Abstract
Arenaviruses cause acute hemorrhagic fevers with high mortality. Entry of the virus into the host cell is mediated by the viral envelope glycoprotein, GPC. In contrast to other class I viral envelope glycoproteins, the mature GPC complex contains a cleaved stable signal peptide (SSP) in addition to the canonical receptor-binding (G1) and transmembrane fusion (G2) subunits. SSP is critical for intracellular transport of the GPC complex to the cell surface and for its membrane-fusion activity. Previous studies have suggested that SSP is retained in GPC through interaction with a zinc-binding domain (ZBD) in the cytoplasmic tail of G2. Here we used NMR spectroscopy to determine the structure of Junín virus (JUNV) ZBD (G2 residues 445–485) and investigate its interaction with a conserved Cys residue (Cys-57) in SSP. We show that JUNV ZBD displays a novel fold containing two zinc ions. One zinc ion is coordinated by His-447, His-449, Cys-455, and His-485. The second zinc ion is coordinated by His-459, Cys-467, and Cys-469 and readily accepts Cys-57 from SSP as the fourth ligand. Our studies describe the structural basis for retention of the unique SSP subunit and suggest a mechanism whereby SSP is positioned in the GPC complex to modulate pH-dependent membrane fusion.
Keywords: NMR, Protein Structure, Protein-Protein Interactions, Viral Protein, Zinc finger, GPC, Arenavirus, Junin Virus, Virus Envelope Glycoprotein, Zinc-binding Domain
Introduction
Arenavirus species are endemic in rodent populations worldwide (1), and some viruses can be transmitted to humans to cause hemorrhagic fevers with up to 30% mortality (2, 3). It is estimated that several hundred thousand people are infected with Lassa fever virus (LASV)2 annually in western Africa, and thousands succumb to the disease (4, 5). In the Americas, at least four arenavirus species are recognized to cause fatal hemorrhagic fevers: Junín (JUNV), Machupo (MACV), Guanarito (GTOV), and Sabiá (SABV) (2), and new pathogenic species continue to emerge throughout the world (6, 7). Prophylactic vaccines are not currently available and treatment options are extremely limited. The hemorrhagic fever arenaviruses are therefore acknowledged by the United States government to pose a significant threat for public health and biodefense (8).
The arenaviruses are enveloped viruses whose bipartite RNA genome encodes four proteins using an ambisense coding strategy (9). Virus entry into its host cell is initiated by the virus envelope glycoprotein (GPC) binding to a host-cell receptor, transferrin receptor 1 (TfR1) in the pathogenic New World viruses (10) and α-dystroglycan in the Old World viruses (11), upon which the virion particle is endocytosed (12). GPC-mediated fusion of the viral and endosomal membranes is activated by acidic pH in the maturing endosome (13) and proceeds through a series of conformational changes in the G2 ectodomain that culminates in formation of the six-helix bundle structure typical of class I viral fusion proteins (14–18). Upon membrane fusion, the viral ribonucleoprotein core is deposited into the cell cytoplasm for replication.
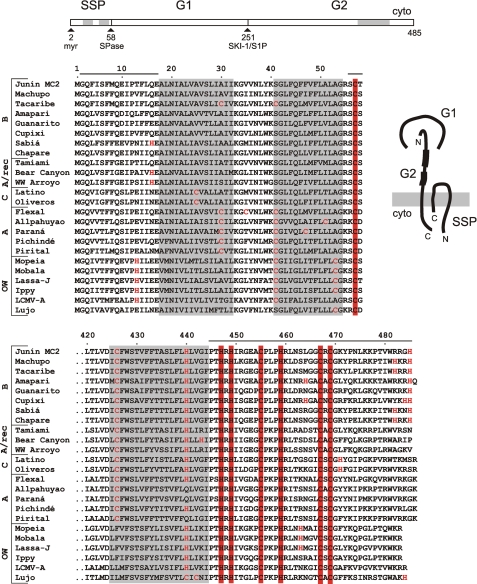
GPC is unique among viral fusion glycoproteins in that the mature complex retains its cleaved stable signal peptide (SSP) associated with the prototypical receptor-binding (G1) and transmembrane fusion (G2) subunits (19, 20) (Fig. 1). SSP is generated by signal peptidase (SPase) cleavage but unlike conventional signal peptides is stable, unusually long (58 amino acids), myristoylated (20), and contains two hydrophobic segments that each span the lipid bilayer (21) (Fig. 1). Both N and C termini of SSP reside in the cytosol. The short cytoplasmic C-terminal tail of SSP contains an absolutely conserved cysteine residue (Cys-57) (Fig. 1) that is required for retention of SSP in the GPC complex (22). Residues flanking Cys-57 are variable among arenaviruses (Fig. 1) and are important for SPase cleavage but not for SSP retention in GPC (22, 23). Interestingly, SSP can be expressed as a separate polypeptide and will associate in trans with a recombinant G1G2 precursor to reconstitute the functional GPC complex (19, 22, 24).
FIGURE 1.
Subunit organization of the arenaviral GPC complex and comparison of SSP and C-terminal G2 amino acid sequences. A schematic drawing of the JUNV GPC open-reading frame is displayed on top. Sequential biosynthetic cleavage events by the cellular signal peptidase (SPase) and SKI-1/S1P protease respectively generate SSP and the mature G1 and G2 subunits. Membrane-spanning domains (hφ1 and hφ2 in SSP (21) and the transmembrane region in G2) are indicated by gray shading. SSP is myristoylated (myr) at glycine 2 (20). A cartoon at the right illustrates the proposed subunit organization and membrane topology of the mature GPC complex. The shaded rectangle depicts the lipid bilayer. The N and C termini of SSP and the C terminus of G2 are located on the cytoplasmic (cyto) side. Thicker lines in the ectodomain of G2 represent heptad-repeat sequences that refold to form the six-helix-bundle structure and drive GPC-mediated membrane fusion (51). Comparisons of the amino acid sequences of SSP as well as the transmembrane and cytoplasmic domains of G2 are shown below the open-reading frame schematic. Arenaviruses are divided into Old World species (OW) and New World species, which are grouped by phylogenetic clades (A, A/recombinant, B, and C) (52). Invariably conserved cysteine and histidine residues in SSP and G2 involved in zinc binding are highlighted by red fill, whereas non-conserved cysteine and histidine residues are in red type. Horizontal lines above the sequences define the JUNV peptides used in this work: the N-terminal cytoplasmic domain of SSP (SSP-N; residues 2–17), the C-terminal cytoplasmic domain of SSP (acetyl-SCT; 56–58), and the cytoplasmic domain of G2 (ZBD; 445–485).
