Abstract
Mitosis is an orchestration of dynamic interaction between chromosomes and spindle microtubules by which genomic materials are equally distributed into two daughter cells. Previous studies showed that CENP-U is a constitutive centromere component essential for proper chromosome segregation. However, the precise molecular mechanism has remained elusive. Here, we identified CENP-U as a novel interacting partner of Hec1, an evolutionarily conserved kinetochore core component essential for chromosome plasticity. Suppression of CENP-U by shRNA resulted in mitotic defects with an impaired kinetochore-microtubule attachment. Interestingly, CENP-U not only binds microtubules directly but also displays a cooperative microtubule binding activity with Hec1 in vitro. Furthermore, we showed that CENP-U is a substrate of Aurora-B. Importantly, phosphorylation of CENP-U leads to reduced kinetochore-microtubule interaction, which contributes to the error-correcting function of Aurora-B. Taken together, our results indicate that CENP-U is a novel microtubule binding protein and plays an important role in kinetochore-microtubule attachment through its interaction with Hec1.
Keywords: CENP-U, Hec1, Aurora-B, Kinetochore, KMN network, CCAN
Introduction
Chromosome segregation during mitosis is orchestrated by dynamic interactions between spindle microtubule and a specialized proteinaceous structure on the chromosome called the kinetochore. Electron microscopic analysis has revealed that the kinetochore exhibits a trilaminar morphology (1): the inner plate, which forms the interface with chromatin; the outer plate, a 50∼60-nm-thick region that forms the interaction surface for spindle microtubules; the central plate, the region between the inner and outer kinetochore that appears less dense (2). Interestingly, centromere structure and function are conserved across eukaryotic kingdom, and its plasticity is regulated epigenetically (3). A combination of comparative genomics and functional proteomics has led to the identification of a large number of new human kinetochore proteins (4–7). Based on their biochemical properties, those kinetochore proteins have classified into several defined subcomplexes with distinct functional properties (8). Among several functionally distinct protein complexes, there are two conserved core complexes; one is the Knl1-Mis12-Ndc80 (KMN)3 network located in the outer plate, which constitutes the core microtubule binding site of the kinetochore (9), and the other is constitutive centromere-associated network (CCAN) located in the inner kinetochore, which makes a contribution to kinetochore specification and assembly (2).
Among KMN network, the best characterized tetrameric Ndc80 complex comprises Hec1, Nuf2, Spc24, and Spc25 (10, 11). Early genetic analyses of the Ndc80 complex in multiple organisms have demonstrated that it is important for stable kinetochore-microtubule attachment, chromosome alignment, and spindle checkpoint activation (11–16). The complex forms a dumbbell-like structure with the globular N-terminal regions of Hec1, Nuf2 at one end possessing the direct microtubule-binding activity and the globular C-terminal regions of Spc24, and Spc25 at the opposite end responsible for the kinetochore localization, separated by a long coiled-coil region (9, 17–23). Recently, mounting evidence indicates that the Ndc80 complex is a direct contact point between the kinetochore and the spindle microtubules (9, 20, 22–27). It was generally believed that Ndc80 complex plays a key role in the robust kinetochore-microtubule interaction. This kind of interaction seems to exhibit a cooperative manner with many relevant proteins involved. The Ndc80 complex along with KNL-1 and the Mis12 complex comprise the KMN network that is believed to function as the core microtubule binding site of the kinetochore. Biochemical reconstitution analyses show that the nine-component network exhibits an enhanced microtubule binding affinity compared with that of each individual components (9). However, it has remained elusive as to how KMN functions in vivo and whether additional kinetochore components cooperate with the Ndc80 complex in the stabilization of kinetochore-microtubule attachment. In the process of stable kinetochore-microtubule attachment formation, the microtubule binding affinity of Hec1 is attenuated by Aurora-B phosphorylation, which provides a potential direct mechanism for eliminating incorrect kinetochore-microtubule attachment (2, 9, 24). In contrast, our recent study demonstrated that phosphorylation by Nek2A increased the affinity of Hec1 for microtubules, thus, providing another regulatory pathway perhaps to stabilize correct kinetochore-microtubule attachment (28).
