Abstract
The EGF receptor is a classical receptor-tyrosine kinase. In the absence of ligand, the receptor adopts a closed conformation in which the dimerization arm of subdomain II interacts with the tethering arm in subdomain IV. Following the binding of EGF, the receptor opens to form a symmetric, back-to-back dimer. Although it is clear that the dimerization arm of subdomain II is central to the formation of receptor dimers, the role of the tethering arm of subdomain IV (residues 561–585) in this configuration is not known. Here we use 125I-EGF binding studies to assess the functional role of the tethering arm in the EGF receptor dimer. Mutation of the three major residues that contribute to tethering (D563A,H566A,K585A-EGF receptor) did not significantly alter either the ligand binding properties or the signaling properties of the EGF receptor. By contrast, breaking the Cys558-Cys567 disulfide bond through double alanine replacements or deleting the loop entirely led to a decrease in the negative cooperativity in EGF binding and was associated with small changes in downstream signaling. Deletion of the Cys571-Cys593 disulfide bond abrogated cooperativity, resulting in a high affinity receptor and increased sensitivity of downstream signaling pathways to EGF. Releasing the Cys571-Cys593 disulfide bond resulted in extreme negative cooperativity, ligand-independent kinase activity, and impaired downstream signaling. These data demonstrate that the tethering arm plays an important role in supporting cooperativity in ligand binding. Because cooperativity implies subunit-subunit interactions, these results also suggest that the tethering arm contributes to intersubunit interactions within the EGF receptor dimer.
Keywords: Allosteric Regulation, Cooperativity, Growth Factors, Signal Transduction, Tyrosine Protein Kinase (Tyrosine Kinase), EGF Receptor
Introduction
The EGF receptor is a receptor-tyrosine kinase composed of an extracellular ligand binding domain, a single α-helical transmembrane domain, and a cytoplasmic tyrosine kinase domain (1). In the absence of ligand, the EGF receptor exists as a monomer, although substantial evidence suggests that it is also present as an inactive predimer (2–5). Binding of EGF induces dimerization of the receptor and leads to the stimulation of its intracellular tyrosine kinase activity (6).
The primary target of this tyrosine kinase is the receptor itself, with phosphorylation occurring in trans on the C-terminal tail of the receptor (7). These phosphotyrosine residues serve as sites for the binding of Src homology 2 and PTB domain-containing proteins that promote the assembly of the signaling complexes that mediate the intracellular effects of EGF (8–11).
The extracellular domain of the EGF receptor is composed of four subdomains referred to as subdomains I–IV (12, 13). Subdomains I and III are homologous and together form the site at which EGF is bound. Subdomains II and IV are also homologous and are regions of high cysteine content. In the absence of ligand, the extracellular domain is held in a closed conformation by interactions between the dimerization arm in subdomain II and the tethering arm in subdomain IV (14). Upon binding ligand, this intramolecular tether is released, and the EGF receptor opens into its extended conformation. In this configuration, the dimerization arm in subdomain II, which had previously been involved in the intramolecular tether, mediates the formation of a back-to-back receptor dimer (15, 16).
Although the function of the subdomain II dimerization arm in EGF receptor dimer formation is well documented (15–17), the role of the tethering arm of subdomain IV remains unclear. The tethering arm is strongly conserved in all ErbB family receptors, even ErbB2, which does not adopt a tethered conformation (18, 19). This suggests that this portion of the extracellular domain plays a role in receptor function beyond simple tethering.
The tethering arm is not present in either of the reported crystal structures of the ligand-occupied EGF receptor (15, 16). Thus, it is not clear how, or whether, this domain interacts with other portions of the extracellular domain in the receptor dimer. Modeling of the position of subdomain IV in the EGF receptor dimer, based on its structure in the tethered receptor monomer, suggests that the subdomains IV cross in the extended, dimerized form of the EGF receptor (14, 18). This conclusion is supported by negative stain electron microscopy (20). Molecular dynamics simulations suggest that the putative crossing could occur at multiple different positions in subdomain IV (21), raising the possibility that subdomain IV-subdomain IV interactions may be highly dynamic.
In the absence of structural data, functional studies of the role of the tethering arm have been pursued. Some studies have suggested that the tethering arm exerts limited control over EGF receptor binding and kinase activity (22). Consistent with this conclusion, small angle x-ray scattering analysis of the soluble extracellular domain of the EGF receptor indicated that the unoccupied EGF receptor retains its closed configuration even when the entire tethering arm is deleted (23). On the other hand, mutagenesis studies have suggested that deletion of the tethering arm alters EGF binding (22, 24, 25) and signal transduction (24). Thus, the biological data on the role of the tethering arm in EGF receptor function are equivocal.
We have recently used a new method for analyzing ligand binding data to show that the binding of EGF to its receptor is negatively cooperative (26, 27). Cooperativity implies subunit-subunit interactions, and indeed, when receptor dimerization is blocked by mutation of the dimerization arm, ligand binding cooperativity is abrogated (26). Therefore, this analysis of ligand binding can be used as a probe for functional subunit-subunit interactions.
Here we use this methodology to explore the role of the tethering arm in the regulation of ligand binding and signal transduction by the EGF receptor. Our data indicate that the tethering arm actively participates in the intersubunit interactions that give rise to the negative cooperativity in EGF binding. In addition, this arm appears to play a key role in the ability of the extracellular domain to regulate the state of activation of the cytoplasmic tyrosine kinase. These findings provide functional evidence that the tethering arm is engaged in receptor-receptor interactions and identify a previously unrecognized role for this subdomain in the regulation of ligand binding allostery.
EXPERIMENTAL PROCEDURES
Materials
The CHO-K1 Tet-On cell line, the pBI Tet vector, and the doxycycline were from Clontech (Mountain View, CA). PfuTurbo DNA polymerase was from Stratagene (La Jolla, CA). Lipofectamine 2000, hygromycin, and Dynabeads Protein A were from Invitrogen. Anti-phosphotyrosine antibodies (PY20) were from BD Transduction Laboratories (San Jose, CA). The EGF receptor was detected using a mixture of antibodies from Cell Signaling (Danvers, MA) and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against Akt and phospho-Akt antibody were from Cell Signaling (Danvers, MA). Anti-Gq antibodies were from Santa Cruz Biotechnology, Inc. The MAPK antibody was from Upstate (Lake Placid, NY), and the phospho-specific MAPK antibody was from Promega (Madison, WI). Anti-transferrin receptor antibodies were from Zymed Laboratories Inc. (San Francisco, CA). Murine EGF was purchased from Biomedical Technologies, Inc. (Stoughton, MA). Na125I was from PerkinElmer Life Sciences. Opti-Prep was from Greiner Bio-One. Sulfo-NHS-LC-Biotin and HRP-conjugated streptavidin were purchased from Thermo-Scientific (Rockford, IL). Polyvinylidene difluoride membranes were from Millipore. The ECL reagents were from GE Healthcare. G418 and all other chemicals were from Sigma.