SSP is required for transport of the G1G2 precursor protein from the endoplasmic reticulum (ER). The interaction with SSP appears to mask endogenous dibasic ER retention/retrieval signals in the cytoplasmic domain of G2 (25) to allow transit through the Golgi compartment, where the G1G2 precursor is cleaved by the cellular SKI-1/S1P protease to form the mature G1 and G2 subunits (Fig. 1) (26–28). As with other class I viral fusion proteins, proteolytic cleavage of the G1G2 precursor is required to render the GPC complex competent for membrane fusion (20, 26, 27).
In addition to its role in GPC transport, SSP association is also essential for the membrane-fusion activity of the GPC complex. In particular, charge mutations at the Lys-33 position in the ectodomain loop of SSP have been shown to systematically modulate the pH at which membrane fusion is activated (24). Fusion deficiencies of Lys-33 mutants can in turn be rescued by specific mutations in the transmembrane domain and membrane-proximal ectodomain of G2 (29). The putative SSP-G2 interface is hypothesized to stabilize the prefusion state of GPC at neutral pH, and trigger the conformational changes leading to membrane fusion at acidic pH (24, 29). Small molecule compounds (30, 31) that target this pH-sensitive interface act to stabilize the prefusion complex against pH-induced activation (32), and thereby prevent virus entry and arenaviral disease.
The retention of SSP in the GPC complex is solely dependent on the transmembrane and cytoplasmic domains of G2; the entire ectodomain of GPC can be replaced with an unrelated sequence without affecting SSP association (25). The cytoplasmic domain of G2 contains an array of six invariant cysteine and histidine residues (Fig. 1), each of which is essential for association with SSP (33). Importantly, a recombinant fusion protein that contains the cytoplasmic domain of JUNV G2 has recently been shown to bind zinc with a dissociation constant (Kd) in the subnanomolar range (33). The three membrane-proximal cysteine and histidine residues in the cytoplasmic domain of G2 (His-447, His-449, and Cys-455) are required for this high-affinity binding, and we speculated that the remaining residues (His-459, Cys-467, and Cys-469) might form a second, intersubunit zinc-binding site in conjunction with Cys-57 to retain and position SSP in the complex (33).
Detailed information regarding the structure and organization of the tripartite GPC complex is currently not available. Although atomic level structure of the soluble G1 subunit, alone and in complex with human TfR1, has recently been reported (34, 35), the structural basis for the critical interactions between SSP and G2 remains unknown. In order to provide a molecular framework for the function of GPC and the role of its unique SSP subunit, we have here determined the structure of the cytoplasmic zinc-binding domain of JUNV G2 by NMR spectroscopy, and investigated its interaction with SSP.
EXPERIMENTAL PROCEDURES
Synthetic Peptides
N-Acetylated synthetic peptides corresponding to the JUNV GPC residues 56–58 (acetyl-SCT) and 445–485 (JUNV zinc-binding domain, JUNV ZBD) were obtained from GenScript Corp. at >98% and >95% purity, respectively. Acetyl-SCT represents the C-terminal cytoplasmic sequence of SSP while JUNV ZBD comprises the complete cytoplasmic sequence of the G2 subunit (Fig. 1). The peptides were used without further purification unless stated otherwise. Concentrations were determined spectrophotometrically at 280 nm for JUNV ZBD and at 214 nm for acetyl-SCT.
Isothermal Titration Calorimetry (ITC)
To reduce metal ion contamination, water and all buffer stock solutions that were used to prepare samples for ITC experiments had been passed through Chelex 100 resin. JUNV ZBD was dialyzed extensively against ITC buffer (20 mm MOPS, pH 8.0, 150 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine (TCEP)) supplemented with 10 mm EDTA to remove any metal ions that might have been carried over from the synthesis steps. The peptide was subsequently dialyzed into ITC buffer to remove EDTA. ITC experiments were performed in a VP-ITC (MicroCal, GE Healthcare) instrument at 25 °C. To minimize the possibility of metal ion contamination in the calorimeter, the ITC cell and syringe were soaked overnight in ITC buffer supplemented with 10 mm EDTA and afterward washed extensively with ITC buffer to remove residual EDTA. 10-μl aliquots of 20 μm ZnCl2 in ITC buffer were injected into the calorimeter cell containing 1.43 ml of 0.85 μm JUNV ZBD in ITC buffer, and the heats of binding were recorded. To investigate the interaction between JUNV ZBD and acetyl-SCT, 10-μl aliquots of 450 μm acetyl-SCT in ITC buffer containing 100 μm ZnCl2 (Zn-ITC buffer) were injected into the calorimeter cell holding 1.43 ml of 50 μm JUNV ZBD that had been dialyzed into Zn-ITC buffer. In control experiments, 10-μl aliquots of 20 μm ZnCl2 in ITC buffer or 450 μm acetyl-SCT in Zn-ITC buffer were injected into the calorimeter cell containing 1.43 ml of ITC or Zn-ITC buffer respectively to measure the heats of dilution. The data were analyzed with the ORIGIN software provided with the ITC instrument.
NMR Spectroscopy and Structure Determination
The samples for NMR experiments contained 0.7 mm JUNV ZBD, 50 mm 2H11-Tris, 2.5 mm 2H16-TCEP, and 4–5 mm ZnSO4 in 90% H2O/10% 2H2O, pH 7.2, or in 2H2O, pH* 7.0 (uncorrected pH meter reading). Air in NMR tubes was replaced with argon for additional protection of cysteine residues from oxidation. Two-dimensional double quantum filtered COSY, TOCSY (supplemental Fig. S1), NOESY (supplemental Fig. S2), E-COSY, and 1H-15N (supplemental Fig. S3), 1H-13C aliphatic (supplemental Fig. S4), and 1H-13C aromatic (supplemental Fig. S5) HSQC spectra at natural abundance were acquired at 25 °C on a Varian 600 MHz NMR System equipped with a cold probe. The data were processed and analyzed with Felix 2004 (Accelrys Inc./Felix NMR Inc.), and sequential assignments were obtained using standard procedures (36) (see supplemental Figs. S1 and S2 for details).
