Abstract
The crystal structure of a heme oxygenase (HO) HugZ from Helicobacter pylori complexed with heme has been solved and refined at 1.8 Å resolution. HugZ is part of the iron acquisition mechanism of H. pylori, a major pathogen of human gastroenteric diseases. It is required for the adaptive colonization of H. pylori in hosts. Here, we report that HugZ is distinct from all other characterized HOs. It exists as a dimer in solution and in crystals, and the dimer adopts a split-barrel fold that is often found in FMN-binding proteins but has not been observed in hemoproteins. The heme is located at the intermonomer interface and is bound by both monomers. The heme iron is coordinated by the side chain of His245 and an azide molecule when it is present in crystallization conditions. Experiments show that Arg166, which is involved in azide binding, is essential for HugZ enzymatic activity, whereas His245, surprisingly, is not, implying that HugZ has an enzymatic mechanism distinct from other HOs. The placement of the azide corroborates the observed γ-meso specificity for the heme degradation reaction, in contrast to most known HOs that have α-meso specificity. We demonstrate through sequence and structural comparisons that HugZ belongs to a new heme-binding protein family with a split-barrel fold. Members of this family are widespread in pathogenic bacteria and may play important roles in the iron acquisition of these bacteria.
Keywords: Enzyme Mechanisms, Heme, Iron Metabolism, Protein Structure, X-ray Crystallography, Helicobacter pylori, Bacterial Infection, Heme Oxygenase, Iron Acquisition
Introduction
One of the essential tasks for pathogenic bacteria to colonize mammalian hosts successfully is the acquisition of iron, an element required by all known life forms. Most bacteria require micromolar levels of iron for survival, but the element is usually not biologically available (1). The low concentration of free iron is caused by two factors: the low solubility (∼10−18 m) of ferric iron at physiological pH, and the sequestration of ferrous iron into porphyrins and carrier molecules such as lactoferrin, transferrin, and ferritins. The latter is a mechanism employed by host cells to restrict microbe growth and to minimize the toxicity from superoxide anions and hydroxyl radicals generated by ferrous via Fenton reactions. To overcome iron shortages, bacteria have developed several mechanisms to import iron in different forms (1).
Heme iron accounts for ∼95% of iron in animal hosts (2), which suggests significant evolutionary pressure for pathogenic bacteria to prefer heme as an iron source, as has been demonstrated for Staphylococcus aureus (3). As a result, pathogenic bacteria have developed sophisticated systems to capture hemes, transfer them into bacterial cells, and degrade them to release free iron (4). Here, we report the 1.8 Å resolution crystal structure of HugZ (Hp0318), a heme oxygenase (HO)4 essential for heme iron utilization by Helicobacter pylori. Wild-type H. pylori can use heme as a sole iron source (5), HugZ mutants, however, are unable to grow on heme iron (6).
It has long been known that in eukaryotic cells, HOs, in conjunction with aerobic electron donors, carry out the degradation of heme through oxidative cleavage. These enzymes are mono-oxygenases that function through oxidative cleavage of the porphyrin rings.
Bacterial HOs were only recently identified from Gram-negative bacteria (7). With the exception of the proteins IsdG and IsdI from S. aureus (8), all of the structurally characterized bacterial and eukaryotic HOs are homologous in sequence. They are all-α-helical monomers with very similar folds (9). They all function and crystallize as monomers. IsdG and IsdI are α/β-type proteins with a ferredoxin-like fold and are not homologous to canonical HOs.
Recently, two new HOs from pathogenic bacteria, Cj1613c from Campylobacter jejuni (10) and HugZ from H. pylori (6), were identified. Both proteins are required for heme iron acquisition by the respective bacteria. HugZ is also required for the adaptive colonization of H. pylori in hosts. Mutation of this protein renders H. pylori unable to grow on heme iron (6). Cj1613c and HugZ share 56% sequence identity and are homologous to a family of smaller proteins involved in heme iron utilization, but they are not homologous to other previously characterized HOs. Interestingly, sequence alignments show that Cj1613c and HugZ have weak sequence similarities with FMN-binding proteins with a split-barrel fold, in contrast to canonical HOs that have all α-helix structures. It was also found that HugZ has a unique δ-meso regiospecificity in the ring-opening reaction of heme (6), whereas other HOs are almost exclusively α-meso regiospecific. The only exception known so far is PigA from Pseudomonas aeruginosa, which produces β,δ-biliverdins. This probably occurs because P. aeruginosa has segregated heme-degrading pathways, one for iron acquisition and one for pigment biosynthesis, and the latter pathway has an HO with α-meso specificity (7). These experimental data indicate that HugZ is a novel HO and its molecular structure and enzymatic mechanism are distinctive from those of the characterized HOs.