Construction of EGF Receptor Mutants and Plasmids
The triple point mutant (D563A,H566A,K585A-EGF receptor) and the two double point mutants, C558A,C567A- and C571A,C593A-EGF receptors, were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) on the pEGFR-WT plasmid that contains the wild type human EGF receptor in the pcDNA5FRT plasmid. To generate the EGF receptor mutants lacking the first (residues 558–567) or the second (residues 571–593) disulfide loop of the tethering arm, a two-step PCR method with internal primer overlap was used with pEGFR-WT as the template. The resulting PCR product was purified and cut with NheI and BstEII and ligated into pcDNA5FRT. The construct was confirmed by sequencing. This construct was then digested with NheI and EcoRV and inserted into the pBI Tet vector to generate the various EGF receptor mutants on the inducible plasmid. The pBI Tet vector product was confirmed by sequencing.
Cells and Tissue Culture
Chinese hamster ovary (CHO)-K1 Tet-On cells were co-transfected with pTK-Hyg and the pBI Tet vector engineered to express wild type or mutant EGF receptors, using Lipofectamine 2000 according to the manufacturer's instructions. Stable clones were isolated by selection in 500 μg/ml hygromycin. Cells were maintained in DMEM containing 10% FetalPlex, 1000 μg/ml penicillin/streptomycin, 100 μg/ml G418, and 100 μg/ml hygromycin. Forty-eight hours before use, 1.1 × 105 cells were seeded into a 35-mm well in medium containing doxycycline at the indicated concentration to induce the expression of EGF receptors.
125I-EGF Synthesis and Binding
125I-EGF was synthesized using the oxidative ICl procedure of Doran and Spar (28). Radioligand binding was performed as described previously (26). Briefly, cells in 6- well dishes were incubated overnight on ice in 3 ml/well Ham's F-12 medium containing 3 mg/ml BSA and 25 mm Hepes, pH 7.0, plus 20–40 pm 125I-EGF and increasing concentrations of unlabeled EGF. The binding medium was aspirated, and the monolayers were washed three times in ice-cold Hanks' balanced salt solution. The cells were dissolved in 1 n NaOH, and the lysates were counted for 125I. Assays were done in triplicate. Nonspecific binding was determined by fitting the raw data to a competition binding model and using the fitted bottom value as nonspecific. Data from all binding isotherms were globally fit to the equation,
 |
where R0 = total concentration of EGF receptors as derived by Wyman and Gill (29), using GraphPad Prism 4.0 as described previously (26).
Signaling Assays and Western Blotting
CHO cells were grown for 48 h in 6-well dishes. For the assay, cells were transferred into Ham's F-12 medium containing 25 mm Hepes, pH 7.2, and 1 mg/ml BSA plus varying concentrations of EGF. After the indicated time, the culture medium was removed, and the cells were washed in ice-cold PBS. The monolayers were scraped into radioimmune precipitation assay buffer (150 mm NaCl, 10 mm Tris, pH 7.2, 0.1% SDS, 1% Triton X-100, 17 mm deoxycholate, and 2.7 mm EDTA) containing 20 mm p-nitrophenyl phosphate, 1 mm sodium orthovanadate, and protease inhibitors. Equal amounts of protein were separated by SDS-PAGE and then transferred to polyvinylidene difluoride membranes. Western blots were blocked with 10% nonfat milk, and specific proteins were detected by chemiluminescence.
Cell Surface Biotinylation
CHO cells were grown to confluence in 6-well dishes. Cultures were washed twice with ice-cold PBS, pH 8.0. Cell surface proteins were biotinylated by incubating the cells in 1 mg/ml sulfo-NHS-LC-biotin in PBS for 45 min at 4 °C. Unreacted biotin was quenched and removed by three washes of ice-cold PBS supplemented with 100 mm glycine. The cells were then washed once with ice-cold PBS and scraped into radioimmune precipitation assay buffer. Cell lysates containing 140 μg of protein were incubated for 3 h at 4 °C with 10 μl of packed Dynabeads Protein A to which had been bound 2 μg of an anti-EGF receptor antibody. The unbound fractions were collected after centrifugation to pellet the Dynabeads. The protein-bead complexes were washed three times with lysis buffer. Bound EGF receptors were eluted by boiling for 10 min in a 1:1 mixture of radioimmune precipitation assay buffer and SDS gel sample buffer. Samples were analyzed by SDS-PAGE and transferred to polyvinylidene difluoride membranes for Western blotting as described above. Biotinylated EGF receptors were detected by ECL after incubating the blots with HRP-conjugated streptavidin.
Isolation of Lipid Rafts
Lipid rafts were prepared according to the method of Macdonald and Pike (30). This is a detergent-free preparation that involves the isolation of rafts on a continuous, linear density gradient of Opti-Prep. The 12-ml gradients were fractionated into 18 equal volume fractions, beginning at the top of the gradient. Equal aliquots of each fraction were separated by polyacrylamide gel electrophoresis and analyzed by Western blotting for EGF receptors, Gq, and the transferrin receptor. The amount of EGF receptor present in each gradient fraction was quantitated by densitometry using Image J software. Fractions 1–5 are considered raft fractions.
RESULTS
Releasing the Tether
The tethering “arm” of subdomain IV of the EGF receptor has been defined as the region between residues 561 and 585 (14). This region is composed of two different disulfide loops, one between Cys558 and Cys567 and the other between Cys571 and Cys593 (Fig. 1). Residues from both loops of the tethering arm form hydrogen bonds with residues in the dimerization arm in subdomain II of the EGF receptor (14). Asp563, His566, and Lys585 in subdomain IV have been identified as the main residues that participate in the tethering interactions (14).
FIGURE 1.
Structure of the EGF receptor. The structure at the top shows the tethered form of the EGF receptor (Protein Data Bank entry 1NQL) (14). The EGF ligand is shown in magenta. The disulfide bonds in subdomain IV are shown in yellow. The tethering arm disulfide loops are shown in blue (residues 558–567) or red (residues 571–593). The bottom structure shows an expanded view of the tether itself highlighting the interactions between the dimerization arm (in gray) and the two disulfide loops in the tethering arm.
To assess the role of tethering in EGF receptor function, we first characterized the D563A,H566A,K585A-EGF receptor triple point mutant with respect to its signaling and ligand binding functions. Mutation of these three residues to alanine alters the major residues that participate in tethering. If the tethering interactions are important for receptor function, the biological properties of this receptor should be altered. As shown in Fig. 2, the wild type EGF receptor and the D563A,H566A,K585A-EGF receptor supported a comparable level of EGF-stimulated receptor autophosphorylation and exhibited similar EC50 values. Despite the loss of all major tethering interactions, no ligand-independent kinase activity was apparent. This is consistent with the observations of Matoon et al. (22), who found no increase in basal autophosphorylation in a series of tethering arm mutations. The wild type and D563A,H566A,K585A-EGF receptors also mediated similar levels of activation of MAPK and Akt over the same concentration range. Thus, releasing all of the major tethering interactions had a negligible effect on the signaling properties of the receptor.