Structures were initially calculated with XPLOR-NIH 2.22 (37) using only distance restraints derived from the two-dimensional NOESY spectrum. Calculations were also performed with Aria1.2 (38) in conjunction with CNS 1.0 (39), using crosspeak volumes from the two-dimensional NOESY spectrum with manual assignments either retained, or discarded. No zinc atoms or constraints for zinc coordination were included in these preliminary calculations. All calculation protocols yielded similar structures that contained two distinct clusters of residues posed to coordinate zinc. In subsequent structure calculations, geometrical constraints were included to coordinate zinc ions by these two clusters. The constraints were based on typical bond lengths and angles reported for tetrahedral protein zinc-binding sites (40). In addition, χ1 was restrained to 60° ± 30° for residues that exhibited small 3JHαHβ(< 5 Hz) for both Hβ atoms as judged from E-COSY, and hydrogen bond restraints were introduced for a short anti-parallel β-sheet (His-447—His-449, Val-480—Arg-482) that was observed in the preliminary structures and supported by manually assigned NOE pattern. Eight independent Aria runs were performed to evaluate convergence and reproducibility. The calculations employed a total of 8 hydrogen bond restraints, 4 χ1 dihedral angle restraints, and 1377 volumes from two-dimensional NOESY spectrum. Manual assignments of NOESY crosspeaks were retained. Default parameters and options were generally used for the calculations. Each run yielded twenty structures, seven of which were refined in explicit water. Structures in one run exhibited slightly higher energy and were not considered further. Three refined structures with the lowest energy from each of the other seven runs were pooled together, resulting in an ensemble of 21 structures. The statistics for the ensemble is summarized in Table 1. Inclusion of the hydrogen bond and zinc coordination restraints had negligible effect on the calculated structures, as judged by comparison of the final ensemble with preliminary models that did not employ such restraints.
TABLE 1.
Structural statistics for JUNV ZBD
| Number of NOE distance restraints in the last ARIA iteration | |
| Unambiguous | 762 ± 2 |
| Ambiguous | 87 ± 2 |
| RMSD from experimental restraints | |
| NOE distance restraints (Å) | 0.018 ± 0.004 |
| Dihedral angle restraints (°) | 0 ± 0 |
| Number of experimental restraint violations | |
| NOE violations > 0.5 Å | 0 ± 0 |
| NOE violations > 0.3 Å | 0.6 ± 0.7 |
| Dihedral angle violations > 5° | 0 ± 0 |
| RMSD from idealized geometrya | |
| Bonds (Å) | 0.0043 ± 0.0003 |
| Angles (°) | 0.61 ± 0.04 |
| Impropers (°) | 1.7 ± 0.1 |
| Distribution of ϕ, ψ dihedral angles in Ramachandran plot (56) | |
| The most favored regions (%) | 68.7 |
| Additional allowed regions (%) | 24.1 |
| Generously allowed regions (%) | 3.7 |
| Disallowed regions (%) | 3.5b |
| RMSD of residues 445–485 from mean coordinatesc | |
| Backbone atoms (N, Cα, C) (Å) | 0.86 ± 0.21 |
| Heavy atoms (Å) | 1.31 ± 0.14 |
a Idealized covalent geometry is based on the parallhdg5.3.pro parameters (38).
b Residues that exhibit ϕ, ψ dihedral angles in the disallowed regions of Ramachandran plot in at least one structure include Thr-446, Arg-451, Ala-454, Arg-460, Ser-463, Arg-468, Cys-469, Lys-471, Leu-475, Lys-476, and Lys-477. Nine structures contain no residues with ϕ, ψ dihedral angles in the disallowed regions.
c Mean coordinates were obtained by averaging coordinates of the 21 calculated structures, which were first superposed using backbone atoms (N, Cα, C) of residues 445–485.
NMR Binding Studies
The interaction of acetyl-SCT with JUNV ZBD was investigated by NMR spectroscopy. A stock solution that contained 14 mm acetyl-SCT, 50 mm 2H11-Tris, and 10 mm 2H16-TCEP in 90% H2O/10% 2H2O, pH 7.1 was kept under argon to help preserve Cys-57 in the reduced state. Small aliquots of this acetyl-SCT stock solution were added with a Hamilton syringe to a screw-cap NMR tube with septum (Wilmad LabGlass) that contained 0.4 mm JUNV ZBD, 50 mm 2H11-Tris, 2.5 mm 2H16-TCEP, and 1.5 mm ZnSO4 in 90% H2O/10% 2H2O, pH 7.1 under argon. One-dimensional 1H NMR spectrum was recorded upon each addition to monitor progress of the titration. Finally, two-dimensional double quantum-filtered COSY, TOCSY, and NOESY spectra were acquired for a sample that contained 0.8 mm JUNV ZBD, 1.5 mm acetyl-SCT, 50 mm 2H11-Tris, 3.3 mm 2H16-TCEP, and 2 mm ZnSO4 in 90% H2O/10% 2H2O, pH 7.1. These spectra were used to obtain resonance assignments and to identify intermolecular contacts in the JUNV ZBD:acetyl-SCT complex.
Expression of Recombinant JUNV GPC
In these studies, SSP and the G1G2 precursor derived from the pathogenic JUNV strain MC2 (41) were translated as two separate polypeptides that associate in trans to reconstitute the native GPC complex (19, 22, 24); this obviates potential concerns regarding SPase cleavage. Vero cells were co-transfected with pcDNA3.1 (Invitrogen)-based plasmids that encoded CD4sp-GPC (a GPC precursor in which SSP is replaced by the conventional signal peptide of CD4) and SSP-term (in which a stop codon was introduced following the C-terminal SSP amino acid Thr-58) (24, 33). Expression was achieved using the bacteriophage T7 promoter present in the pcDNA3.1 vectors and infection by a recombinant vaccinia virus expressing the T7 polymerase (vTF7–3) (42), as described previously (20). Mutations in GPC were introduced with the QuikChange mutagenesis kit (Stratagene) and verified by DNA sequencing.
For the analysis of SSP incorporation into the GPC complex, cultures were metabolically labeled using 50 μCi/ml of 35S-ProMix (GE Healthcare), starting at 6 h after transfection and continuing overnight (20). Cells were lysed in cold Tris-saline buffer (50 mm Tris-HCl and 150 mm NaCl, pH 7.5) containing 1% Triton X-100 nonionic detergent and protease inhibitors (1 μg/ml each of aprotinin, leupeptin, and pepstatin), and the GPC complex was immunoprecipitated using the JUNV G1-specific monoclonal antibody QC03-BF11 (43) and protein A-Sepharose (Sigma). The antibody was obtained from the United States Centers for Disease Control and Prevention through the NIH Biodefense and Emerging Infections Research Resources Program. In some experiments, the isolated glycoprotein was additionally deglycosylated with peptide:N-glycosidase F (PNGase F, New England Biolabs) to better resolve the mature G1 and G2 subunits. Proteins were analyzed by polyacrylamide gel electrophoresis (PAGE) using NuPAGE 4–12% Bis-Tris gels (Invitrogen) and the recommended sample buffer containing lithium dodecylsulfate and reducing agent. Radiolabeled proteins were visualized with a Fuji FLA-3000G imager and the extent of SSP co-precipitation was quantitated with ImageGauge software (Fuji). 14C-methylated Rainbow proteins (GE Healthcare) served as molecular size markers.