CENP-U (also known as CENP-50/PBIP1) is a component of CCAN due to its co-localization with CENP-A throughout the cell cycle and co-purification with CENP-A nucleosomes in vertebrate cells (2, 6, 7, 29). On the basis of in vivo phenotype analyses and biochemical studies, CENP-U along with CENP-O, CENP-P, CENP-Q, and CENP-R are identified as one subclass of CCAN named CENP-O class proteins (2, 6, 7, 29). In chicken cells, CENP-U is not essential for viability but is required for the prevention of premature sister chromatid separation during recovery from spindle damage (30, 31). In human cells, depletion of CENP-U can cause a mitotic defect in chromosome attachment without affecting the spindle assembly checkpoint (6). During cell cycle progression, CENP-U function may be regulated by post-translational modification. In recent studies, CENP-U (PBIP1) is identified as a phosphorylation substrate of Plk1, and the phosphorylation-dependent CENP-U-Plk1 interaction is required for Plk1 recruitment to interphase and mitosis kinetochore (31, 32).
Although the previous studies indicate that CENP-U is important for chromosome segregation, the precise molecular mechanisms remain less well characterized. To gain new insight into CENP-U function in mitosis, we carried out a new search for proteins that interact with CENP-U using yeast two-hybrid assay. This screen has identified Hec1 as one of the several dozen positive clones. Our biochemical characterization validated the interaction between CENP-U and Hec1 and mapped the regions of this interaction. Functional analyses revealed that CENP-U is required for stable kinetochore-microtubule attachment in vivo, and CENP-U cannot only bind microtubules directly but also displays a cooperative microtubule binding activity with Hec1 in vitro. Furthermore, the CENP-U-Hec1 interaction is under Aurora-B modulation. We propose that CENP-U interacts with Hec1 to stabilize kinetochore-microtubule attachment, which provides a novel link between the KMN-Aurora-B pathway and the CCAN pathway.
MATERIALS AND METHODS
Molecular Cloning
GFP-tagged and GST-tagged full-length CENP-U and CENP-U shRNA was kindly provided by Dr. Kyung S. Lee (National Institutes of Health). FLAG-tagged CENP-U was cloned by inserting the PCR product into the p3XFLAG-myc-CMV-24 vector (Sigma) with BglII and SalI digestion. To generate GFP-tagged CENP-U deletion mutants, PCR-amplified cDNAs were cloned into pEGFP-C1 (Clontech) vector by BglII and SalI. BglII-SalI-digested CENP-U D6 fragment was also inserted into pET-28a(+) (Novagen), pMAL-c2 (New England Biolabs, MA), and pGEX-6P-1 (Amersham Biosciences) digested with BamHI and SalI. GFP-tagged and GST-tagged non-phosphorylating and phospho-mimicking CENP-U mutants were created by standard PCR methods as described previously (33).
Yeast Two-hybrid Assay
Yeast two-hybrid assays were performed as previously described (34). Briefly, bait CENP-U was inserted into the BamHI-EcoRI sites of pGBKT7 to create a fusion with aa 1–147 of the Gal4 DNA-binding domain. The resultant pGBKT7/CENP-U was transformed into strain AH109 along with the GAL4 reporter plasmid pCL and the negative control plasmid pGBKT7-Lam. Protein expression was validated by Western blot using Gal4 and an anti-CENP-U antibody. Specificity of the interaction was independently verified by retransforming the candidate cDNAs back into AH109 along with BD-CENP-U. Those cDNAs that from colonies grew up on Leu−, Trp−, His−, Ade− SD plates were sequenced. N and C termini of Hec1 were inserted into pGBKT-7 vector. AD-CENP-U and these BD-Hec1 deletion mutants were co-transformed into AH109 yeast strain to map out interacting region of Hec1 with CENP-U.
Protein Expression and Purification
GST-tagged full-length CENP-U (wild type and non-phosphorylating) expressed in Rosetta (DE3) pLysS was induced at a higher cell density (optical density ≈ 1.5) at 30 °C for 2 h. His-tagged, GST-tagged, and MBP-tagged CENP-U D6 expressed in Rosetta (DE3) pLysS were induced at a standard cell densities (optical density ≈ 0.6) at 30 °C for 4∼6 h. Hec1 and Ndc80 complexes were expressed in bacteria as fusion proteins and purified as described (18).
GST Pulldown Assay
The GST-CENP-U fusion protein-bound glutathione beads were incubated with 293T cell lysate containing FLAG-tagged Ndc80 components for 2 h at 4 °C. After the incubation, the beads were washed 3 times with PBS containing 0.5% Triton X-100 and twice with PBS and boiled in SDS-PAGE sample buffer. The bound proteins were then separated on 8% SDS-PAGE. Separated proteins were then transferred onto nitrocellulose membrane for Western blot with FLAG antibody. Two domain mapping experiments were conducted in the similar procedure.