In light of the important role played by HugZ in H. pylori pathogenicity and the unique sequence and regiospecificity of this protein among HOs, it was of interest to elucidate the three-dimensional structure of HugZ. We have solved the crystal structure of HugZ complexed with hemin and refined the structure at 1.8 Å resolution. The HugZ structure reported here is distinctive from that of either the canonical HOs or IsdG/IsdI. It adopts a split-barrel fold often observed in FMN-binding proteins, as would be suggested by sequence homology. Like other split-barrel proteins, HugZ exists as a dimer in solution and in crystals. Two hemes are located at the intermonomer interface of HugZ and are bound by both monomers. The heme iron is coordinated by the side chain of His245. Comprehensive structural and sequence comparisons show that HugZ represents a new heme-binding split-barrel protein family. Based on structural and mutagenesis analyses, we demonstrate that Arg166 is the critical residue for HugZ enzymatic mechanisms, whereas the heme-coordinating residue His245 is not, in contrast to the essential roles of its counterparts in other HOs.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization
The purification and crystallization of HugZ with a His6 tag have been reported previously (6, 11). In brief, the pET22b plasmid containing the HugZ gene with a His6 tag at the C terminus was transformed into Escherichia coli BL21(DE3) strain competent cells. The cells were grown in LB medium at 310 K until the A600 reached 0.8 and were then induced with 10 μm isopropyl-1-thio-β-d-galactopyranoside. The bacteria were grown at 295 K for an additional 12 h and harvested by centrifugation. Cells were suspended in lysis buffer containing 50 mm phosphate at pH 8.0, 300 mm NaCl, 10 mm imidazole, and 1 mm phenylmethylsulfonyl fluoride and lysed by ultrasonication. The lysate was centrifuged, and the supernatant was applied to a nickel-nitrilotriacetic acid column (Novagen) previously equilibrated with lysis buffer. The column was washed extensively, and HugZ was eluted with elution buffer (50 mm phosphate buffer, pH 8.0, 300 mm NaCl, and 250 mm imidazole). Imidazole in the eluate was removed by using a HiTrap desalting column (GE Healthcare). Hemin solution (5 mm in dimethyl sulfoxide) was added into HugZ to a final hemin:HugZ ratio of 2:1. The mixture was incubated at 277 K overnight with rocking. After centrifugation, the soluble fraction was applied onto a HiLoad 16/60 Superdex 200 gel-filtration column (GE Healthcare) preequilibrated with 20 mm Tris, pH 8.0, buffer containing 5 mm sodium azide. The peak fractions were collected and assayed by SDS-PAGE. The fractions containing pure dimeric HugZ-hemin complex were pooled and concentrated by ultrafiltration for crystallization. Crystals were obtained using the hanging-drop vapor-diffusion method by mixing 1 μl of 25 mg/ml HugZ-hemin solution (in 20 mm Tris, pH 8.0, 5 mm sodium azide) and 1 μl of reservoir solution consisting of 16% (w/v) PEG 3350, 3% Tacsimate, 0.1 m imidazole, pH 7.5, and 12% methanol.
Data Collection and Structure Determination
Diffraction datasets were collected at Beamline 17A of Photon Factory, KEK, Japan, as described previously (11). In summary, a single SeMet derivatized crystal of the HugZ-hemin complex was used to collect a diffraction dataset to 1.8 Å resolution at 1.000 Å and three additional multiwavelength anomalous dispersion datasets at 2.3Å resolution at 0.97912Å (peak), 0.97931Å (inflection), and 0.96408Å (high energy remote point). The crystal structure of the HugZ-hemin complex was solved by the multiwavelength anomalous dispersion method using the 2.3 Å resolution datasets. Thirty selenium atoms with occupancies above 0.5 were located by using the program ShelxD (12). The program SHARP (13) was used to refine the heavy atom parameters and to calculate the initial phase. Density modification, including histogram matching, solvent flattening, and noncrystallographic symmetry averaging, was performed using the program RESOLVE (14). The initial structure model was automatically constructed using the program Arp/wArp (15) with 82% of the sequence docked into the electron density map. The remainder of the structure was manually built using the program COOT (16). This structure was refined using the program CNS (17) against the 1.8 Å resolution dataset collected at 1.0Å wavelength. Throughout the refinement, 5% of the reflections were randomly chosen and set aside for the calculation of Rfree and were not used in the refinements. After the first round of simulated annealing and group B-factor refinement, the Rwork and Rfree values decreased to 0.2941 and 0.3072, respectively. According to the σ-A weighted difference maps, six hemin molecules were placed in the appropriate positions. Then, iterative cycles between CNS refinement and manual rebuilding were performed, during which 6 azide anions, 22 ethylene glycol molecules, and 1289 water molecules were added to the model, until the Rwork and Rfree reached 0.192 and 0.222, respectively. The refinement statistics are summarized in Table 1. Figs. 1, 2, 4, and 6 were prepared using the program PyMOL (18).
TABLE 1.