FIGURE 2.
Signal transduction by the wild type EGF receptor and the D563A,H566A,K585A-EGF receptor mutant. Cells expressing the wild type or mutant EGF receptor were treated with concentrations of doxycycline to give approximately equivalent levels of EGF receptor expression. Cultures were treated with the indicated concentration of EGF for 5 min. Lysates were prepared and analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies.
It has long been recognized that there is heterogeneity in the affinity of EGF for binding to its receptor (22, 25, 31–34). We have recently shown that this heterogeneity can best be explained by a model that involves negative cooperativity in an aggregating system (26). In this model (Fig. 3A), the EGF receptor is present in a pre-existing equilibrium between monomers and dimers. EGF can then bind to three species: the monomer (described by the association constant, K11), the first site on the dimer (described by K21), and the second site on the dimer (described by K22). Heterogeneity in EGF binding affinity arises when EGF binds with a different affinity to these different species. Because the position of the monomer-dimer equilibrium depends on the concentration of EGF receptors on the cell surface, the position of the saturation binding isotherm will shift as the concentration of EGF receptors increases if the monomeric and dimeric species exhibit different affinities for EGF.
FIGURE 3.
125I-EGF binding to wild type EGF receptor and the D563A,H566A,K585A-EGF receptor mutant. A, model for EGF binding in an aggregating system. Unoccupied receptor subunits are shown as open circles. E indicates a bound EGF molecule. Equilibrium association constants are indicated. B and C, 125I-EGF binding isotherms for cells expressing wild type EGF receptors (B) or D563A,H566A,K585A-EGF receptors (C). Receptors were expressed on a tetracycline-inducible plasmid in CHO cells. Cells were grown for 48 h in the presence of increasing concentrations of doxycycline followed by analysis of the binding of 125I-EGF as described under “Experimental Procedures.” Fitted values for the equilibrium constants are given in the lower right corner of B and C.
Using this model as the basis for our analysis, the 125I-EGF binding properties of the wild type and D563A,H566A,K585A-EGF receptor were compared. Fig. 3B shows the saturation binding isotherms in CHO cells expressing increasing concentrations of wild type EGF receptors. For this receptor, the saturation binding isotherms shift from left to right as the number of cell surface EGF receptors increases. Global fitting of the data from all six binding curves yields the parameters given in the lower right corner of Fig. 3B. All parameters are given as association constants. The results are consistent with our previous studies and indicate that the wild type EGF receptor exhibits negative cooperativity (26, 27). Ligand binds to the first site on the dimer with about a 7-fold higher affinity than to the second site on the dimer.
Fig. 3C shows the saturation binding isotherms in CHO cells expressing increasing concentrations of the D563A,H566A,K585A-EGF receptor. As was the case for the wild type receptor, the binding isotherms shift from left to right as the number of cell surface EGF receptors increases. Global fitting of the data indicates that, like the wild type receptor, the D563A,H566A,K585A-EGF receptor exhibits negative cooperativity in which binding to the first site on the dimer occurs with an affinity that is about 7-fold higher than that for binding to the second site on the dimer. The affinity of EGF for binding to either of the sites on the mutant dimer is slightly increased as compared with the wild type receptor, consistent with the reported increase in affinity of the soluble extracellular domain of this mutant (14, 17). These data suggest that simply releasing the tether has relatively little effect on ligand binding of the D563A,H566A,K585A-EGF receptor and, in particular, does not alter the allosteric properties of the receptor.
Disulfide Loop Mutants in the Tethering Arm
The results with the D563A,H566A,K585A-EGF receptor suggest that selectively altering residues involved in tethering interactions does not affect EGF receptor function. Nonetheless, the tether region is highly conserved among ErbB family receptors. To further examine the role of the subdomain IV tethering arm in EGF receptor function, two different types of mutations were made (Fig. 4A). In one, Cys558 and Cys567 (C558A,C567A-EGF receptor) or Cys571 and Cys593 (C571A,C593A-EGF receptor) were substituted with alanines to release the disulfide bond while retaining all of the other residues in the respective loops. In a second type of mutant, the entire disulfide loop was deleted from cysteine to cysteine, yielding the Δ558–567-EGF receptor and the Δ571–591-EGF receptor. These constructs were stably transfected into Tet-On CHO cells.
FIGURE 4.
Expression of EGF receptor tethering arm mutants. A, sequence of the tethering arm mutants characterized in these studies. B and C, cells expressing the 558–567 loop mutants or the 571–593 loop mutants were subjected to cell surface biotinylation as described under “Experimental Procedures,” and lysates were prepared using radioimmune precipitation assay buffer. Lysates were analyzed by SDS-PAGE and Western blotting with an anti-EGF receptor antibody (left panels). Aliquots of the lysates were immunoprecipitated (IP) with an anti-EGF receptor antibody and analyzed by SDS-PAGE and Western blotting (IB) with an anti-EGF receptor antibody (middle panels) or HRP-conjugated streptavidin (right panels).
Both of the 558–567 loop mutants (Fig. 4B) and both of the 571–593 loop mutants (Fig. 4C) were expressed in CHO cells and could be identified in cell lysates by Western blotting with an anti-EGF receptor antibody (left panels). However, all four mutant receptors were expressed as a pair of bands as opposed to the single band observed in cells expressing the wild type EGF receptor. Because alterations in the two disulfide loops could affect the intracellular transport or trafficking of the mutant receptors, a cell surface biotinylation experiment was performed to determine which of the forms of the mutant receptors were present on the cell surface.
CHO cells expressing wild type or one of the four mutant EGF receptors were subjected to cell surface biotinylation with the membrane-impermeable reagent, Sulfo NHS-LC-biotin. Cells were lysed, and the EGF receptor was immunoprecipitated with an anti-EGF receptor antibody. The immunoprecipitates were analyzed by SDS-PAGE and Western blotting with an anti-EGF receptor antibody (middle panels of Fig. 4, B and C) or horseradish peroxidase-conjugated streptavidin (right panels of Fig. 4, B and C). As can be seen from the figure, the immunoprecipitates contained both the higher and lower molecular weight forms of the mutant EGF receptors. However, in all cases, only the higher molecular weight form was recognized by streptavidin, indicating that it had been biotinylated. These data indicate that for all four disulfide loop mutants, the high molecular weight form is present on the cell surface, whereas the low molecular weight form is in an intracellular compartment. Therefore, for all subsequent comparisons of receptor levels and phosphorylation done by Western blotting, only the higher molecular weight band was considered.