In the case of reconstitution of CENP-U-Hec1 interaction in vitro, GST-Nuf2/Hec1 purified on glutathione beads were used as an affinity matrix for absorbing His tagged CENP-U D6 in the buffer containing PBS, pH 7.4, 0.5% Triton X-100, 1 mm PMSF for 2 h at 4 °C. The beads were washed 3 times with PBS plus 1% Triton X-100 and once with PBS, then boiled for 5 min in SDS-PAGE sample buffer. Proteins were resolved by SDS-PAGE for Coomassie Brilliant Blue staining.
Co-immunoprecipitation
293T cells were transfected with GFP-Hec1 or pEGFP-C1 plus FLAG-CENP-U plasmids individually. 36 h post-transfection, the cells were extracted using lysis buffer (50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm EGTA, 0.5% Triton X-100, 1 mm PMSF, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A). Lysates were clarified by centrifugation at 16,000 × g for 10 min at 4 °C and then incubated with FLAG-M2-agarose beads (Sigma) at 4 °C for 2 h. These beads were washed 3 times with lysis buffer and once with PBS free of Triton X-100 and then boiled in sample buffer followed by fractionation on SDS-PAGE. Separated proteins were then transferred onto nitrocellulose membrane and probed for GFP and FLAG, respectively.
Antibodies and RNA Interference
The following antibodies were used: anti-FLAG mouse serum (Sigma), anti-GFP mAb (BD Biosciences), anti-Hec1 (9G3) mAb (Abcam), anti-MBP rabbit Ab (New England Biolabs), human anti-centromere antibody (ACA), anti-tubulin antibody DM1A (Sigma) (35). Hec1 siRNA was obtained as described previously (36) and introduced into HeLa cells using Lipofectamine 2000 (Invitrogen). Plasmid based shRNA used to repress CENP-U were transfected into HeLa cells together with mcherry-H2B as a marker of transfection at a ratio of 3:1 using Lipofectamine 2000 (Invitrogen). Cells were examined by immunocytochemistry 48–72 h after initiation of transfection.
Cell Culture and Synchronization
HeLa cells (American Type Culture Collection, Manassas, VA) were maintained as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37 °C with 8% CO2. Cells were synchronized at G1/S with 5 mm thymidine for 12∼16 h and then washed with PBS 5 times and cultured in thymidine-free medium for 10 h. In some cases, after 8 h of release, 10 μm MG132 was added into medium for 2–3 h.
Immunofluorescence Microscopy and Kinetochore Distance Measurement
Cells were seeded onto sterile, acid-treated 12-mm coverslips in 24-well plates (Corning Glass Works, Corning, NY). The next day, the cells were transfected with 1 μl of Lipofectamine 2000 premixed with plasmids or siRNAs described above. If not specified, 48 h after transfection, cells were rinsed with PHEM buffer (100 mm PIPES, 20 mm HEPES, pH 6.9, 5 mm EGTA, 2 mm MgCl2, and 4 m glycerol) and permeabilized for 1 min with PHEM plus 0.1% Triton X-100 before fixation in freshly prepared 3.7% formaldehyde for 5 min. After rinsing 3 times in PBS, cells were blocked with PBST (0.05% Tween 20 in PBS) with 1% bovine serum albumin (Sigma) followed by incubation with various primary antibodies in a humidified chamber for 1 h. After three washes in PBST, primary antibodies were visualized by fluorescein isothiocyanate (FITC) or rhodamine-conjugated goat anti-mouse or rabbit IgG. DNA was stained with DAPI (4′,6-diamidino-2-phenylindole). Slides were examined under a DeltaVision deconvolution microscopy (Applied Precision Inc.).
Deconvolution images were collected using a DeltaVision wide-field deconvolution microscope system built on an Olympus IX-70 inverted microscope base as previously described (37). For imaging, a 100 × 1.35 NA lens was used, and optical sections were taken at intervals of 0.25 μm. Images were processed using DeltaVision Softworx software. The distance between sister kinetochores marked with ACA was measured as described (36, 37).
