., Liang Q., Molkentin J. D., Meloche S., Nemer M. (2001) Genes Dev. 15, 2702–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McMullen J. R., Shioi T., Zhang L., Tarnavski O., Sherwood M. C., Dorfman A. L., Longnus S., Pende M., Martin K. A., Blenis J., Thomas G., Izumo S. (2004) Mol. Cell. Biol. 24, 6231–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Molkentin J. D., Dorn G. W., 2nd (2001) Annu. Rev. Physiol. 63, 391–426 [DOI] [PubMed] [Google Scholar]
- 15. Oudit G. Y., Sun H., Kerfant B. G., Crackower M. A., Penninger J. M., Backx P. H. (2004) J. Mol. Cell. Cardiol. 37, 449–471 [DOI] [PubMed] [Google Scholar]
- 16. Wilkins B. J., Molkentin J. D. (2004) Biochem. Biophys. Res. Commun. 322, 1178–1191 [DOI] [PubMed] [Google Scholar]
- 17. Zechner D., Thuerauf D. J., Hanford D. S., McDonough P. M., Glembotski C. C. (1997) J. Cell Biol. 139, 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang T., Brown J. H. (2004) Cardiovasc. Res. 63, 476–486 [DOI] [PubMed] [Google Scholar]
- 19. Sugden P. H. (2003) J. Mol. Cell. Cardiol. 35, 871–886 [DOI] [PubMed] [Google Scholar]
- 20. Shubeita H. E., McDonough P. M., Harris A. N., Knowlton K. U., Glembotski C. C., Brown J. H., Chien K. R. (1990) J. Biol. Chem. 265, 20555–20562 [PubMed] [Google Scholar]
- 21. Ito H., Hirata Y., Hiroe M., Tsujino M., Adachi S., Takamoto T., Nitta M., Taniguchi K., Marumo F. (1991) Circ. Res. 69, 209–215 [DOI] [PubMed] [Google Scholar]
- 22. Akazawa H., Komuro I. (2003) Circ. Res. 92, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 23. Liang Q., Molkentin J. D. (2002) J. Mol. Cell. Cardiol. 34, 611–616 [DOI] [PubMed] [Google Scholar]
- 24. Liang Q., Wiese R. J., Bueno O. F., Dai Y. S., Markham B. E., Molkentin J. D. (2001) Mol. Cell. Biol. 21, 7460–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morimoto T., Hasegawa K., Kaburagi S., Kakita T., Wada H., Yanazume T., Sasayama S. (2000) J. Biol. Chem. 275, 13721–13726 [DOI] [PubMed] [Google Scholar]
- 26. Wang J., Paradis P., Aries A., Komati H., Lefebvre C., Wang H., Nemer M. (2005) Mol. Cell. Biol. 25, 9829–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yanazume T., Hasegawa K., Morimoto T., Kawamura T., Wada H., Matsumori A., Kawase Y., Hirai M., Kita T. (2003) Mol. Cell. Biol. 23, 3593–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang Q., De Windt L. J., Witt S. A., Kimball T. R., Markham B. E., Molkentin J. D. (2001) J. Biol. Chem. 276, 30245–30253 [DOI] [PubMed] [Google Scholar]
- 29. Aries A., Paradis P., Lefebvre C., Schwartz R. J., Nemer M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oka T., Maillet M., Watt A. J., Schwartz R. J., Aronow B. J., Duncan S. A., Molkentin J. D. (2006) Circ. Res. 98, 837–845 [DOI] [PubMed] [Google Scholar]
- 31. Bisping E., Ikeda S., Kong S. W., Tarnavski O., Bodyak N., McMullen J. R., Rajagopal S., Son J. K., Ma Q., Springer Z., Kang P. M., Izumo S., Pu W. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14471–14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Debrus S., Rahbani L., Marttila M., Delorme B., Paradis P., Nemer M. (2005) Mol. Cell. Biol. 25, 8669–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ardati A., Nemer M. (1993) EMBO J. 12, 5131–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xing W., Zhang T. C., Cao D., Wang Z., Antos C. L., Li S., Wang Y., Olson E. N., Wang D. Z. (2006) Circ. Res. 98, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 35. Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 36. Oh J., Wang Z., Wang D. Z., Lien C. L., Xing W., Olson E. N. (2004) Mol. Cell. Biol. 24, 8519–8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Charron F., Paradis P., Bronchain O., Nemer G., Nemer M. (1999) Mol. Cell. Biol. 19, 4355–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sohal D. S., Nghiem M., Crackower M. A., Witt S. A., Kimball T. R., Tymitz K. M., Penninger J. M., Molkentin J. D. (2001) Circ. Res. 89, 20–25 [DOI] [PubMed] [Google Scholar]
- 39. Petrich B. G., Molkentin J. D., Wang Y. (2003) FASEB J. 17, 749–751 [DOI] [PubMed] [Google Scholar]
- 40. Georges R., Nemer G., Morin M., Lefebvre C., Nemer M. (2008) Mol. Cell. Biol. 28, 4052–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morin S., Paradis P., Aries A., Nemer M. (2001) Mol. Cell. Biol. 21, 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Houser S. R., Piacentino V., 3rd, Mattiello J., Weisser J., Gaughan J. P. (2000) Trends Cardiovasc. Med. 10, 101–107 [DOI] [PubMed] [Google Scholar]
- 43. Clerk A., Cullingford T. E., Fuller S. J., Giraldo A., Markou T., Pikkarainen S., Sugden P. H. (2007) J. Cell. Physiol. 212, 311–322 [DOI] [PubMed] [Google Scholar]
- 44. Clouthier D. E., Hosoda K., Richardson J. A., Williams S. C., Yanagisawa H., Kuwaki T., Kumada M., Hammer R. E., Yanagisawa M. (1998) Development 125, 813–824 [DOI] [PubMed] [Google Scholar]
- 45. Kurihara Y., Kurihara H., Oda H., Maemura K., Nagai R., Ishikawa T., Yazaki Y. (1995) J. Clin. Invest. 96, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dhaun N., Goddard J., Kohan D. E., Pollock D. M., Schiffrin E. L., Webb D. J. (2008) Hypertension 52, 452–459 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y., Goss A. M., Cohen E. D., Kadzik R., Lepore J. J., Muthukumaraswamy K., Yang J., DeMayo F. J., Whitsett J. A., Parmacek M. S., Morrisey E. E. (2008) Nat. Genet. 40, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang H., Lu M. M., Zhang L., Whitsett J. A., Morrisey E. E. (2002) Development 129, 2233–2246 [DOI] [PubMed] [Google Scholar]
- 49. Keijzer R., van Tuyl M., Meijers C., Post M., Tibboel D., Grosveld F., Koutsourakis M. (2001) Development 128, 503–511 [DOI] [PubMed] [Google Scholar]
- 50. Lepore J. J., Mericko P. A., Cheng L., Lu M. M., Morrisey E. E., Parmacek M. S. (2006) J. Clin. Invest. 116, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lepore J. J., Cappola T. P., Mericko P. A., Morrisey E. E., Parmacek M. S. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 309–314 [DOI] [PubMed] [Google Scholar]
- 52. Heineke J., Auger-Messier M., Xu J., Oka T., Sargent M. A., York A., Klevitsky R., Vaikunth S., Duncan S. A., Aronow B. J., Robbins J., Crombleholme T. M., Molkentin J. D. (2007) J. Clin. Invest. 117, 3198–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nelson T. J., Balza R., Jr., Xiao Q., Misra R. P. (2005) J. Mol. Cell. Cardiol. 39, 479–489 [DOI] [PubMed] [Google Scholar]
- 54. Zhang X., Azhar G., Chai J., Sheridan P., Nagano K., Brown T., Yang J., Khrapko K., Borras A. M., Lawitts J., Misra R. P., Wei J. Y. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H1782–H1792 [DOI] [PubMed] [Google Scholar]