GPC-mediated Cell-Cell Fusion
A vaccinia virus-based β-galactosidase fusion-reporter assay (44) was used to characterize the ability of GPC to mediate pH-dependent cell-cell fusion (20, 24). Briefly, Vero cells expressing GPC and infected by vTF-7 (which expresses bacteriophage T7 polymerase) (42) were co-cultured in 96-well microculture dishes with target cells infected with a recombinant vaccinia virus bearing the β-galactosidase gene under control of the T7 promoter (vCB21R-lacZ) (44). As previously described, cytopathic effect of vaccinia virus infection and the potential for cross-infection between effector and target cells in this assay were obviated through use of the vaccinia virus replication inhibitors cytosine arabinoside and rifampin (20, 24). Membrane fusion was initiated by incubation of the co-culture in medium that had been adjusted to pH 5.0 using HEPES and PIPES buffers. After 10 min at 37 °C, the cultures were restored to neutral medium and grown for 5 h to allow for expression of the β-galactosidase fusion-reporter gene, which was then quantitated using the chemiluminescent GalactoLite Plus substrate (Applied Biosystems) and a Tropix TR717 microplate luminometer.
RESULTS
The Cytoplasmic Domain of G2 Contains Two Zinc-binding Sites and Interacts with SSP
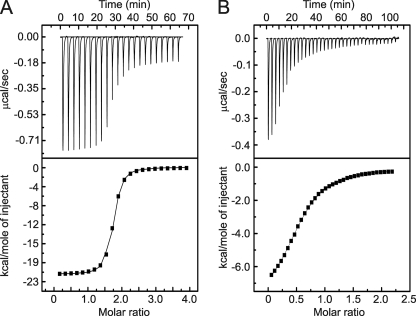
Genetic studies demonstrated that SSP retention in GPC depends on an array of six invariant cysteine and histidine residues in the cytoplasmic domain of G2 (33), as well as a conserved cysteine residue (Cys-57) in SSP (22). Using a recombinant fusion protein containing the cytoplasmic domain of G2, we also showed that three of the G2 residues are involved in high-affinity zinc binding. We speculated that the other three residues in G2 and Cys-57 in SSP might form a second zinc-binding site bridging the two subunits (33). To investigate this model, we determined the stoichiometry of zinc binding to a synthetic peptide comprising the entire cytoplasmic domain of JUNV G2 (residues 445–485; JUNV ZBD). Isothermal titration calorimetry (ITC) experiments demonstrated that JUNV ZBD binds 1.8 mol equivalents of Zn2+ with Kd in the nm range (Fig. 2A and Table 2). This result is consistent with the cytoplasmic domain of G2 containing two zinc-binding sites.
FIGURE 2.
Calorimetric titrations of JUNV ZBD with Zn2+ and acetyl-SCT. A, binding of Zn2+ to JUNV ZBD. B, binding of acetyl-SCT to JUNV ZBD in the presence of Zn2+. Representative ITC traces and binding curves fitted to the data are shown in the upper and lower panels, respectively. The experimental results demonstrate that JUNV ZBD contains two zinc-binding sites. Resolution of the subnanomolar binding event is precluded by the relatively high concentration of the peptide required to accurately measure the heats released upon binding. Hence, the data are dominated by the enthalpy and Kd of the low-affinity binding site.
TABLE 2.
Thermodynamic parameters for interaction of JUNV ZBD with Zn2+ and acetyl-SCT determined by ITC at 25 °C
| Ligand | Stoichiometry (N) | Dissociation constant (Kd) | Enthalpy (−ΔH) |
|---|---|---|---|
| Zn2+ | 1.81 ± 0.05a | 19.2 ± 0.4 nma | 21 ± 1 kcal mol−1a |
| Acetyl-SCT | 0.86 ± 0.04a | 23.5 ± 0.3 μma | 8.7 ± 0.3 kcal mol−1a |
a The values represent averages and standard deviations of duplicate experiments.
Because the penultimate Cys-57 residue in SSP is essential for SSP-G2 association in the GPC complex, we proposed that it might interact with the cytoplasmic domain of G2. To probe this hypothesis, we examined the binding of a synthetic tripeptide corresponding to the C-terminal cytoplasmic sequence of SSP (residues 56–58; acetyl-SCT) to JUNV ZBD in the presence of a saturating concentration of Zn2+. ITC studies demonstrated that acetyl-SCT interacts with JUNV ZBD with ∼1:1 stoichiometry and Kd ∼ 20 μm (Fig. 2B, Table 2). The moderate binding affinity likely reflects the lack of membrane anchorage in our peptide model of SSP-G2 interaction.
Solution Structure of JUNV ZBD
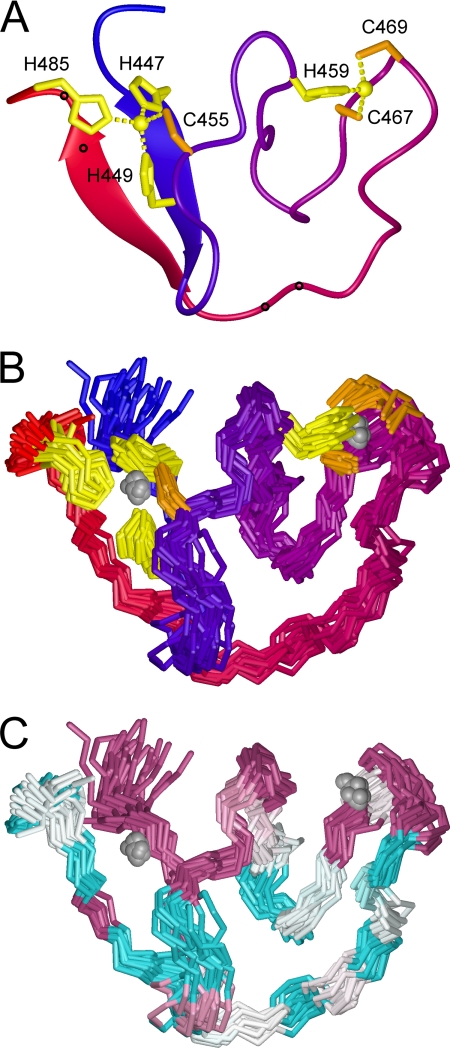
To provide structural context for the interaction between the cytoplasmic domain of G2 and SSP, we undertook to investigate the zinc-bound form of JUNV ZBD by NMR spectroscopy. Consistent with the ITC results, JUNV ZBD required two mole equivalents of Zn2+ to yield NMR spectra with well dispersed signals typical of folded proteins (see supplemental Fig. S6 for details). The two-dimensional NMR spectra of zinc-bound JUNV ZBD were amenable to analysis (supplemental Fig. S1–S5), and we proceeded with resonance assignment and determination of JUNV ZBD solution structure. ARIA calculations based on NOESY-derived restraints converged to a single fold and revealed that JUNV ZBD indeed contains two distinct clusters of residues posed to coordinate zinc. Cluster I includes His-447, His-449, Cys-455, and His-485. All three histidines are oriented to coordinate zinc via their Nϵ2 atoms. Cluster II includes His-459, Cys-467, and Cys-469, with the histidine positioned to coordinate zinc via Nδ1. A water molecule is presumed to serve as the fourth ligand in this cluster. The backbone of JUNV ZBD forms several switchback loops that provide a scaffold for the two zinc-binding sites on one side of the structure; the C-terminal 15 amino acids wrap around the other side and bring the C terminus close to the N terminus. The proximal arrangement of the N and C termini is stabilized by a short anti-parallel β-sheet (His-447—His-449, Val-480—Arg-482) and by participation of the C-terminal His-485 in zinc cluster I. The partitioning of the histidine and cysteine residues between the two clusters is consistent with the conclusions of previous equilibrium binding studies using recombinant fusion proteins (33). The refined three-dimensional model of JUNV ZBD containing two zinc ions is presented in Fig. 3, A and B.