In Vitro Kinase Assay
In vitro phosphorylation and kinase assays were conducted as described (38). His-tagged Aurora-B kinase was expressed in bacteria and purified by Ni2+-nitrilotriacetic acid beads. The kinase reactions were performed in 50 μl of 1× kinase buffer (25 mm HEPES, pH 7.2, 5 mm MgSO4, 1 mm DTT, 50 mm NaCl, 2 mm EGTA) containing 1 μl eluted Aurora-B kinase, 5 μl of GST bead-bound GST-CENP-U proteins, 5 μCi of [γ-32P]ATP, and 50 μm ATP. The mixtures were incubated at 30 °C for 30 min. The reactions were stopped with 5× SDS sample buffer and separated by SDS-PAGE. The gel was stained with Coomassie Brilliant Blue, dried, and conducted autoradiography subsequently with x-ray film.
Microtubule Co-sedimentation Assay
The early steps of MBP-CENP-U D6 purification was instructed by New England Biolabs manual. But for the last steps, the beads were rinsed with BRB80 buffer followed by elusion with BRB80 buffer containing 10 mm maltose. For microtubule binding reactions, MBP-CENP-U D6 in BRB80 buffer containing 10 mm maltose were used directly, whereas Hec1/Nuf2 were diluted into BRB80 buffer before use, and the proteins were precleared at 90,000 rpm for 10 min in a TLA100.3 rotor. 90 μl of the supernatant was mixed with 10 μl of microtubules diluted in BRB80 to result in a final microtubule concentration of 1 μm. Reactions were incubated at room temperature for 20 min, pipetted onto 100 μl of BRB80 + 40% glycerol, and pelleted for 10 min at 80,000 rpm in a TLA100.3 rotor at 25 °C. For the supernatant sample, 100 μl was kept from the top of the tube. For the pellet sample, the remaining supernatant was removed, and 100 μl of BRB80 containing 10 mm CaCl2 was added to the pellet for 10 min on ice. Equivalent amounts of supernatant and pellet fractions were separated on SDS-PAGE followed by staining with Coomassie Brilliant Blue and the brightness/contrast and levels were adjusted to maximize dynamic range. Equivalent amounts of supernatant and pellet fractions were also immunoblotted (9).
RESULTS
Hec1 Is a Novel Binding Partner of CENP-U
Previous studies have shown that CENP-U is a constitutive centromere component important for proper chromosome segregation (6, 7, 30, 32). However, the precise molecular mechanisms underlying CENP-U function in centromere plasticity have remained largely elusive. To illustrate the molecular regulation of CENP-U in mitosis, the full-length of CENP-U was chosen as bait to screen the HeLa cDNA library using GAL4 yeast two-hybrid system as previously described (34). Among 36 positive clones sequenced, one positive clone encodes the C terminus of Hec1 (amino acids 486–604).
To verify the specificity of interaction between CENP-U and Hec1 and further define the domain required for this interaction, we cloned CENP-U cDNA into pGAD T7 vector. According to the structure analysis of Hec1, N- and C-terminal fragments were generated as illustrated in Fig. 1A. The BD-tagged Hec1 full-length and deletion mutants were co-transformed with AD-tagged CENP-U (where AD represents the GAL4 DNA activation domain) into yeast cells, and β-galactosidase activity (lacZ reporter) was used as readout of protein-protein interaction. Consistent with the outcome of our initial yeast two-hybrid screen, human Hec1 binds to CENP-U via its C-terminal coiled-coil region (Fig. 1A). Thus, our data suggest that human Hec1 is a novel binding partner for CENP-U.
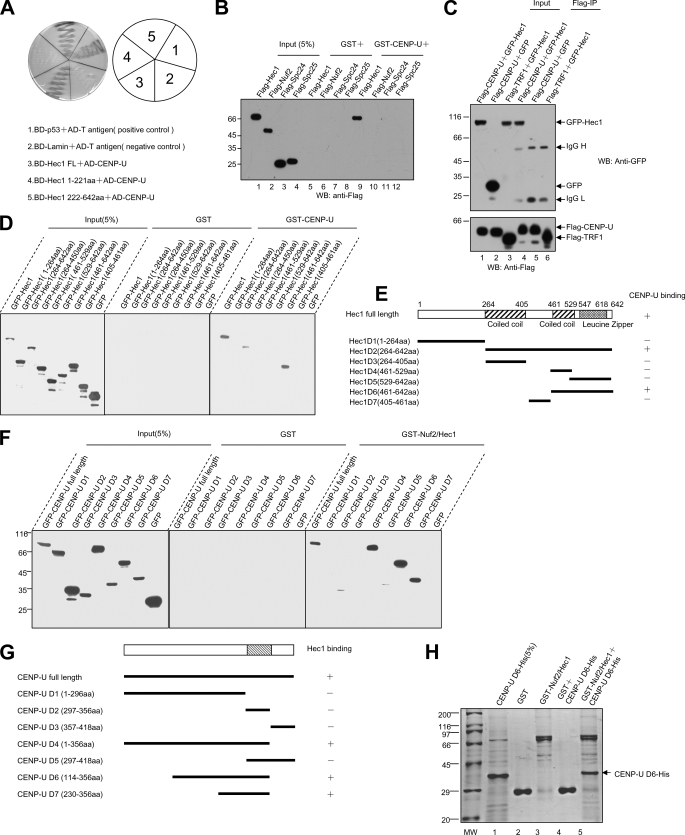
FIGURE 1.