Binding and Kinase Activity of the 558–567 Disulfide Loop Mutants
Fig. 5, A and B, show the results of 125I-EGF binding studies performed on CHO cells expressing increasing levels of EGF receptors in which the 558–567 disulfide was either broken (C558A,C567A-EGF receptor) or deleted (Δ558–567-EGF receptor). The binding patterns for these two mutants are significantly different from those of the wild type receptor (Fig. 3B). In the C558A,C567A-EGF receptor (Fig. 5A), the binding isotherms are very close together but move from right to left with increasing receptor concentration. Global fitting of all of the binding isotherms yields a set of parameters that indicates that negative cooperativity has been modestly weakened in this mutant. There is only about a 3-fold difference in affinity of EGF for binding to the first site versus the second site on the dimer. Binding to the Δ558–567-EGF receptor (Fig. 5B) shows a similar effect. Although cooperativity is still present, the affinity difference between the first and second sites on the dimer has been reduced to only 2-fold. Thus, alterations in the 558–567 disulfide bond lead to small but noticeable changes in the ligand binding properties of the EGF receptor, suggesting that this region of the receptor makes a limited contribution to the receptor-receptor interactions that mediate ligand binding cooperativity.
FIGURE 5.
125I-EGF binding to the 558–567 disulfide loop EGF receptor mutants. 125I-EGF binding isotherms for receptors expressing C558A,C567A-EGF receptors (A) or Δ558–567-EGF receptors (B). Receptors were expressed on a tetracycline-inducible plasmid in CHO cells. Cells were grown for 48 h in the presence of increasing concentrations of doxycycline followed by analysis of the binding of 125I-EGF as described under “Experimental Procedures.” Fitted values for the equilibrium constants are given in the lower right corner of A and B.
Although these two 558–567 disulfide loop mutants bound EGF with good affinity and were expressed at high levels on the cell surface, both were severely impaired with respect to their ability to mediate receptor autophosphorylation. Fig. 6A shows the dose-response curves for EGF-stimulated receptor autophosphorylation in the wild type, C558A,C567A-, and Δ558–567-EGF receptors. The results are quantitated in the graph at the right. As is clear from the figure, both the C558A,C567A-EGF receptor and the Δ558–567-EGF receptor exhibited relatively weak kinase activity as compared with the wild type receptor. For the average of four separate experiments, the maximal level of autophosphorylation in the mutants was significantly less that of the wild type receptor (35 ± 14 and 24 ± 7% of wild type for C558A,C567A- and Δ558–567-EGF receptors, respectively, both p < 0.05). However, their EC50 values did not differ significantly from that of wild type.
FIGURE 6.
Signal transduction by the wild type EGF receptor and the 558–567 disulfide loop EGF receptor mutants. Cells expressing the wild type or mutant EGF receptors were treated with concentrations of doxycycline to give approximately equivalent levels of EGF receptor expression. Cultures were treated with the indicated concentration of EGF for 5 min. Lysates were prepared and analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies. Panel A, phosphotyrosine and EGF receptor; panel B, phospho-MAPK and MAPK; panel C, phospho-Akt and Akt. The right panels provide quantitation of the Western blots with phosphorylation normalized to the amount of that specific protein present. A replicate experiment is shown in supplemental Fig. 1.
Although it exhibited a significantly reduced level of EGF-induced autophosphorylation, stimulation of the Δ558–567-EGF receptor mediated essentially wild type levels of activation of both MAPK (Fig. 6B) and Akt (Fig. 6C). The observation that downstream signaling via MAPK was much less severely affected than the tyrosine kinase activity of the receptor is consistent with previous studies that suggest that relatively few EGF receptors must be activated to achieve full activation of these kinases (35). Stimulation of cells expressing the C558A,C567A-EGF receptor resulted in somewhat lower levels of activation of MAPK and Akt than wild type (62 ± 9 and 50 ± 12%, respectively, n = 2). Thus, this receptor was capable of transmitting a signal across the membrane although with somewhat reduced efficacy.
Binding and Kinase Activity of the 571–593 Disulfide Loop Mutants
The 125I-EGF saturation binding isotherms for the EGF receptors with mutations in the 571–593 disulfide loop are shown in Fig. 7, A and B. It is apparent that breaking versus deleting the disulfide loop resulted in dissimilar effects on the ligand binding properties of the receptors. As shown in Fig. 7A, the C571A,C593A-EGF receptor, in which the disulfide loop has been broken, shows a pattern somewhat similar to that seen in the wild type receptor. The binding isotherms shift from left to right with increasing concentrations of EGF receptor. However, the final fitted parameters are substantially different from those of the wild type receptor. Specifically, the affinity of EGF for binding to either site on the dimer has been significantly reduced. In particular, the affinity of EGF for the second site on the receptor dimer is at least 2 orders of magnitude lower in the mutant than in the wild type receptor. Thus, this mutant exhibits extreme negative cooperativity.
FIGURE 7.
125I-EGF binding to the 571–793 disulfide loop EGF receptor mutants. A, 125I-EGF binding isotherms for receptors expressing C571A,C593A-EGF receptor. B, 125I-EGF binding isotherms for receptors expressing 571–593-EGF receptors. Receptors were expressed on a tetracycline-inducible plasmid in CHO cells. Cells were grown for 48 h in the presence of increasing concentrations of doxycycline followed by analysis of the binding of 125I-EGF as described under “Experimental Procedures.” Fitted values for the equilibrium constants are given in the lower right corner of A and B.
The saturation binding isotherms for the Δ571–593-EGF receptor (Fig. 7B), in which the second disulfide loop was deleted, showed no shift with increasing concentrations of EGF receptors. The data fit to a model with a single affinity of ∼300 pm for both the monomeric and dimeric species. Thus, deletion of the 571–593 disulfide loop appears to abrogate the receptor-receptor interactions necessary for ligand binding cooperativity and results in a receptor that has a uniform and relatively high affinity for EGF.
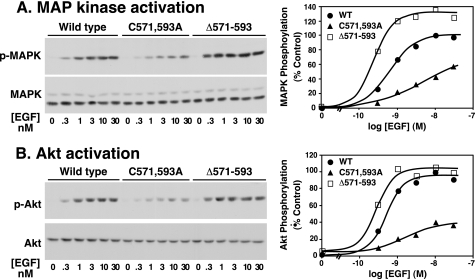
Despite these significant changes in ligand binding properties, both the C571A,C593A-EGF receptor and the Δ571–593-EGF receptor underwent EGF-stimulated receptor autophosphorylation. Fig. 8A shows the dose response to EGF for the wild type, C571A,C593A-, and Δ571–593-EGF receptors. Although it lacks ligand binding cooperativity, the Δ571–593-EGF receptor exhibited about 60% of wild type kinase activity. Consistent with its higher ligand binding affinity, the Δ571–593-EGF receptor exhibited an EC50 for EGF that was slightly better than that of the wild type receptor (see the legend to Fig. 8 for details). The C571A,C593A-EGF receptor exhibited essentially wild type levels of EGF-stimulated receptor autophosphorylation and an EC50 value that was not significantly different from wild type. However, in contrast to all of the other receptors, the C571A,C593A-EGF receptor consistently showed a significant basal kinase activity.
FIGURE 8.