FIGURE 3.
Solution structure of JUNV ZBD. A, ribbon model of JUNV ZBD. The color changes smoothly from blue at the N terminus to red at the C terminus. Zinc ions are depicted as spheres, and cysteine and histidine side chains that coordinate the zinc ions are colored orange and yellow, respectively. Small circles along the ribbon mark the positions of dibasic ER retention/retrieval motifs (Lys-476—Lys-477 and Arg-482—Arg-483). B, 21 backbone traces of JUNV ZBD are shown superimposed. The coloring scheme is the same as in panel A. C, superimposed backboned traces of JUNV ZBD are colored according to residue conservation among arenaviruses (Fig. 1). Highly conserved residues are purple, variable residues are cyan, and white indicates average conservation (53, 54). The images were prepared with Molmol 2K.1 (55).
Zinc-binding domains are extremely rare in viral glycoproteins, the only other reported example being a dual CCHC-type classical zinc finger in the cytoplasmic tail of the G1 envelope glycoprotein of bunyaviruses (45). JUNV ZBD shows no structural similarity to the bunyavirus zinc-binding domain. VAST (46) and SSM (47) searches identified a number of three-dimensional protein structures with topologies similar to JUNV ZBD (supplemental Fig. S7). However, none of these structures contains two zinc-binding sites and hence the significance of these distant similarities is obscure.
Residues on the zinc-binding face of the JUNV ZBD structure are highly conserved among arenaviruses (Fig. 3C). In contrast, the C-terminal 15 amino acids that wrap around the other side of the structure are highly variable, with the exception of the strictly conserved Trp-481 (Fig. 1). Interestingly, Trp-481 is positioned in the C-terminal β-strand of the short anti-parallel β-sheet that holds the N and C termini together (Fig. 3C and supplemental Fig. S8). The ring of Trp-481 is on the side of the β-sheet opposite from zinc cluster I, where it is sandwiched between the side chains of highly conserved Thr-446 and Arg-448 from the N-terminal β-strand (supplemental Fig. S8). This arrangement suggest that despite the generally low degree of sequence conservation in the C-terminal third of the domain, the short β-sheet and the packing of Trp-481 against residues from the N-terminal β-strand are likely to be universal features of arenaviral GPCs.
Functional Analysis of His-485 GPC Mutants
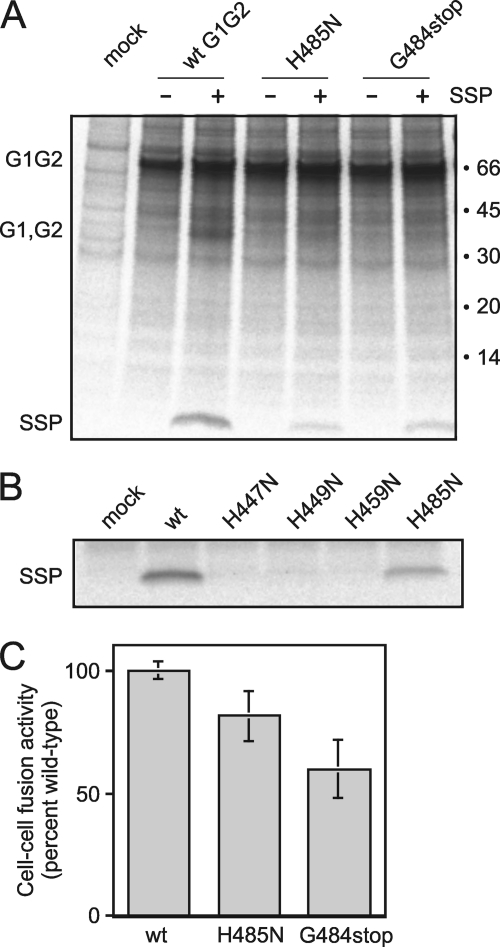
Six of the seven residues that coordinate zinc (His-447, His-449, Cys-455, His-459, Cys-467, and Cys-469) are strictly conserved in all arenaviruses (Fig. 1) and required for association with SSP (25, 33). It was therefore surprising to discover that the C-terminal histidine in JUNV (His-485), which is not conserved, participates in zinc cluster I. A C-terminal histidine residue is found in all clade B New World arenaviruses but is absent in other New World clades and most Old World arenaviruses (Fig. 1). Our finding that the C-terminal His-485 participates in zinc cluster I raised the question whether this residue is required for GPC structure and function in the New World clade B arenaviruses. To investigate the role of His-485 in the context of the intact JUNV GPC complex expressed in mammalian cells, we replaced the histidine with asparagine (H485N), a substitution that typically impairs zinc binding (33, 48, 49). Another mutant was constructed in which the C-terminal glycine and histidine were removed, leaving a C-terminal Arg-483 (G484stop). Immunoprecipitation of the resulting GPC complexes using a monoclonal antibody specific to G1 revealed that both H485N and G484stop mutants included considerably less SSP than did wild-type GPC (∼20 and 30% of wild-type levels, respectively) (Fig. 4A). The mutants also contained little of the mature G1 and G2 glycoproteins, which co-migrate as a heterogeneous smear of ∼35 kDa (Fig. 4A). The paucity of mature subunits was confirmed by enzymatic deglycosylation of the immunoprecipitated complexes, which allows clear resolution of the G1 and G2 polypeptides (not shown) (20). The decrease in proteolytic maturation in the mutants is consistent with specific retention of the G1G2 precursor in the ER in the absence of SSP association (25). Overall, the effects on SSP binding were significant but less extreme than those observed for asparagine mutations at the other zinc-binding histidines, His-447, His-449, or His-459 (Fig. 4B) (33), or for serine mutations at any of the three zinc-binding cysteines in G2 or at Cys-57 in SSP (22, 33).