Identification and characterization of a novel CENP-U-Hec1 interaction. A, yeast cells were co-transformed with a CENP-U prey construct and the indicated Hec1 bait constructs (Hec1 full-length, aa 1–221, and aa 222–642). An example of such an experiment in which cells were selected on supplemented minimal plates lacking uracil, tryptophan, leucine, and histidine is presented. B, a GST pulldown assay was used to determine the potential components of the Ndc80 complex that can interact with CENP-U. Bacterial recombinant GST-CENP-U purified on glutathione beads was used as an affinity matrix for absorbing FLAG-tagged Hec1, Nuf2, Spc24, and Spc25 from human embryonic kidney 293T cells as described under “Materials and Methods.” GST protein bound agarose beads were used as a control. After washing, proteins bound to agarose beads were boiled in sample buffer and analyzed by Western blot (WB) using FLAG antibody. Note that only FLAG-Hec1 can be pulled down by GST-CENP-U. Lanes 1–4, cell lysates of FLAG-tagged Hec1, Nuf2, Spc24, and Spc25; lanes 5–8, GST control; lanes 9–12, proteins retained on GST-CENP-U affinity beads. C, CENP-U forms a complex with Hec1 in vivo. Anti-FLAG immunoprecipitates (FLAG-IP) from lysates of 293T cells expressing FLAG-CENP-U or FLAG-TRF1 together with (GFP)-Hec1 were prepared, and the anti-GFP and anti-FLAG blotting was verified co-precipitation of GFP-Hec1 (upper) and FLAG-CENP-U (lower). Lanes 1, 2, and 3 represent the cell lysates before incubating with anti-FLAG antibody; lanes 4, 5, and 6 represent the anti-FLAG immunoprecipitates. D, a GST pulldown assay was used to determine the regions of Hec1 responsible for CENP-U binding. GST-CENP-U purified on glutathione beads were used as an affinity matrix for absorbing GFP-tagged Hec1 full-length and the indicated deletion mutants from 293T cells. GST protein-bound agarose beads were used as a control. Anti-GFP blotting revealed that Hec1 full-length, aa 264–642, and aa 461–642 can be pulled down by GST-CENP-U. The left nine lanes represent cell lysates expressing the indicated Hec1 proteins and a GFP control; the middle nine lanes represent GST control; the right nine lanes represent proteins retained on GST-CENP-U affinity beads. E, shown is a schematic drawing of Hec1 deletion mutants. +, positive; −, negative. F, a GST pulldown assay was used to determine the regions of CENP-U responsible for Hec1 binding. GST-Nuf2/Hec1 purified on glutathione beads were used as an affinity matrix for absorbing GFP-tagged CENP-U full-length and the indicated deletion mutants from 293T cells. GST protein-bound agarose beads were used as a control. Anti-GFP blotting revealed that CENP-U full-length, aa 1–356, aa 114–356, and aa 230–356 can be pulled down by GST- Nuf2/Hec1. The left nine lanes represent cell lysates expressing the indicated CENP-U proteins and a GFP control; the middle nine lanes represent GST control; the right nine lanes represent proteins retained on GST- Nuf2/Hec1 affinity beads. G, shown is a schematic drawing of CENP-U deletion mutants. +, positive; −, negative. H, reconstitution of CENP-U-Hec1 interaction using bacterially expressed recombinant proteins is shown. GST-Nuf2/Hec1 purified on glutathione beads were used as an affinity matrix for absorbing His-tagged CENP-U D6. GST protein bound agarose beads were used as a control. After washing, proteins bound to agarose beads were boiled in sample buffer and fractionated on a SDS-PAGE gel and analyzed by Coomassie Blue staining. An aliquot of purified His-CENP-U D6 was loaded on an adjacent well as a positive control. The arrow (lane 5) indicates His-CENP-U D6 absorbed by GST-Nuf2/Hec1.