Data collection, phasing, and structural refinement statistics
Statistics of data collection in this table were previously reported (11) and are included here for completeness and clarity.
| Space group | P212121 | |||
| Cell dimensions | ||||
| a, Å | 88.40 | |||
| b, Å | 139.37 | |||
| c, Å | 152.97 | |||
| Data collection | Native | Peak | Inflection | Remote |
| Wavelength, Å | 1.000 | 0.97912 | 0.97931 | 0.96408 |
| Res. range, Å | 48–1.80 | 58–2.32 | 58–2.32 | 58–2.32 |
| Total reflections | 1,117,733 | 810,952 | 406,245 | 406,378 |
| Unique reflections | 165,702 | 82,219 | 82,283 | 82,233 |
| Completeness, %a | 94.9 (72.8) | 100 (100) | 100 (99.9) | 99.9 (99.9) |
| Rsym (I),a % | 4.9 (17.2) | 6.0 (12.7) | 5.0 (11.5) | 5.2 (12.7) |
| I/σa (I) | 8.6 (4.3) | 7.8 (5.2) | 9.3 (5.7) | 9.0 (5.3) |
| Refinement | ||||
| Resolution (Å) | 47.87-1.80 | |||
| No. reflections | 165,278 | |||
| Rwork/Rfree | 19.2/22.2 | |||
| No. atoms | ||||
| Protein | 11,850 | |||
| Heme | 258 | |||
| Water | 1,289 | |||
| B-factors, Å2 | ||||
| Protein | 26.0 | |||
| Heme | 21.6 | |||
| Water | 32.0 | |||
| r.m.s.d. | ||||
| Bond lengths, Å | 0.01 | |||
| Bond angles, ° | 1.5 | |||
| Ramachandran plot | ||||
| Most favored regions | 91.2% | |||
| Additional allowed regions | 8.6% | |||
| Generously allowed regions | 0.2% | |||
a Values in parentheses are for the outermost resolution shells: 1.90–1.80 Å for the native dataset and 2.45–2.32 Å for the multiwavelength anomalous dispersion datasets.
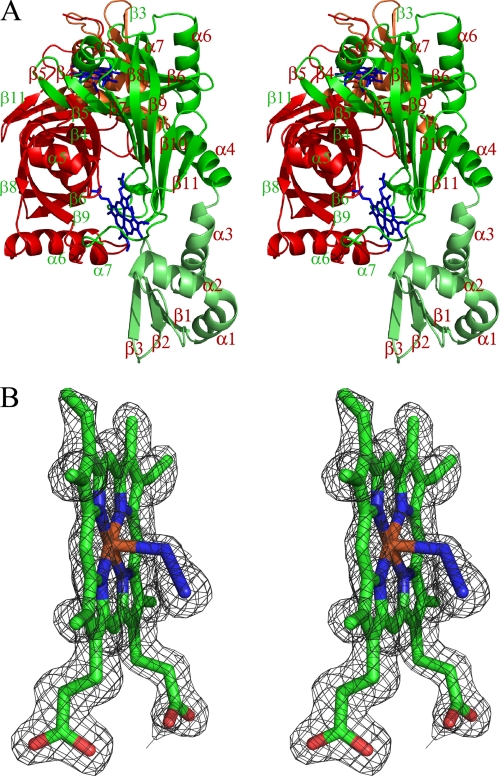
FIGURE 1.
A, stereoview of the HugZ dimer. The two monomers are displayed in red and green, respectively. The N domains are in darker colors. The heme and azide moieties are shown as stick models. The dimer is viewed along the barrel of the red monomer whereas the barrel of the green monomer is on the plane. B, stereoview of the σA-weighted 2 Fo − F c omit map around one of the bound hemin and azide moieties, contoured at 1.5σ. The map is superimposed on the final model of the ligands.
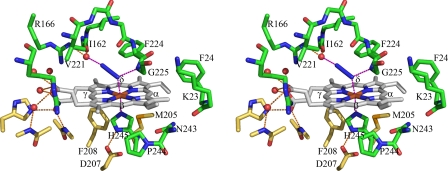
FIGURE 2.
Stereoview of the heme binding pocket of HugZ and HugZ-heme-azide interactions. Carbon atoms from different HugZ monomers are colored green and yellow, respectively.
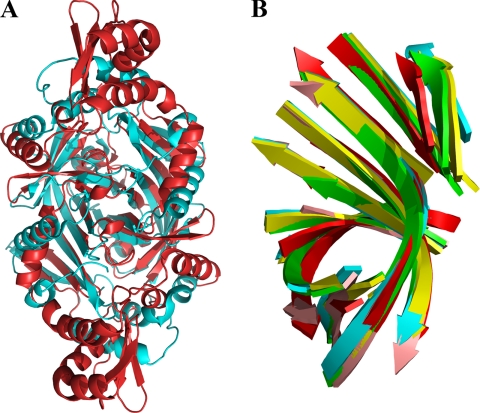
FIGURE 4.
Structural similarities between HugZ and FMN-binding split-barrel proteins. A, superposition of HugZ and a randomly selected FMN-binding split-barrel protein (Protein Data Bank code 1TY9). HugZ is colored red, and the FMN-binding protein is cyan. Heme and FMN are removed for clarity. B, superposition of β-strand substructures of the HugZ monomer (red, C-terminal domain only) with four randomly chosen FMN-binding split-barrel proteins (1CI0, 1DNL, 1NRG, and 1RFE).
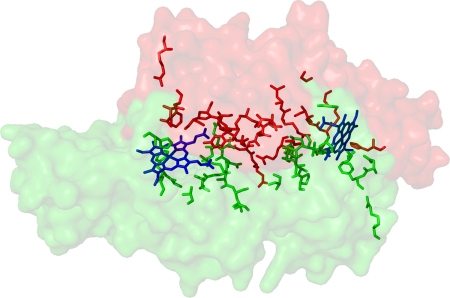
FIGURE 6.