FIGURE 4.
Mutations at His-485 in GPC affect SSP association and membrane-fusion activity. A, cells expressing wild-type (wt) and mutant (H485N and G484stop) forms of CD4sp-GPC (G1G2 precursor) alone or in trans with wild-type SSP-term (+SSP) were metabolically labeled and GPC was immunoprecipitated using the G1-specific monoclonal antibody QC03-BF11. Uncleaved G1G2 precursor and the heterogeneous smear of the mature glycoproteins (G1, G2) are marked, as is the co-precipitated SSP. Positions of the molecular weight standards and their sizes in kilodaltons are indicated at right. The high level of the uncleaved G1G2 precursor is typical in cells overexpressing GPC (20). Proteolytic cleavage of the G1G2 precursor is dependent on SSP association and is therefore compromised in the GPC mutants. B, lack of SSP incorporation in GPC bearing asparagine mutations at the three previously identified zinc-binding histidine residues (H447N, H449N, and H459N) (33) is compared with the partial defect in H485N. C, cell-cell fusion activity of wt and mutant GPC at pH 5.0 was determined using a vaccinia virus-based β-galactosidase fusion reporter assay (44) and chemiluminescence, as described under “Experimental Procedures.” All measurements were from eight replicates and the average extent of fusion was normalized to that of wt GPC. The figure represents the average and standard deviation of seven independent determinations.
The reduction in SSP association in the H485N and G484stop mutants was reflected in a corresponding, albeit smaller, decrease in the ability of the mutant GPCs to mediate pH-dependent cell-cell fusion (∼80 and ∼60% of wild-type levels, respectively; Fig. 4C). The lesser effect of the mutations on cell-cell fusion likely reflects nonlinearities in the respective assays. The modest fusion deficiency in the H485N and G484stop mutants is, again, in contrast to the absolute defect (≤ 5% of wild-type level) engendered by mutations at the other zinc-binding histidine and cysteine residues (33). Thus, we conclude that the C-terminal His-485 is important but not essential for SSP association in the GPC complex.
Interaction between JUNV ZBD and Acetyl-SCT
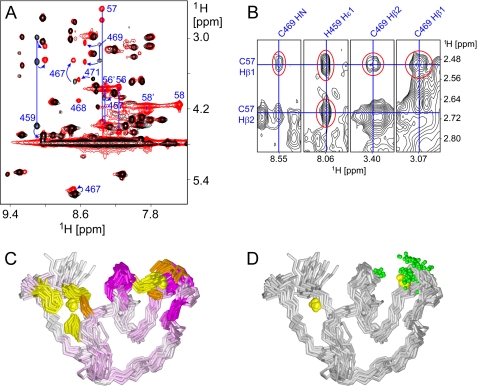
To gain structural insight into the interaction of JUNV ZBD with acetyl-SCT, we used NMR spectroscopy. When acetyl-SCT was added to JUNV ZBD, new signals from certain JUNV ZBD atoms appeared in the one-dimensional 1H NMR spectrum. As the titration progressed, the signals from free JUNV ZBD diminished in intensity while those from the acetyl-SCT-bound form grew stronger (supplemental Fig. S9), consistent with slow exchange between the free and bound forms. Only a small number of JUNV ZBD residues exhibited clearly distinct chemical shifts when bound to acetyl-SCT (Fig. 5A). This suggests that the conformational changes upon binding are minimal and rather localized, and that the affected residues are part of or proximal to the binding site. Hence, a rough picture of the binding site can be generated by mapping the acetyl-SCT-induced spectral perturbations on the three-dimensional structure of JUNV ZBD (Fig. 5C). The chemical shift perturbation data indicate that acetyl-SCT interacts with zinc cluster II, thus providing direct evidence for a zinc-mediated interaction between G2 and SSP. Because the zinc ion in cluster II of JUNV ZBD is coordinated by only three protein residues (His-459, Cys-467, and Cys-469), it can readily accommodate Cys-57 from SSP as the fourth ligand.
FIGURE 5.
Interaction of JUNV ZBD with acetyl-SCT. A, numerous crosspeak shifts are noticeable in the superimposed amide-aliphatic regions of two-dimensional TOCSY spectra of JUNV ZBD in the absence of acetyl-SCT (black) and JUNV ZBD in the presence of a 2-fold molar excess of acetyl-SCT (red). The most conspicuous crosspeak shifts are marked with blue arrows and residue numbers. Signals from both bound and free acetyl-SCT peptide are only present in the latter spectrum and are also labeled with residue numbers (56–58 and 56′-58′, respectively). Several blue lines that connect crosspeaks from the same residues are included for clarity. B, crosspeaks between JUNV ZBD and acetyl-SCT are observable in two-dimensional NOESY spectrum of JUNV ZBD in the presence of a 2-fold molar excess of acetyl-SCT. Several strips from the NOESY spectrum are shown with the intermolecular signals highlighted in red and chemical shifts that pass through the crosspeaks labeled in blue. C, superimposed backbone traces of JUNV ZBD are colored according to 1H chemical shift changes of individual residues upon binding of the acetyl-SCT peptide. HN chemical shifts were used where available; Hδ shifts were used for proline residues while Hα shifts were substituted for other residues without observable HN signals. Color intensity is proportional to the observed change. Zinc ions are depicted as yellow spheres, and cysteine and histidine side chains that coordinate the zinc ions are colored orange and yellow, respectively. Note that all significant chemical shift changes are localized to zinc cluster II, thus marking it as the acetyl-SCT-binding site. D, superimposed backbone traces of JUNV ZBD are shown with green spheres representing all atoms that exhibit unambiguous NOEs to the acetyl-SCT peptide. The orientation of the molecules is the same as in panel C. All atoms in JUNV ZBD for which unambiguous intermolecular NOEs were identified are localized at zinc cluster II.