Because Hec1 is a component of Ndc80 complex, we conducted a GST pulldown assay to determine whether CENP-U can bind other components of this complex. To this end, FLAG-tagged Ndc80 complex components (Hec1, Nuf2, Spc24, and Spc25) were transfected into 293T cells, respectively. 36 h after transfection, each cell lysate was incubated with bacterial-expressed GST-CENP-U, and binding proteins were analyzed by Western blotting. As shown in Fig. 1B, Hec1 can be brought down by CENP-U, whereas Nuf2, Spc24, and Spc25 failed to bind with CENP-U. This data demonstrate the specificity of the CENP-U-Hec1 interaction.
To examine whether CENP-U interacts with Hec1 in vivo, we co-transfected 293T cells with FLAG-CENP-U and GFP-Hec1, and then FLAG-CENP-U protein was immuno-purified with an anti-FLAG M2 affinity gel. Subsequent immunoblot with anti-GFP antibody confirmed the presence of GFP-Hec1 in FLAG-CENP-U immunoprecipitates, suggesting that CENP-U and Hec1 forms a cognate complex in vivo (Fig. 1C, lane 4). To exclude the possibility of nonspecific interaction, an irrelevant flag-tagged TRF1 was used as a negative control (Fig. 1C, lane 6).
To delineate the minimal region of Hec1 required for CENP-U binding, GFP-tagged Hec1 and its deletion mutants as illustrated in Fig. 1E were expressed in 293T cells, and each lysate was subsequently incubated with GST-CENP-U. As shown in Fig. 1D, Hec1 full-length, Hec1 D2 (aa 264–642), and Hec1 D6 (aa 461–642) could interact with CENP-U as judged by Western blot analysis of the GFP tag. Thus our biochemical study confirmed that Hec1 binds to CENP-U via its C-terminal, and the minimal region is aa 461–642, validating our results obtained from yeast two-hybrid assay.
In a similar way we then attempted to map the regions of CENP-U, which bind to Hec1. Based on protein secondary structure prediction, we generated a series of GFP-tagged CENP-U deletion mutants as illustrated in Fig. 1G. Among the several reactions, CENP-U full-length, CENP-U D4 (aa 1–356), CENP-U D6 (aa 114–356), and CENP-U D7 (aa 230–356) could be pulled down by GST-Nuf2/Hec1 (Fig. 1F). In summary, our data suggest that the minimal region of CENP-U responsible for Hec1 binding is aa 230–356. The coiled coil domain of CENP-U (aa 297–356) is necessary but not sufficient for the interaction.
To test whether CENP-U binds directly to Hec1 in vitro, GST-tagged Nuf2/Hec1 and His-tagged CENP-U D6 (aa 114–356) were expressed and purified from bacteria. As predicted, our GST pulldown assay showed that His-tagged CENP-U D6 is pulled down by GST-Nuf2/Hec1, demonstrating a direct interaction between Hec1 and CENP-U (Fig. 1H). Thus, we conclude that CENP-U interacts with Hec1 in vitro and in vivo, and the interface responsible for this interaction consists of aa 230–356 of CENP-U and aa 461–642 of Hec1.
CENP-U Is Required for Stable Kinetochore-Microtubule Attachment
Given the physical interaction between CENP-U and Hec1 established above, we sought to examine their spatiotemporal distribution profiles and interrelationship during cell cycle. To this end, HeLa cells stably expressing GFP-CENP-U were fixed and stained with antibody against Hec1. Because CENP-U is a constitutive centromere component, it was readily apparent that CENP-U signal appears in interphase kinetochores. However, no Hec1 kinetochore signal can be detected in the same cell (Fig. 2Aa). Upon cells enter into mitosis, Hec1 signals progressively accumulated and became most prominent at prometaphase kinetochores, with clear co-localization with CENP-U (Fig. 2A, b–e). Therefore, the co-localization of CENP-U and Hec1 occurred during the progression from prophase to anaphase. The spatial vicinity provides the physical foundation for the functional relationship between CENP-U and Hec1.