Most of the conserved residues (stick models) of the heme-binding split-barrel fold proteins are distributed on the intermonomer interface and the heme binding pockets of the HugZ dimer. Two HugZ monomers are shown as masks with different colors.
HugZ Mutant Purifications
Genes for HugZ mutants were prepared by site-directed mutagenesis using His6-tagged HugZ plasmid as a template. The tag-less genes were ligated into the pET22b vector. A truncated HugZ mutant gene with only the C domain (residues 82–251) was cloned from the pET22b-HugZ plasmid using PCR methods. The DNA was digested with NdeI and XhoI and ligated into the pET22b vector for protein expression.
Eight hundred milliliters of E. coli strain BL21 (DE3) cells expressing HugZ mutants were grown as described above for wild-type HugZ and were lysed by ultrasonication. After centrifugation, the supernatant (∼20 ml) was combined with 4.86 g of ammonium sulfate to a concentration of 40% saturation. The solution was cleared by centrifugation. Another 6.36 g of ammonium sulfate was added to 80% saturation, and the solution was centrifuged at 16,000 rpm for 20 min to precipitate the protein. The protein was resolubilized in 5 ml of a buffer containing 20 mm Tris-HCl at pH 8.9 and was desalted by a HiTrap desalting column (GE Healthcare) with the same buffer. Desalted protein sample was loaded onto a HiTrap Q column (GE Healthcare) and was eluted in a 0–150 mm NaCl gradient in 20 mm Tris-HCl, pH 8.9, buffer and used for heme binding and enzymatic activity assays.
Enzymatic Activity Assay
We used the previously published protocol for HO activity assays on HugZ and its mutants (6). In brief, protein and heme were added into 1 ml of solution containing 400 units of bovine liver catalase (Worthington), 20 mm Tris-HCl, pH 7.6, and 150 mm NaCl to a final concentration of 10 μm. Catalase was included in the assays to suppress nonenzymatic H2O2-mediated heme degradation (10). The binding of heme to protein was monitored by measuring the peak absorptions using a U-2010 Spectrophotometer (Hitachi) until the reading stabilized. Twenty microliters of 1 m l-ascorbic acid was then added into the protein-heme solution to start the region. Absorptions were recorded at 2-min intervals between 340 nm and 800 nm for 40 min. All measurements were made at 298 K using a circulating water bath.
RESULTS
Overall Structure of HugZ-Hemin
HugZ-hemin crystallizes as a trimer of dimers. Although interactions between dimers are typical for crystal packing of proteins of similar sizes, those between monomers within dimers were more numerous, indicating that the dimeric form is the natural state of HugZ. Size-exclusion chromatography shows that HugZ exists exclusively as dimers, in the presence or absence of heme.
Each asymmetric unit contains one trimer of HugZ dimers. The full length of the HugZ molecule is 251 residues. The N-terminal one or two residues and C-terminal two or three residues of each monomer are disordered in the structure and are not included in the model. All of the other residues have good electron density. Structures of all six monomers in the asymmetric unit are essentially identical, with a r.m.s.d. of 0.3–0.4 Å for all Cα atoms. Subsequent discussions will not distinguish between monomers and dimers and will apply to all of them.
The two monomers in a dimer are related by a local 2-fold axis (Fig. 1A). Each monomer consists of two domains. The N-terminal residues 1–82 form a simple α/β-type domain, in which a three-stranded anti-parallel β-sheet (formed by β1–3) is stacked against three α-helices (α1–3). The remaining residues form the larger C domain. Two C-domains from two dimers form a typical split-barrel fold. The C-domain is almost exclusively responsible for HugZ dimer formation; only Glu56 from the N domain forms direct hydrogen bonds with Thr201 from the other monomer.
The C domain of the HugZ monomer consists of 8 β-strands (β4–11) and 4 α-helices (α4–7) (Fig. 1A). Strands β4–β9 are anti-parallel and form a closed, distorted β-barrel. Strands β8 and β9 are much longer than the other four strands and extend out from one end of the barrel to form an antiparallel β-sheet with β10 and β11. Helix α4 is packed against this β-sheet and blocks one opening of the β-barrel. Another short α-helix, α5, blocks the other opening of the β-barrel. Helices α6 and α7 are packed against the β-barrel on the side. The HugZ dimer is formed mostly by packing the β-barrels from two monomers on their sides, at an angle of ∼90°. Two crevices are formed at the intermonomer interface between the β-sheet formed by strands β8–β11 from one monomer and helix α7 from another, and these crevices serve as the binding sites for heme.
HugZ-Heme Interactions
When an electron density map was calculated with the experimental multiwavelength anomalous dispersion phases, and the primary sequence of HugZ was fitted in, two groups of strong peaks were observed at the monomer-monomer interface of each dimer that could not be accounted for by HugZ itself. Upon inspection of their sizes and shapes, each of the peaks was determined to be due to a bound heme molecule. Subsequent refinements with heme at both locations resulted in very good geometry and B-factors for both heme molecules (Fig. 1B).