Autophosphorylation of the wild type EGF receptor and the 571–593 disulfide loop EGF receptor mutants. A, cells expressing the wild type or mutant EGF receptors were treated with concentrations of doxycycline to give approximately equivalent levels of EGF receptor expression. Cultures were treated with the indicated concentration of EGF for 5 min. Lysates were prepared and analyzed by SDS-PAGE followed by Western blotting with anti-phosphotyrosine antibodies (top) or anti-EGF receptor antibodies (bottom). The lower graph provides quantitation of the Western blots with phosphorylation normalized to the amount of EGF receptor present. For the mutant receptors, only the higher molecular weight cell surface form of the receptor was considered. A replicate experiment is shown in supplemental Fig. 2. Maximal autophosphorylation of the Δ571–593-EGF receptor was 57 ± 7% of wild type levels (n = 5, p < 0.05). Maximal autophosphorylation of the C571A,C593A-EGF receptor was 114 ± 23% of wild type (n = 5, not significant). The EC50 of the wild type receptor for EGF was 3.7 ± 1.5 nm (n = 5) but was 0.76 ± 0.25 nm for the Δ571–593-EGF receptor (n = 5, p < 0.05 as compared with wild type) and was 5.4 ± 3 nm (n = 5, not significant) for the C571A,C593A-EGF receptor. B, cells expressing wild type or mutant EGF receptors were treated with increasing concentrations of doxycycline to induce receptor expression. Cultures were treated with or without 25 nm EGF for 5 min. Lysates were prepared and analyzed by SDS-PAGE followed by Western blotting with anti-phosphotyrosine (pTyr) antibodies (top panels) or anti-EGF receptor antibodies (bottom panels).
To further explore the ligand-independent kinase activity in the C571/593A-EGF receptor, cells expressing the wild type EGF receptor as well as the C571A,C593A-EGF receptor and the Δ571–593-EGF receptor were treated with increasing concentrations of doxycycline to induce increasing levels of receptor expression. The ability of EGF to stimulate receptor autophosphorylation was then assessed in all three cell lines. The results are shown in Fig. 8B.
As expected, increasing the level of expression of the wild type receptor resulted in visibly increased levels of EGF-stimulated autophosphorylation. However, no increase in basal autophosphorylation of the receptor was seen even at the highest levels of receptor expression. This pattern was also observed in the Δ571–593-EGF receptor, although extremely high levels of receptor expression could not be achieved using this mutant. By contrast, even at very low levels of receptor expression, the C571A,C593A-EGF receptor exhibited some basal autophosphorylation. As the level of receptor expression increased, the extent of basal autophosphorylation increased such that at very high levels of expression, receptor autophosphorylation became entirely ligand-independent. In addition, both the higher molecular weight cell surface form of the receptor and the lower molecular weight intracellular form of the receptor became phosphorylated. This indicates that phosphorylation of the C571A,C593A-EGF receptor can occur intracellularly and is consistent with the ligand-independent nature of its phosphorylation at high levels of expression.
As shown in Fig. 9, A and B, the Δ571–593-EGF receptor, which lacked cooperativity, mediated essentially wild type (or better) levels of MAPK and Akt activation. Consistent with its higher binding affinity, cells expressing the Δ571–593-EGF receptor showed a greater sensitivity to EGF for activating MAPK and Akt (see the legend to Fig. 9 for details). By contrast, the C571A,C593A-EGF receptor, which exhibited strong negative cooperativity, showed a modestly decreased activation of MAPK and Akt with a reduced sensitivity to EGF. Thus, the absence of negative cooperativity was associated with greater sensitivity to EGF, whereas high negative cooperativity was associated with reduced sensitivity of downstream pathways to EGF.
FIGURE 9.
Signal transduction by the wild type EGF receptor and the 558–567 disulfide loop EGF receptor mutants. Cells expressing the wild type or mutant EGF receptors were treated with concentrations of doxycycline to give approximately equivalent levels of EGF receptor expression. Cultures were treated with the indicated concentration of EGF for 5 min. Lysates were prepared and analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies. The right panels provide quantitation of the Western blots with phosphorylation normalized to the amount of that specific protein present. A replicate experiment is shown in supplemental Fig. 2. In two separate experiments, maximal activation of MAPK (A) by the C571A,C593A-EGF receptor was 62 ± 5% of wild type (p < 0.05) and was 122 ± 4% of wild type for the Δ571–593-EGF receptor (not significant). Maximal activation of Akt (B) by the C571A,C593A-EGF receptor was 41 ± 2% of wild type (p < 0.05) and was 101 ± 3.5% of wild type for the Δ571–593-EGF receptor (not significant). The EC50 values for activation of MAPK were 0.47 ± 0.15, 2.3 ± 1, and 0.13 ± 0.035 nm for wild type, C571A,C593A-EGF, and Δ571–593A-EGF receptors, respectively. The EC50 values for activation of Akt were 0.48 ± 0.04, 1.2 ± 0.3, and 0.12 ± 0.04 nm for wild type, C571A,C593A-EGF, and Δ571–593A-EGF receptors, respectively.
Raft Localization of Tethering Arm Mutants
The EGF receptor has been shown to partition into membrane microdomains known as lipid rafts (36–39). Stimulation of cells with EGF leads to the migration of the EGF receptor out of membrane rafts and into the bulk plasma membrane (36). The most membrane-proximal 60 amino acids of the extracellular domain have been implicated in the ability of the EGF receptor to partition into membrane rafts (40). Because this region includes the tethering arm, we assessed the effect of our tethering arm mutations on the ability of EGF to partition into rafts. The results are shown in Fig. 10.
FIGURE 10.
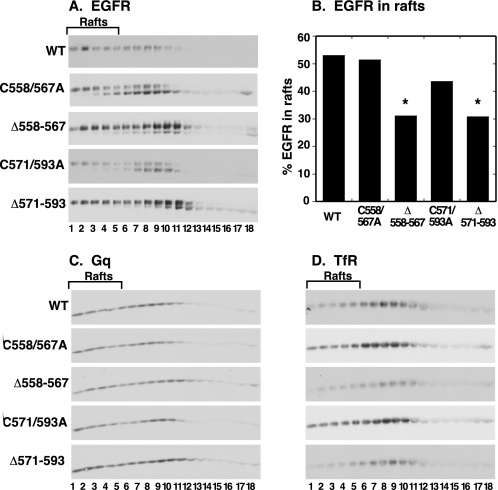
Raft localization of wild type and tethering arm mutant EGF receptors. Cells expressing the wild type or mutant EGF receptors were treated with concentrations of doxycycline to give approximately equivalent levels of EGF receptor expression. Membrane rafts were prepared using the non-detergent method of Macdonald and Pike (30). Equal aliquots of each gradient fraction were separated on SDS gels and blotted for EGF receptors (A), Gq (C), or transferrin receptor (D). B shows the quantitation of the fraction of EGF receptors in the raft fractions (fractions 1–5). For the mutant receptors, only the higher molecular weight cell surface form of the receptor was considered in this analysis. The experiment was repeated three times, and the results were used for analysis of the significance of the observed differences. *, value is different from wild type at a level of p < 0.05.