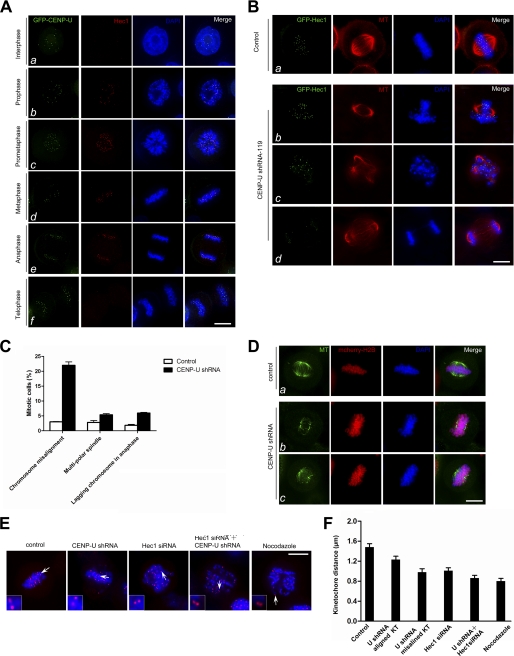
FIGURE 2.
Depletion of CENP-U causes unstable kinetochore-microtubule attachment. A, co-localization of CENP-U and Hec1 during mitosis is shown. This montage represents optical images collected from HeLa cells stably expressing GFP-CENP-U double-stained for Hec1 (red) and DAPI (blue). CENP-U co-localizes with Hec1 on kinetochore from prophase to anaphase (b–e). Bar, 10 μm. B, suppression of CENP-U causes multiple mitotic defects. HeLa cells were transfected with CENP-U shRNA-119 along with GFP-Hec1 as a marker of transfection and examined for spindle and DNA morphology. After 48 h transfection, cells were fixed and stained with tubulin (red) and DAPI (blue). The following phenotypes were observed in the absence of CENP-U: chromosome misalignment (b), multipolar spindle (c), chromosome bridge in anaphase (d). Bar, 10 μm. C, shown is quantification of the phenotypes observed above. An average of 150 GFP-Hec1-positive mitotic cells from three separate experiments was surveyed. Error bars represent S.E.; n = 3 preparations. D, suppression of CENP-U results in destabilization of spindle microtubules. HeLa cells were transfected with CENP-U shRNA along with mcherry-tubulin as a marker of transfection for 48 h followed by a 3-h MG132 treatment and examined for spindle stability in response to cold treatment. Cold-treated control and CENP-U-depleted cells were stained for tubulin (green) and DAPI (blue). The kinetochore microtubules became unstable due to cold treatment in the absence of CENP-U (b and c). Bar, 10 μm. E, shown is an immunofluorescence assay of control siRNA-treated HeLa cells (scramble), CENP-U shRNA-treated cells, Hec1 siRNA-treated cells, Hec1 siRNA- and CENP-U shRNA-treated cells, and nocodazole-treated cells. After 48 h transfection, cells were fixed and stained for ACA (red) and DNA (Blue). Bar, 10 μm. F, statistic analyses of ACA distance in the aforementioned siRNA- or shRNA-treated cells are shown. The basic principle of measuring the ACA double-dot distance is to choose those couples whose ACA spots have almost similar fluorescence intensity, which indicates the ACA couple localize at the same focal plane. Each value was calculated from 150 kinetochores (KT) selected from 20 different cells.
We next investigated the potential role of CENP-U in mitosis. To this end, HeLa cells were transiently transfected to express GFP-Hec1 with premixed CENP-U shRNA. Infection of HeLa cells with CENP-U shRNA (shRNA-119 or shRNA-129) effectively suppressed the CENP-U expression without altering the levels of Hec1 (see supplemental Fig. S1). We conducted three independent experiments and surveyed about 150 GFP-Hec1-positive cells. Spindle and DNA morphology were examined in mitotic cells by immunofluorescence. Whereas the distribution pattern of GFP-Hec1 was not affected by depletion of CENP-U, which is in agreement with previous studies (6), cells deficient in CENP-U suffered a severe mitotic defect. In control cell population, only 3% cells exhibited aberrant kinetochore attachments such as unaligned chromosomes. In contrast, 22% of CENP-U-suppressed metaphase cells displayed misaligned chromosomes (Fig. 2, B and C). The percentage of two mitotic defects (multi-poles and sister chromatid bridges) was also dramatically increased in CENP-U depleted cells (Fig. 2, B and C). Using a shRNA targeted to a different region of CENP-U mRNA (shRNA-129), we obtained essentially the same phenotypes (supplemental Fig. S2), demonstrating the critical role of CENP-U in chromosome plasticity.