To obtain further information about the interaction between JUNV ZBD and acetyl-SCT, two-dimensional NOESY spectrum of JUNV ZBD in the presence of excess acetyl-SCT was inspected for intermolecular NOEs. Eleven unambiguous NOEs between JUNV ZBD and acetyl-SCT were identified (Fig. 5B and supplemental Table S1). Ten of these are between Cys-57 in acetyl-SCT and His-459 or Cys-469 in JUNV ZBD, and one is between Thr-58 and Cys-469 (Fig. 5D). The NOESY data suggest that the interaction is mediated primarily by Cys-57 while contact with the rest of the acetyl-SCT peptide is minimal. This is consistent with genetic studies showing that mutations at positions 56 and 58 of SSP are well tolerated (22).
DISCUSSION
Based on the overall high degree of sequence and functional homology among arenaviral GPCs, it is likely that the cytoplasmic G2 domains of other species resemble JUNV in their structural organization. However, the lack of a C-terminal histidine in other species brings up the question of how these viruses might complete tetrahedral coordination of zinc in cluster I. None of the other cytosolic histidines is sufficiently well conserved to stand out as an obvious alternative for the C-terminal histidine, and the clade A New World species possess no additional cysteine or histidine in their cytoplasmic sequences (Fig. 1). The nonconserved histidines found in G2 of some species, His-463 in several Old World viruses (numbered by reference to JUNV GPC) or His-471 in clade C New World viruses, could not participate in zinc coordination without major structural rearrangements relative to the JUNV ZBD structure. His-440 (C440 in Lujo virus) in the transmembrane domain of G2 is relatively well conserved and close to the cytoplasm (Fig. 1) and might be in a position to participate in zinc cluster I. However, the H440A mutant of JUNV GPC displays membrane-fusion activity comparable to wild type (32), suggesting that His-440 is not required for SSP association, at least in the presence of C-terminal histidine. This line of reasoning leaves us with no obvious candidate for the fourth zinc ligand in cluster I of viruses that lack the C-terminal histidine, and it raises the possibility that some arenaviral species may coordinate zinc in cluster I by only three residues, in a manner analogous to zinc cluster II prior to SSP association.
Our model for SSP incorporation in the GPC complex does not include potential structural contributions from the membrane and membrane-spanning regions of GPC. It is likely that the cytoplasmic domains of both SSP and G2 are constrained upon exiting from the membrane and by interactions among the three membrane-embedded segments. Indeed, a single amino acid deletion within the C-terminal hydrophobic sequence of SSP prevents SSP retention in the GPC complex (21), suggesting that the interactions among the membrane-embedded segments and/or the positioning of the C terminus of SSP relative to the lipid bilayer are crucial for association of SSP with G2. The model also does not consider potential contributions from the N-terminal cytoplasmic region of SSP. NMR studies using a peptide corresponding to the N-terminal cytoplasmic sequence of SSP (SSP-N; residues 2–17) did not, however, reveal any interaction with JUNV ZBD (see supplemental Fig. S10 for details). Previous studies have also shown that myristoylation at the N terminus of SSP does not appear to affect SSP retention in GPC (20, 21, 50). This suggests that neither the cytoplasmic N-terminal region of SSP nor the myristoyl moiety is involved in direct interaction with G2.
Our paradoxical finding that the C-terminal His-485 in JUNV ZBD coordinates zinc in cluster I yet is neither essential for membrane-fusion activity nor conserved among arenaviruses suggests that our present understanding of the interaction between SSP and G2 is necessarily incomplete. Likewise, the minimal structural changes induced by acetyl-SCT binding to JUNV ZBD in our experiments do not suggest a structural mechanism whereby SSP association is able to overcome the dibasic amino acid ER retention/retrieval motifs in G2 to allow transport of the G1G2 precursor through the Golgi and to the cell surface (25). The two dibasic motifs (Lys-476—Lys-477 and Arg-482—Arg-483) are positioned on the opposite face of JUNV ZBD from zinc cluster II, on the variable C-terminal segment that wraps around the structure (Fig. 3A) and are unaffected by acetyl-SCT binding.
In conclusion, our studies provide the first structural view of the unusual cytoplasmic zinc-binding domain in the arenavirus envelope glycoprotein complex. The face of the domain that contains the two zinc-binding clusters is well conserved among arenaviruses, and cluster II serves as a binding site for Cys-57 in SSP. It is likely that the zinc-mediated anchoring of SSP contributes to positioning the ectodomain loop of SSP relative to the G2 ectodomain to modulate membrane fusion (26, 29). Small molecule compounds that target the pH-sensitive SSP-G2 interface in the ectodomain of GPC have been shown to inhibit pH-induced activation of membrane fusion and prevent virus entry (32). Structural studies of the intact membrane-associated GPC complex represent an important, albeit daunting, challenge for future research.
Supplementary Material
This work was supported by National Institutes of Health Research Grant R01 AI074818 (to J. H. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10 and Table S1.
The atomic coordinates and structure factors (code 2LOZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LASV
- Lassa fever virus
- JUNV
- Junín virus
- ER
- endoplasmic reticulum
- GPC
- arenavirus envelope glycoprotein
- GTOV
- Guanarito virus
- ITC
- isothermal titration calorimetry
- Kd
- dissociation constant
- MACV
- Machupo virus
- PNGase F
- peptide:N-glycosidase F
- RMSD
- root mean square deviation
- SABV
- Sabiá virus
- SPase
- signal peptidase
- SSP
- stable signal peptide
- TCEP
- tris(2-carboxyethyl)phosphine
- TfR1
- transferrin receptor 1
- ZBD
- zinc-binding domain.