For these studies, the membrane rafts were prepared by a non-detergent, OptiPrep gradient method reported previously (30). In these preparations, fractions 1–5 represent the low density raft fractions of the gradient. Fig. 10A shows the distribution of the various EGF receptors across the OptiPrep gradients. The data are quantitated in Fig. 10B. Because only the higher molecular weight species of each of the mutants appears to be located on the cell surface (see Fig. 4), only the partitioning of this form was considered. As shown in Fig. 10B, ∼50% of the wild type EGF receptor was recovered in the low density fractions of the gradient. A similar fraction of the cell surface C558A,C567A-EGF receptor was found in the raft fractions, indicating that releasing this disulfide bond did not affect the ability of the EGF receptor to partition into membrane rafts. By contrast, deletion of the 558–567 disulfide loop resulted in a receptor that was significantly less localized to membrane rafts. A similar pattern was seen for the mutants in the 571–593 disulfide loop. Releasing the disulfide loop (the C571A,C593A-EGF receptor) had a modest but not significant effect on the partitioning of the EGF receptor into membrane rafts. However, deletion of the 571–593 disulfide loop consistently resulted in a significant decrease in the fraction of the cell surface EGF receptor recovered in the low density raft fractions. These differences cannot be attributed to differences among the gradients because the distribution of Gq (Fig. 10C), another raft marker, as well as the transferrin receptor (Fig. 10D), a plasma membrane marker, did not differ significantly among the various receptor mutants. These data are consistent with the involvement of sequences in the tethering arm in directing the partitioning of the EGF receptor into membrane rafts.
DISCUSSION
The tethering arm of the EGF receptor is known to be involved in interactions that stabilize the receptor in its closed configuration (14). However, this arm does not appear to be required for the maintenance of the tethered conformation because its removal does not lead to the adoption of the extended conformation (23). The fact that the sequence of this domain is relatively well conserved in ErbB family receptors, including the non-tethered ErbB2 (18, 19), suggests that it plays a role in some receptor function other than tethering. We therefore sought evidence for a role of this domain in activity-related functions of the EGF receptor. Because the subdomains IV are predicted to cross in the EGF receptor dimer (14, 18, 21), we looked for an indication that the tethering arm participated in intersubunit interactions.
To do this, we took advantage of our recent finding that the binding of EGF to its receptor is negatively cooperative (26, 27). Because cooperativity arises from subunit-subunit interactions, changes in cooperativity are an indication of alterations in subunit-subunit interactions. We can detect such changes by examining the binding of EGF in cells expressing increasing levels of EGF receptors (26, 27). Our data reported here provide clear evidence that the tethering arm contributes significantly to subunit-subunit interactions within the EGF receptor dimer.
Our characterization of the D563A,H566A,K585A-EGF receptor is consistent with previous studies showing no effect of the removal of the tethering interactions on EGF receptor-mediated signaling (22). Interestingly, our binding analysis showed essentially no difference in the affinity of EGF for binding to the wild type or D563A,H566A,K585A-EGF receptor monomers. This implies that ligand binding heterogeneity is not due to differences in the affinity of EGF for tethered versus untethered monomers. However, the D563A,H566A,K585A-EGF receptor did show an ∼3-fold increase in affinity for binding to either site on the dimer as compared with wild type receptor. This suggests that removal of the tethering interactions does reduce the energy barrier that must be overcome for the receptor to adopt the open conformation.
Subdomain IV of the EGF receptor contains 10 disulfide bonds. They form a spine down the center of this rodlike domain (see Fig. 1). The tethering arm is composed of two disulfide loops, 558–567 and 571–593. In this work, we either deleted each disulfide loop independently or broke each disulfide bond by replacing the relevant cysteines with alanines. If residues in a loop were involved in receptor-receptor interactions, then deletion of those residues should abrogate those interactions. By contrast, if the disulfide bond were important largely because of the structural stability it provided, then deletion of the residues in the loop should have a relatively modest effect on receptor function.
Both deletion and release of the 558–567 disulfide loop resulted in receptors in which the negative cooperativity associated with binding of EGF to the second site on the dimer was modestly reduced in the mutant as compared with the wild type receptor. As a result, the affinity of EGF for the monomer and both sites on the dimer is similar, and consequently, the binding isotherms lie close together. These data suggest that the 558–567 disulfide loop participates in a limited fashion in the receptor-receptor interactions that give rise to the observed cooperativity in EGF binding.
In contrast to what was seen with the 558–567 loop mutations, deletion of the 571–593 disulfide loop (the Δ571–593-EGF receptor) resulted in the complete loss of cooperativity in EGF binding. This suggests that residues in this loop are directly involved in the intersubunit interactions that give rise to binding cooperativity in the EGF receptor dimer. Interestingly, in the absence of cooperativity, the Δ571–593-EGF receptor shows a relatively high affinity for EGF. This implies that the interactions mediated by the 571–593 disulfide loop serve to reduce the affinity of a high affinity binding site. This is consistent with the observation of Elleman et al. (41), who showed that the affinity of EGF for a soluble EGF receptor (residues 1–501) fragment was higher than for the full-length (residues 1–621) EGF receptor extracellular domain. Because we could not express the Δ571–593-EGF receptor at extremely high levels, it is possible that some small degree of cooperativity is retained that we cannot detect. However, the characteristics of the C571A,C593A-EGF receptor mutant provide additional support for the importance of the 571–591 disulfide loop of the tethering arm in ligand binding cooperativity.
Breaking the 571–593 disulfide bond in the C571A,C593A-EGF receptor mutant also substantially altered the ligand binding properties of the receptor. The affinity of EGF for binding to the first site on the dimer was somewhat lowered. However, binding to the second site on the receptor dimer was markedly reduced compared with the wild type receptor so that the receptor exhibited an extreme form of negative cooperativity. K22, the association constant for the binding of EGF to the second site on the dimer, was reduced to the point that this site would be unable to bind EGF, even at pharmacologic doses of the growth factor. As a result, doubly occupied dimers would never form, and at high concentrations of EGF, the dimer would dissociate into monomers that possess a more favorable binding affinity for ligand. Together with the effect of deletion of the 571–593 disulfide loop, these findings indicate that this loop plays a major role in the subunit-subunit interactions that underlie cooperative ligand binding in the EGF receptor. These interactions appear to be independent of the tethering function of this domain because negative cooperativity was essentially normal in the D563A,H566A,K585A-EGF receptor triple point mutant.
Our binding data provide functional evidence that the tethering arm participates in intersubunit interactions in EGF receptor dimers. Tests of the signaling activity of the tethering arm mutants suggest that this domain is also important for transducing the EGF binding signal across the cell membrane. For both mutants in the 558–567 loop, there was a significant decrease in EGF-stimulated receptor autophosphorylation as compared with the wild type receptor. Thus, although alterations in this loop result in relatively minor changes in ligand binding cooperativity, the integrity of this loop is clearly necessary for transmission of the EGF binding signal across the membrane to the kinase domain.