The two hemes are near the edge of and are almost parallel to the central β-sheet of one HugZ monomer, and they are bound by residues from both monomers. Their propionic groups point toward the center of the HugZ dimer. They form four direct and five water-mediated hydrogen bonds with HugZ. In addition, the hemes form numerous van der Waals interactions with hydrophobic groups of HugZ. Residues Phe224 and Gly225, which form a short loop between β9 and β10 of the central β-sheet, run parallel to the heme group, and their main chain atoms and the side chain of Phe224 are stacked against the heme near the α-meso position. The opposite side of the heme is stacked against the side chains of Val133, Met205, Phe208, Asn243, and Pro244. From the lateral directions, the hemes form van der Waals interactions with the side chains of Phe24, Thr201, and Ile202.
A cavity formed between the heme and the central β-sheet is an obvious binding pocket of dioxygen. When the crystals were obtained under aerobic conditions in the presence of azide, an azide molecule occupied this cavity and coordinated the iron (see below).
Interactions between Azide and HugZ-Hemin
Even though hemin was used to complex HugZ, a slow degradation of the protoporphyrin IX ring was still observed. The color of HugZ-hemin crystals changed from dark red to green and lost their ability to diffract x-rays over time. This process could be retarded by including compounds such as azide and thiocyanate in crystallization conditions. HugZ-hemin crystals obtained in the presence of 5 mm azide diffracted to higher resolution. During structure refinement, an azide molecule was located at the hemin site, and it coordinated the iron.
The Fe-N bond lengths in Fe-N3 moieties range from 2.23 to 2.34 Å, and the Fe-N-N bond angles range from 116.8 to 122.7°. N1, the terminal atom of the azide that coordinates the iron atom, accepts a hydrogen bond from the Gly225 main chain NH group, whereas N3 accepts a hydrogen bond from a well defined water molecule, which, in turn, is hydrogen-bonded to the side chain of Arg166 and the main chain carbonyl oxygen of Ile162 (Fig. 2). A hydrogen bond from a water molecule to the terminal atoms of the ligands coordinating the central iron is a common theme of HO-heme-ligand complex structures. This water molecule is proposed to position the dioxygen molecule in the correct orientation and to relay protons during the reaction. In the HugZ structure, a nearby crevice connects this water molecule to the outside environment, and it may serve as the channel for proton transfer.
As observed in the structures of canonical HOs, the imidazole plane of His245 is parallel to one of the meso axes (meso α-γ axis). Interestingly, the bound azide is almost parallel to one of the N-Fe-N axes of the heme and thus at an angle of ∼45° to the His245 imidazole plane. This is in contrast to what has been observed in canonical HO structures in which the ligands (O2, NO, or azide) are perpendicular to the imidazole plane. This difference may arise from the fact that the two hydrogen bonds with azide make the current azide position energetically more favorable. Both hydrogen bonds would be disrupted if the azide was positioned perpendicular to the imidazole plane (Fig. 2).
Mutagenesis Analyses on the Roles of Arg166 and His245
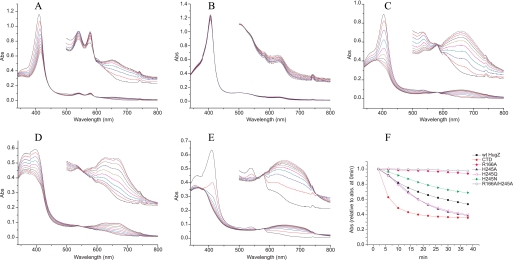
Residues His245 and Arg166 are involved in heme iron coordination and in azide binding (Fig. 2), respectively, so they could play critical roles in the HO enzymatic mechanisms of HugZ. To investigate their importance, we prepared H245A, H245Q, H245N, R166A, and H245A/R166A mutants and assayed their activities.
The R166A mutation completely abolished the HO activity of HugZ (Fig. 3B). We propose that Arg166 is essential in maintaining the water network at the heme binding site of HugZ. This network is required for the binding of O2 to heme iron, the first step of heme hydroxylation, by donating a hydrogen bond to O2. It is probably also required for the transfer of protons between the HugZ active site and the aqueous environment during hydroxylation of the protoporphyrin IX ring.
FIGURE 3.
A–E, enzymatic activity assays of wild-type HugZ (A), R166A mutant (B), H245A mutant (C), H245N mutant (D), and the C-terminal domain truncate mutant of HugZ (CTD) (E). The spectra were recorded at an interval of 4 min for 40 min over the wavelength range 340–800 nm. The spectra between 500 nm and 800 nm are magnified ×5 in insets. F, enzymatic activities of HugZ and its mutants, measured by the change of maximum absorptions over time relative to their respective maximum absorptions at 0 min.
Mutation of His245, however, did not abolish HO activity. All three mutants of His245 are able to degrade heme, as indicated by the disappearance of the peak at 410 nm, and to produce biliverdin, as indicated by the strong product absorption peak at 680 nm (Fig. 3). The existence of well defined isosbestic points in the absorption spectra indicates that the degradation of heme and the formation of biliverdin occur simultaneously and represent the same process. In contrast, in canonical HOs such as human HO-1 (19) and Corynebacterium diphtheriae Hmu O (20, 21), mutation of the heme ligand histidine residues completely abolishes biliverdin formation.