REFERENCES
- 1. Buchmeier M. J., de la Torre J. C., Peters C. J. (2007) in Arenaviridae: The Viruses and Their Replication, Fields Virology (Knipe D. M., Howley P. M. eds), Lippincott, Williams & Wilkins [Google Scholar]
- 2. Peters C. J. (2002) Curr. Top. Microbiol. Immunol. 262, 65–74 [DOI] [PubMed] [Google Scholar]
- 3. McCormick J. B., Fisher-Hoch S. P. (2002) Curr. Top. Microbiol. Immunol. 262, 75–109 [DOI] [PubMed] [Google Scholar]
- 4. McCormick J. B., Webb P. A., Krebs J. W., Johnson K. M., Smith E. S. (1987) J. Infect. Dis. 155, 437–444 [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization (2005) Fact Sheet N°197 [Google Scholar]
- 6. Delgado S., Erickson B. R., Agudo R., Blair P. J., Vallejo E., Albariño C. G., Vargas J., Comer J. A., Rollin P. E., Ksiazek T. G., Olson J. G., Nichol S. T. (2008) PLoS Pathol. 4, e1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briese T., Paweska J. T., McMullan L. K., Hutchison S. K., Street C., Palacios G., Khristova M. L., Weyer J., Swanepoel R., Egholm M., Nichol S. T., Lipkin W. I. (2009) PLoS Pathog. 5, e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rotz L. D., Khan A. S., Lillibridge S. R., Ostroff S. M., Hughes J. M. (2002) Emerg. Infect. Dis. 8, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meyer B. J., De la Torre J. C., Southern P. J. (2002) Curr. Top. Microbiol. Immunol. 262, 139–157 [DOI] [PubMed] [Google Scholar]
- 10. Radoshitzky S. R., Abraham J., Spiropoulou C. F., Kuhn J. H., Nguyen D., Li W., Nagel J., Schmidt P. J., Nunberg J. H., Andrews N. C., Farzan M., Choe H. (2007) Nature 446, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao W., Henry M. D., Borrow P., Yamada H., Elder J. H., Ravkov E. V., Nichol S. T., Compans R. W., Campbell K. P., Oldstone M. B. A. (1998) Science 282, 2079–2081 [DOI] [PubMed] [Google Scholar]
- 12. Borrow P., Oldstone M. B. A. (1994) Virology 198, 1–9 [DOI] [PubMed] [Google Scholar]
- 13. Di Simone C., Zandonatti M. A., Buchmeier M. J. (1994) Virology 198, 455–465 [DOI] [PubMed] [Google Scholar]
- 14. York J., Berry J. D., Ströher U., Li Q., Feldmann H., Lu M., Trahey M., Nunberg J. H. (2010) J. Virol. 84, 6119–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eschli B., Quirin K., Wepf A., Weber J., Zinkernagel R., Hengartner H. (2006) J. Virol. 80, 5897–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skehel J. J., Wiley D. C. (2000) Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 17. White J. M., Delos S. E., Brecher M., Schornberg K. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison S. C. (2005) Adv. Virus Res. 64, 231–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eichler R., Lenz O., Strecker T., Eickmann M., Klenk H. D., Garten W. (2003) EMBO Rep. 4, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. York J., Romanowski V., Lu M., Nunberg J. H. (2004) J. Virol. 78, 10783–10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agnihothram S. S., York J., Trahey M., Nunberg J. H. (2007) J. Virol. 81, 4331–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. York J., Nunberg J. H. (2007) Virology 359, 72–81 [DOI] [PubMed] [Google Scholar]
- 23. von Heijne G. (1985) J. Mol. Biol. 184, 99–105 [DOI] [PubMed] [Google Scholar]
- 24. York J., Nunberg J. H. (2006) J. Virol. 80, 7775–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Agnihothram S. S., York J., Nunberg J. H. (2006) J. Virol. 80, 5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beyer W. R., Pöpplau D., Garten W., von Laer D., Lenz O. (2003) J. Virol. 77, 2866–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kunz S., Edelmann K. H., de la Torre J. C., Gorney R., Oldstone M. B. A. (2003) Virology 314, 168–178 [DOI] [PubMed] [Google Scholar]
- 28. Lenz O., ter Meulen J., Klenk H. D., Seidah N. G., Garten W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12701–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. York J., Nunberg J. H. (2009) J. Virol. 83, 4121–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolken T. C., Laquerre S., Zhang Y., Bailey T. R., Pevear D. C., Kickner S. S., Sperzel L. E., Jones K. F., Warren T. K., Lund S. A., Kirkwood-Watts D. L., King D. S., Shurtleff A. C., Guttieri M. C., Deng Y., Bleam M., Hruby D. E. (2006) Antiviral Research 69, 86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larson R. A., Dai D., Hosack V. T., Tan Y., Bolken T. C., Hruby D. E., Amberg S. M. (2008) J. Virol. 82, 10768–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. York J., Dai D., Amberg S. M., Nunberg J. H. (2008) J. Virol. 82, 10932–10939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. York J., Nunberg J. H. (2007) J. Virol. 81, 13385–13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bowden T. A., Crispin M., Graham S. C., Harvey D. J., Grimes J. M., Jones E. Y., Stuart D. I. (2009) J. Virol. 83, 8259–8265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abraham J., Corbett K. D., Farzan M., Choe H., Harrison S. C. (2010) Nat. Struct. Mol. Biol. 17, 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons [Google Scholar]
- 37. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 38. Linge J. P., Habeck M., Rieping W., Nilges M. (2003) Bioinformatics 19, 315–316 [DOI] [PubMed] [Google Scholar]
- 39. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 40. Alberts I. L., Nadassy K., Wodak S. J. (1998) Protein Sci. 7, 1700–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghiringhelli P. D., Rivera-Pomar R. V., Lozano M. E., Grau O., Romanowski V. (1991) J. Gen. Virol. 72, 2129–2141 [DOI] [PubMed] [Google Scholar]
- 42. Fuerst T. R., Niles E. G., Studier F. W., Moss B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanchez A., Pifat D. Y., Kenyon R. H., Peters C. J., McCormick J. B., Kiley M. P. (1989) J. Gen. Virol. 70, 1125–1132 [DOI] [PubMed] [Google Scholar]
- 44. Nussbaum O., Broder C. C., Berger E. A. (1994) J. Virol. 68, 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Estrada D. F., Boudreaux D. M., Zhong D., St. Jeor S. C., De Guzman R. N. (2009) J. Biol. Chem. 284, 8654–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gibrat J. F., Madej T., Bryant S. H. (1996) Curr. Opin. Struct. Biol. 6, 377–385 [DOI] [PubMed] [Google Scholar]
- 47. Krissinel E., Henrick K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 48. Ma L., Tibbitts T. T., Kantrowitz E. R. (1995) Protein Sci. 4, 1498–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lesburg C. A., Huang C., Christianson D. W., Fierke C. A. (1997) Biochemistry 36, 15780–15791 [DOI] [PubMed] [Google Scholar]
- 50. Saunders A. A., Ting J. P., Meisner J., Neuman B. W., Perez M., de la Torre J. C., Buchmeier M. J. (2007) J. Virol. 81, 5649–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. York J., Agnihothram S. S., Romanowski V., Nunberg J. H. (2005) Virology 343, 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Charrel R. N., Feldmann H., Fulhorst C. F., Khelifa R., de Chesse R., de Lamballerie X. (2002) Biochem. Biophys. Res. Commun. 296, 1118–1124 [DOI] [PubMed] [Google Scholar]
- 53. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]
- 54. Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. (2004) Bioinformatics 20, 1322–1324 [DOI] [PubMed] [Google Scholar]
- 55. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 56. Laskowski R. A., Rullmann J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.