Deletion of the 571–593 disulfide loop had a modest effect on the level of EGF-stimulated receptor autophosphorylation and had essentially no impact on the activation of MAPK and Akt. In fact, the activation of both of these kinases was slightly enhanced in Δ571–593-EGF receptor-expressing cells. This may be due to the absence of allosteric regulation in this receptor, which probably mediates desensitization of the receptor (27). Ablation of receptor desensitization would lead to enhanced signaling. Interestingly, the C571A,C593A-EGF receptor, in which the disulfide bond had been broken, retained nearly normal levels of tyrosine kinase activity but became ligand-independent at high concentrations of receptor. This finding is reminiscent of the finding of Sorokin (42), who observed constitutive tyrosine kinase activity when he placed a 40-amino acid flexible linker after position 618 in the extracellular domain of the EGF receptor.
Jura et al. (43) have recently suggested that one role of the extracellular domain of the EGF receptor might be to hold the transmembrane helices apart in the predimerized form of the receptor to prevent the formation of the activating asymmetric kinase dimers. Our data are consistent with this view. Release of the Cys571-Cys593 disulfide bond would certainly increase the flexibility of this portion of subdomain IV. The loss of conformational rigidity could prevent the extracellular domains from holding the transmembrane helices and their attendant kinase domains apart, resulting in constitutive activation of the kinase domain. Simply deleting the entire loop (the Δ571–593-EGF receptor) would not generate such flexibility and, indeed, did not give rise to ligand-independent kinase activation.
Despite the fact that the C571A,C593A-EGF receptor exhibited an elevated level of basal autophosphorylation, it did not support ligand-independent activation of MAPK or Akt. In fact, the activation of these two downstream pathways by EGF was only about half as great in cells expressing the C571A,C593A-EGF receptor as in cells expressing the wild type receptor. It is possible that the constitutive tyrosine kinase activity leads to partial desensitization of the receptor or that other adaptations are made by the cells to limit signaling. Alternatively, the apparent inability of this receptor to form doubly occupied dimers, as judged by the very low value of K22, may selectively impair its ability to stimulate these downstream signaling events.
Membrane rafts are low density, cholesterol- and sphingolipid-enriched microdomains present in many cell types (44). Residues 557–617 of the EGF receptor have previously been shown to contain targeting information necessary to localize the receptor to membrane rafts (40). We therefore assessed the ability of our tethering arm mutants to partition into rafts. Releasing either disulfide bond failed to significantly affect the ability of the EGF receptor to partition into the low density membrane fraction. However, wholesale deletion of either loop resulted in a 40% decrease in the fraction of the EGF receptor that was recovered in low density membranes.
Exactly how these disulfide loops serve to target the EGF receptor to membrane rafts is unclear. The extracellular domain of the EGF receptor has been shown to interact with gangliosides, particularly GM32 (45–48), that partition into lipid rafts. Thus, it is possible that these disulfide loops interact with gangliosides, thereby directing the receptor into membrane microdomains. Alternatively, in the tethered monomer, the two loops could support the formation of a specific conformation of the receptor that exposes a targeting signal located elsewhere in the receptor.
Together, our findings suggest that in the human EGF receptor dimer, the two subdomains IV interact functionally to enable ligand binding cooperativity and regulate signal transduction. The observation that the 571–593 disulfide loop is the most important portion of the tethering arm is in agreement with the molecular dynamics simulations of Kästner et al. (21), who predicted that residues in this loop would be involved in all three predicted “crossing modes” of the subdomains IV in EGF receptor dimers. In this context, it is interesting to note that mutations at four different positions in the 571–593 disulfide loop, none of which are involved in tethering, have been reported to be associated with the formation of tumors in humans (49). Examination of the properties of these receptors may provide additional insight into the role of this loop in regulating EGF receptor function.
Alvarado et al. (50) recently reported the structural basis of negative cooperativity in the Drosophila EGF receptor. In the absence of ligand, the Drosophila EGF receptor can form a symmetric dimer in which the subdomains IV cross (51). Upon binding of SpitzEGF, the structure adopts an asymmetric conformation in which the binding sites for SpitzEGF are structurally distinct in the two subunits. In this asymmetric dimer, the subdomains IV move apart from each other, indicating that the optimization of ligand binding leads to a conformational change in a domain far removed from the actual binding site. Given the homology between the human and Drosophila EGF receptors, it is not unlikely that ligand binding in the human EGF receptor would also be associated with changes in the position of subdomain IV.
Our data suggest the following model for ligand binding cooperativity in the EGF receptor. Binding of EGF to a receptor monomer (or the first site on a preassembled dimer) leads to the formation of a singly occupied EGF receptor dimer. In this dimer, which exhibits high affinity for EGF, subdomain IV interactions are weak or non-existent, possibly due to a lateral separation of the domains. In this configuration, the asymmetric kinase dimer could form, leading to activation of the tyrosine kinase activity. Binding of the second ligand induces a conformational change that promotes subdomain IV-mediated interactions. These interactions restrict the conformations accessible to the ligand binding site, leading to a reduction in EGF binding affinity. Deletion of the 571–593 loop removes these limitations, allowing the dimer to assume a conformation that has high affinity for both ligands. Release of the Cys571-Cys593 disulfide bond facilitates these subdomain IV-subdomain IV interactions, ultimately making it impossible for a second ligand to bind to the dimer. The closer approximation of the fourth subdomains in the doubly occupied extracellular domain dimer may force a change in the configuration of the intracellular kinase dimers, causing the conversion of one asymmetric dimer into the reciprocal one or possibly inducing the complete dissociation of the dimer.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM064491 (to L. J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GM3
- ganglioside GM3.