H245N mutation degraded, but did not abolish, HugZ activity. H245A and H245Q mutants, on the other hand, have almost identical activities, as both are more active than wild-type HugZ. This indicated that the coordination of the heme iron by the side chain of His245 is not required for the HugZ enzymatic mechanism. The release of the C-terminal region from the heme binding site in these mutants would result in more exposed heme binding sites. This would then facilitate the entry of heme and oxygen and the exit of the products and increase the activity of the enzyme in the cases of the H245A and H245Q mutants. We propose that the H245N mutant could still coordinate the heme iron because the size of an asparagine side chain is very similar to that of a histidine. As a result, the heme moiety in H245N is proposed to be similarly exposed compared with that in the wild-type protein, but the iron is less optimally coordinated, by an amide group instead of an imidazole group, and thus the activity is diminished compared with wild-type HugZ.
To investigate the possibility that His245 mutants could hydroxylate heme with O2 coordinating the heme iron from the opposite side of Arg166, we prepared a H245A/R166A double mutant and found it to be inactive (Fig. 3F). This indicated that the degradation of heme can only occur from the Arg166 side and that the His245 mutants are bona fide HOs.
Structural and Sequence Comparisons Reveal HugZ as a Member of a New Protein Family
Prior to structure determination, weak sequence similarities were observed between HugZ and proteins from the FMN-binding split-barrel superfamily. This was intriguing because this superfamily consisted almost exclusively of FMN-binding proteins involved in electron transfer reactions. There are two subfamilies within this superfamily: the PNP oxidase-like subfamily and the NADH:FMN oxidoreductase-like subfamily. Structural comparison revealed that the structure of HugZ is very similar to that of the members of the PNP oxidase-like subfamily. Indeed, of 21 known structures in this subfamily, as identified by the program SCOP (22), 20 can be superimposed onto the HugZ monomer with r.m.s.d. values of 1.4–3.0Å for 100–150 Cα atoms (Fig. 4A). The substructures of the β-strands are especially well conserved. If only 69 residues from eight β-strands of the C-domain are used, the r.m.s.d. values are between 0.7 and 1.9 Å (Fig. 4B). Only the C domain of HugZ can be superimposed with PNP oxidase-like proteins. A survey with DALI did not reveal any structures homologous to the N domain.
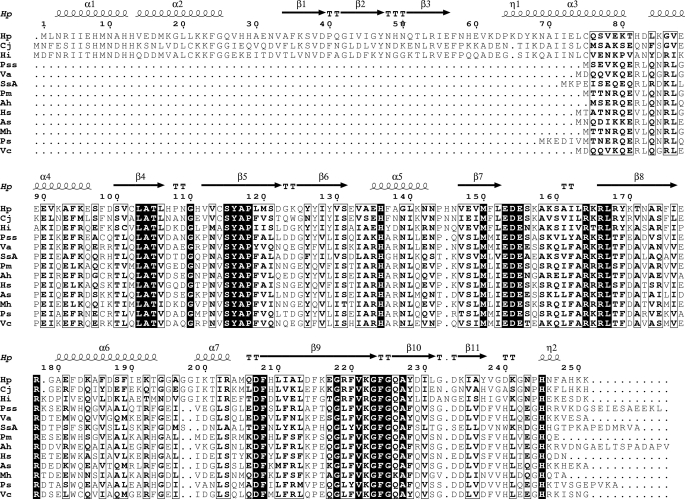
A BLAST search identified two homologous proteins with similar molecular sizes: Cj1613c from C. jejune and hypothetical protein HI0854 from Haemophilus influenzae, with sequence identities of 56 and 47%, respectively, to HugZ. Cj1613c has been identified as an HO (10), but HI0854 has not been characterized. HugZ also shows significant sequence homology to a series of smaller proteins from various pathogenic bacteria (Fig. 5). These proteins are ∼160 residues in lengths and are part of the heme utilization systems of the respective bacteria. Functional characterizations showed, however, that they are heme-binding proteins but not HOs (23, 24). They lack the N domain but retain the C domain of HugZ. Mapping of the strictly conserved residues onto the HugZ structure shows that most of these residues are involved in dimer formation or heme binding (Fig. 6), suggesting that these proteins, although significantly smaller, should still form dimers and bind heme.
FIGURE 5.
Sequence alignment of HugZ, Cj1613c, HI0854, and some of the smaller heme-utilization proteins. Residues in bold font with black background are completely conserved. Residues in bold font with white background are highly conserved. Proteins used in the alignment are (from top to bottom): H. pylori HugZ, C. jejuni protein Cj1613c, H. influenzae protein HI0854, Photobacterium sp. SKA34 HugZ, Vibrio alginolyticus protein V12G01-06051, Shewanella sp. ANA-3, Pasteurella multocida subsp. multocida str. Pm70 PM0299, Aeromonas hydrophila subsp. hydrophila ATCC 7966 HutZ, Histophilus somni HutZ, Aliivibrio salmonicida LFI1238 HuvZ, Mannheimia hemolytica PHL213 HugZ, Plesiomonas shigelloides HugZ, and Vibrio cholerae HutZ.
Residues involved in heme binding and azide binding in HugZ are highly conserved among these proteins. For example, Arg166 and His245 are completely conserved. Residues involved in van der Waals interactions with the protoporphyrin ring, such as Val133, Ile202, Phe208, Phe224, Gly225, and Ala227, and residues that are not in direct contact with heme but that line the binding pocket, such as Val221, Gly223, and Ala227, are either completely conserved or are only replaced once by similar residues. This suggests that the heme binding pockets in these proteins are very similar to what we observe in HugZ, even though they do not have HO activities. The lack of the N-terminal domain would result in a more exposed heme than in the current HugZ structure but would still be comparable with that in canonical HO structures. Based on these observations, we propose that Cj1613c, HI0854, and the smaller heme utilization proteins have three-dimensional structures very similar to that of HugZ and are members of the FMN-binding split barrel superfamily. They form a new, heme-binding subfamily, alongside the PNP oxidase-like and the NADH:FMN oxidoreductase-like subfamilies.
The seemingly apparent correlation between the existence of N domain and HO activities for this subfamily, however, was proved false: we purified a truncated HugZ mutant with only the C domain and assayed its HO activities, and we found it to be more active than wild-type HugZ (Fig. 3E). This is probably because the more exposed heme binding site facilitates the entry of substrates and the exit of products. Therefore, the relationship between the N domain and the HO activity of HugZ needs further investigation.
DISCUSSION
The difference between the overall folding of HugZ and that of other HOs is obvious, but some of the unique properties of HugZ, such as the δ-meso specificity and the roles played by the heme iron-coordinating histidine residue, can be very well understood by localized structural features, i.e. the difference in the protein-heme interactions between HugZ and other HOs.
Unique Heme Binding Site and the Role of His245
In canonical HOs, the heme moieties are sandwiched between two α-helices, and between a β-sheet and an α-helix in IsdG/IsdI. The heme iron-coordination histidine residues of these proteins are located in α-helices. These heme binding sites are rather rigid because of the rigid nature of the secondary structures involved. In fact, the heme-bound and the heme-free structures of human HO-1 and IsdG are essentially identical. In these enzymes, the heme iron-coordinating histidine residues are essential for their enzymatic activities. Mutation of the human HO-1 residue His25, which coordinates the heme iron, resulted in a total loss of enzymatic activity. This loss could be reversed by the addition of imidazole (25). These results clearly demonstrated that the coordination of the heme by an imidazole moiety, either from a histidine residue or from added imidazole, is essential for the enzymatic mechanism of human HO-1.
HugZ, on the other hand, has a flexible C-terminal loop region, which contains the heme iron-coordinating residue His245 as part of the binding site. Thus, the heme binding site of HugZ is more flexible. Our mutagenesis studies on His245 showed that this residue is not required for the catalytic activities of HugZ. In the cases of H245A and H245Q, the mutants are actually more active than the wild-type enzyme, so this residue is not part of the enzymatic mechanism of HugZ. We propose that His245 is only responsible for the recognition and binding of heme by HugZ. Without this histidine residue to bind the iron and to “close off” the heme binding site, the heme would be much more exposed, resulting in low binding affinity and specificity. His245 is proposed to contribute to the affinity and specificity of HugZ toward heme. The flexible nature of the C terminus is proposed to facilitate the release of biliverdin after the hydroxylation of heme and the removal of iron. The exact role of this residue is the subject of a future research project.
Structural Basis of δ-Meso Specificity
Because azide coordinates heme iron with an sp2 hybrid orbital, the Fe-N-N unit has a bond angle of 120°. A similar structural feature was observed in other HOs in which iron is coordinated by O2 or O2 analogs, and it was proposed to be the determinant of the regiospecificity of HOs (26, 27). In most HO structures, the bound O2 (or its analogs) points toward, and the terminal atom comes into van der Waals contact with, the α-meso carbon. Correspondingly, these HOs hydroxylate heme at the α-meso position. In HugZ, however, the azide tilts toward and bisects pyrrole IV, implying γ- and/or δ-meso regiospecificity. HPLC characterization of heme-degrading products of HugZ showed that only biliverdin-IXδ was present (6), verifying the unique δ-meso regiospecificity of HugZ.
It has been demonstrated that in canonical HOs, regiospecificity is achieved by physically blocking all the meso carbon atoms except for the one to be hydroxylated (28). In the HugZ-hemin-azide complex structure, the α- and δ-meso carbon atoms are blocked by the main chain of Gly225 and the side chain of Phe224, respectively, whereas the α- and γ-meso carbon atoms are relatively accessible. However, the δ-meso carbon atom can be easily made accessible by rotation around the Cβ-Cγ bond of Phe224 and by swinging the side chain of this residue to a nearby vacant position. In contrast, blocking of the meso carbon positions in canonical HOs is achieved exclusively by main chain groups. Consequently, these meso positions cannot be unblocked by local structural adjustments.