REFERENCES
- 1. Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J., Downward J., Mayes E. L., Whittle N., Waterfield M. D., Seeburg P. H. (1984) Nature 309, 418–425 [DOI] [PubMed] [Google Scholar]
- 2. Clayton A. H., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., Burgess A. W. (2005) J. Biol. Chem. 280, 30392–30399 [DOI] [PubMed] [Google Scholar]
- 3. Moriki T., Maruyama H., Maruyama I. N. (2001) J. Mol. Biol. 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 4. Tao R. H., Maruyama I. N. (2008) J. Cell Sci. 121, 3207–3217 [DOI] [PubMed] [Google Scholar]
- 5. Yu X., Sharma K. D., Takahashi T., Iwamoto R., Mekada E. (2002) Mol. Biol. Cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yarden Y., Schlessinger J. (1987) Biochemistry 26, 1443–1451 [DOI] [PubMed] [Google Scholar]
- 7. Yarden Y., Schlessinger J. (1987) Biochemistry 26, 1434–1442 [DOI] [PubMed] [Google Scholar]
- 8. Citri A., Yarden Y. (2006) Nat. Rev. Mol. Cell Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 9. Jorissen R. N., Walker F., Pouliot N., Garrett T. P., Ward C. W., Burgess A. W. (2003) Exp. Cell Res. 284, 31–53 [DOI] [PubMed] [Google Scholar]
- 10. Olayioye M. A., Neve R. M., Lane H. A., Hynes N. E. (2000) EMBO J. 19, 3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 12. Bajaj M., Waterfield M. D., Schlessinger J., Taylor W. R., Blundell T. (1987) Biochim. Biophys. Acta 916, 220–226 [DOI] [PubMed] [Google Scholar]
- 13. Lax I., Burgess W. H., Bellot F., Ullrich A., Schlessinger J., Givol D. (1988) Mol. Cell Biol. 8, 1831–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferguson K. M., Berger M. B., Mendrola J. M., Cho H. S., Leahy D. J., Lemmon M. A. (2003) Mol. Cell 11, 507–517 [DOI] [PubMed] [Google Scholar]
- 15. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2002) Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 16. Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J. H., Saito K., Sakamoto A., Inoue M., Shirouzu M., Yokoyama S. (2002) Cell 110, 775–787 [DOI] [PubMed] [Google Scholar]
- 17. Dawson J. P., Berger M. B., Lin C. C., Schlessinger J., Lemmon M. A., Ferguson K. M. (2005) Mol. Cell Biol. 25, 7734–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho H. S., Mason K., Ramyar K. X., Stanley A. M., Gabelli S. B., Denney D. W., Jr., Leahy D. J. (2003) Nature 421, 756–760 [DOI] [PubMed] [Google Scholar]
- 19. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Kofler M., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2003) Mol. Cell 11, 495–505 [DOI] [PubMed] [Google Scholar]
- 20. Mi L. Z., Grey M. J., Nishida N., Walz T., Lu C., Springer T. A. (2008) Biochemistry 47, 10314–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kästner J., Loeffler H. H., Roberts S. K., Martin-Fernandez M. L., Winn M. D. (2009) J. Struct. Biol. 167, 117–128 [DOI] [PubMed] [Google Scholar]
- 22. Mattoon D., Klein P., Lemmon M. A., Lax I., Schlessinger J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dawson J. P., Bu Z., Lemmon M. A. (2007) Structure 15, 942–954 [DOI] [PubMed] [Google Scholar]
- 24. Saxon M. L., Lee D. C. (1999) J. Biol. Chem. 274, 28356–28362 [DOI] [PubMed] [Google Scholar]
- 25. Walker F., Orchard S. G., Jorissen R. N., Hall N. E., Zhang H. H., Hoyne P. A., Adams T. E., Johns T. G., Ward C., Garrett T. P., Zhu H. J., Nerrie M., Scott A. M., Nice E. C., Burgess A. W. (2004) J. Biol. Chem. 279, 22387–22398 [DOI] [PubMed] [Google Scholar]
- 26. Macdonald J. L., Pike L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macdonald-Obermann J. L., Pike L. J. (2009) J. Biol. Chem. 284, 13570–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doran D. M., Spar I. L. (1980) J. Immunol. Methods 39, 155–163 [DOI] [PubMed] [Google Scholar]
- 29. Wyman J., Gill S. J. (1990) Binding and Linkage: Functional Chemistry of Biological Macromolecules, pp. 203–236, University Science Books, Mill Valley, CA [Google Scholar]
- 30. Macdonald J. L., Pike L. J. (2005) J. Lipid Res. 46, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 31. King A. C., Cuatrecasas P. (1982) J. Biol. Chem. 257, 3053–3060 [PubMed] [Google Scholar]
- 32. Magun B. E., Matrisian L. M., Bowden G. T. (1980) J. Biol. Chem. 255, 6373–6381 [PubMed] [Google Scholar]
- 33. Rees A. R., Gregoriou M., Johnson P., Garland P. B. (1984) EMBO J. 3, 1843–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shoyab M., De Larco J. E., Todaro G. J. (1979) Nature 279, 387–391 [DOI] [PubMed] [Google Scholar]
- 35. Schoeberl B., Eichler-Jonsson C., Gilles E. D., Müller G. (2002) Nat. Biotechnol. 20, 370–375 [DOI] [PubMed] [Google Scholar]
- 36. Mineo C., Gill G. N., Anderson R. G. (1999) J. Biol. Chem. 274, 30636–30643 [DOI] [PubMed] [Google Scholar]
- 37. Pike L. J., Miller J. M. (1998) J. Biol. Chem. 273, 22298–22304 [DOI] [PubMed] [Google Scholar]
- 38. Ringerike T., Blystad F. D., Levy F. O., Madshus I. H., Stang E. (2002) J. Cell Sci. 115, 1331–1340 [DOI] [PubMed] [Google Scholar]
- 39. Roepstorff K., Thomsen P., Sandvig K., van Deurs B. (2002) J. Biol. Chem. 277, 18954–18960 [DOI] [PubMed] [Google Scholar]
- 40. Yamabhai M., Anderson R. G. (2002) J. Biol. Chem. 277, 24843–24846 [DOI] [PubMed] [Google Scholar]
- 41. Elleman T. C., Domagala T., McKern N. M., Nerrie M., Lönnqvist B., Adams T. E., Lewis J., Lovrecz G. O., Hoyne P. A., Richards K. M., Howlett G. J., Rothacker J., Jorissen R. N., Lou M., Garrett T. P., Burgess A. W., Nice E. C., Ward C. W. (2001) Biochemistry 40, 8930–8939 [DOI] [PubMed] [Google Scholar]
- 42. Sorokin A. (1995) Oncogene 11, 1531–1540 [PubMed] [Google Scholar]
- 43. Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. (2009) Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pike L. J. (2006) J. Lipid Res. 47, 1597–1598 [DOI] [PubMed] [Google Scholar]
- 45. Bremer E. G., Schlessinger J., Hakomori S. (1986) J. Biol. Chem. 261, 2434–2440 [PubMed] [Google Scholar]
- 46. Liu Y., Su Y., Wiznitzer M., Epifano O., Ladisch S. (2008) Glycobiology 18, 593–601 [DOI] [PubMed] [Google Scholar]
- 47. Miljan E. A., Meuillet E. J., Mania-Farnell B., George D., Yamamoto H., Simon H. G., Bremer E. G. (2002) J. Biol. Chem. 277, 10108–10113 [DOI] [PubMed] [Google Scholar]
- 48. Wang X., Rahman Z., Sun P., Meuillet E., George D., Bremer E. G., Al-Qamari A., Paller A. S. (2001) J. Invest. Dermatol. 116, 69–76 [DOI] [PubMed] [Google Scholar]
- 49. Forbes S. A., Tang G., Bindal N., Bamford S., Dawson E., Cole C., Kok C. Y., Jia M., Ewing R., Menzies A., Teague J. W., Stratton M. R., Futreal P. A. (2010) Nucleic Acids Res. 38, D652–D657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alvarado D., Klein D. E., Lemmon M. A. (2010) Cell 142, 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alvarado D., Klein D. E., Lemmon M. A. (2009) Nature 461, 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.