The competition for iron has been recognized in recent years as one of the most important host-pathogen interactions during bacterial infections. The rivals involved in this competition, the host iron withholding systems (29) and the bacterial iron acquisition systems (1), have received substantial attention. Determination of the structures of virulence factors involved in bacterial heme iron acquisition, and of heme oxygenases in particular, are essential for our understanding of these vital interactions and may aid in the design of novel antibiotics. The HugZ structure and the identification of a new family of heme-utilization proteins, as reported in this work, may provide us with new targets in our battle against pathogenic bacteria.
Acknowledgments
We thank X. Zhao, R. Zheng, and Y. Han at IBP Core Facilities for technical assistance. The diffraction datasets were collected at Beamline 17A, Photon Factory, KEK, Japan (KEK2008G007).
This work was supported by funding from Minister of Science and Technology of China Grants 2006CB806502, 2006CB910901, 2006AA02A322, and 2009CB522606, National Natural Science Foundation of China Grant 30400019, and Natural Science Foundation Project of Chongqing Grant CSTC 2009BB5166.
- HO
- heme oxygenase
- r.s.m.d.
- root mean square deviation.
REFERENCES
- 1. Wandersman C., Delepelaire P. (2004) Annu. Rev. Microbiol. 58, 611–647 [DOI] [PubMed] [Google Scholar]
- 2. Otto B. R., Verweij-van Vught A. M., MacLaren D. M. (1992) Crit. Rev. Microbiol. 18, 217–233 [DOI] [PubMed] [Google Scholar]
- 3. Skaar E. P., Humayun M., Bae T., DeBord K. L., Schneewind O. (2004) Science 305, 1626–1628 [DOI] [PubMed] [Google Scholar]
- 4. Wilks A., Burkhard K. A. (2007) Nat. Product Rep. 24, 511–522 [DOI] [PubMed] [Google Scholar]
- 5. Dhaenens L., Szczebara F., Van Nieuwenhuyse S., Husson M. O. (1999) Res. Microbiol. 150, 475–481 [DOI] [PubMed] [Google Scholar]
- 6. Guo Y., Guo G., Mao X., Zhang W., Xiao J., Tong W., Liu T., Xiao B., Liu X., Feng Y., Zou Q. (2008) BMC Microbiol. 8, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frankenberg-Dinkel N. (2004) Antioxidants Redox Signal. 6, 825–834 [DOI] [PubMed] [Google Scholar]
- 8. Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. P. (2008) J. Biol. Chem. 283, 30957–30963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Unno M., Matsui T., Ikeda-Saito M. (2007) Nat. Product Rep. 24, 553–570 [DOI] [PubMed] [Google Scholar]
- 10. Ridley K. A., Rock J. D., Li Y., Ketley J. M. (2006) J. Bacteriol. 188, 7862–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang F., Hu Y., Guo Y., Guo G., Zou Q. M., Wang da C. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 376–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. Sect. D 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 13. de La Fortelle E., Bricogne G. (1997) Methods Enzymol. 276, 472–494 [DOI] [PubMed] [Google Scholar]
- 14. Terwilliger T. C. (2000) Acta Crystallogr. Sect. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 16. Emsley P., Cowtan K. (2004) Acta Crystallogr. Sect. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 18. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 19. Liu Y., Moënne-Loccoz P., Hildebrand D. P., Wilks A., Loehr T. M., Mauk A. G., Ortiz de Montellano P. R. (1999) Biochemistry 38, 3733–3743 [DOI] [PubMed] [Google Scholar]
- 20. Chu G. C., Katakura K., Tomita T., Zhang X., Sun D., Sato M., Sasahara M., Kayama T., Ikeda-Saito M., Yoshida T. (2000) J. Biol. Chem. 275, 17494–17500 [DOI] [PubMed] [Google Scholar]
- 21. Wilks A., Moënne-Loccoz P. (2000) J. Biol. Chem. 275, 11686–11692 [DOI] [PubMed] [Google Scholar]
- 22. Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 23. Oldham A. L., Wood T. A., Henderson D. P. (2008) Can. J. Microbiol. 54, 97–102 [DOI] [PubMed] [Google Scholar]
- 24. Wyckoff E. E., Schmitt M., Wilks A., Payne S. M. (2004) J. Bacteriol. 186, 4142–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilks A., Sun J., Loehr T. M., Demontellano P. R. (1995) J. Am. Chem. Soc. 117, 2925–2926 [Google Scholar]
- 26. Sugishima M., Sakamoto H., Higashimoto Y., Omata Y., Hayashi S., Noguchi M., Fukuyama K. (2002) J. Biol. Chem. 277, 45086–45090 [DOI] [PubMed] [Google Scholar]
- 27. Takahashi S., Ishikawa K., Takeuchi N., Ikedasaito M., Yoshida T., Rousseau D. L. (1995) J. Am. Chem. Soc. 117, 6002–6006 [Google Scholar]
- 28. Friedman J., Lad L., Li H., Wilks A., Poulos T. L. (2004) Biochemistry 43, 5239–5245 [DOI] [PubMed] [Google Scholar]
- 29. Ganz T. (2009) Curr. Opin. Immunol. 21, 63–67 [DOI] [PMC free article] [PubMed] [Google Scholar]