The variety of defects observed suggests that the kinetochore-microtubule attachment has become unstable. To test this hypothesis, we performed a cold treatment assay. Aliquots of HeLa cells were transfected with CENP-U shRNA along with mcherry-H2B indicative of positive transfection followed by thymidine synchronization. MG132 was added into fresh medium 8 h after thymidine wash-out. After 3 h of MG132 treatment, the coverslips were changed into prechilled medium (4 °C) for 10 min to destabilize non-kinetochore microtubules. As shown in Fig. 2D, in control cells, the spindle microtubules were stable under cold treatment even though the aster microtubules were depolymerized (Fig. 2Da). In contrast, fewer kinetochore microtubules were retained in CENP-U-depleted cells judged by the immunofluorescence microscopic analysis (Fig. 2D, b and c), demonstrating that CENP-U is required for a stable kinetochore-microtubule attachment.
If CENP-U is essential for accurate kinetochore-microtubule attachment, an unstable attachment in CENP-U-suppressed cells should display an attenuated tension across sister kinetochores. To test this hypothesis, we next measured inter-kinetochore distance between pairs of ACA dots, as the inter-kinetochore distance serves as a faithful indicative of the tension across the kinetochores (37). In this case, shortened distance often reflects aberrant microtubule attachment to the kinetochore, in which less tension is developed across the sister kinetochore. Therefore, we measured ACA distance in 150 kinetochore pairs in which both kinetochores were in the same focal plane in scramble control cells and cells suppressing CENP-U, Hec1, and CENP-U plus Hec1. Nocodazole-treated cells were used as a tensionless-kinetochore control in which kinetochore pairs were presumably under no tension due to the microtubule depolymerization. As shown in Fig. 2E, depletion of CENP-U, Hec1, and CENP-U + Hec1 resulted in errors in chromosome alignment at the equator. Control kinetochore pairs exhibited a separation of 1.48 ± 0.12 μm, whereas the distances between kinetochores were 1.23 ± 0.12 μm (p < 0.001 compared with that of control) in aligned chromosomes in CENP-U-depleted cells, 0.98 ± 0.12 μm (p < 0.001 compared with that of control) in misaligned chromosomes in CENP-U depleted cells, and 1.01 ± 0.10 μm (p < 0.001 compared with that of control) in Hec1-depleted cells (Fig. 2, E and F). Simultaneous depletion of CENP-U and Hec1 resulted in striking shortening of the inter-kinetochore distance (0.86 ± 0.10 μm) (p < 0.001 compared with that of control), suggesting the significant decrease of tension across the sister kinetochore (Fig. 2, E and F). The distance between sister kinetochores in nocodazole-treated cells, in which kinetochore pairs were presumably under no tension, was 0.80 ± 0.10 μm, whereas this distance in CENP-U and Hec1 double-repressed cells was 0.86 ± 0.10 μm (Fig. 2, E and F), indicating that both CENP-U and Hec1 are required for a stable kinetochore-microtubule attachment. Thus, we conclude that CENP-U cooperates with Hec1 in stabilizing kinetochore-microtubule association.
CENP-U and Hec1 Exhibit Cooperative Binding Property to Microtubules
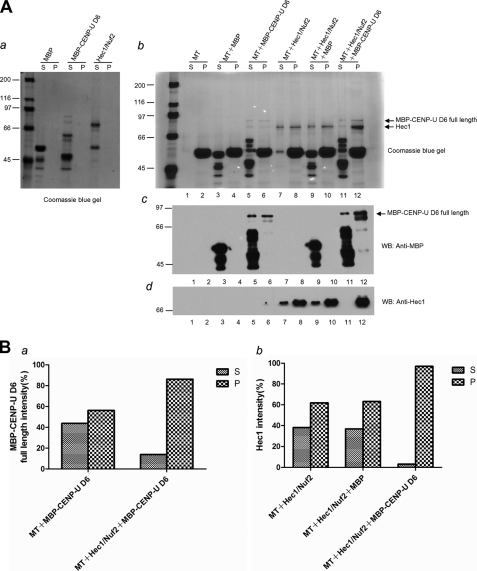
To get insight into the functional effect of the CENP-U-Hec1 interaction, we tested whether CENP-U influences the microtubule binding activity of Hec1 in vitro. To this end, we conducted a microtubule co-sedimentation assay. We were able to purify GST-tagged full-length CENP-U. However, the molecular property of CENP-U prevented robust expression and resulted in some smaller molecular weight products because of either degradation or the use of alternative start codons during translation as shown later in Fig. 4B. What is worse, the difficultly purified full-length CENP-U was unstable in BRB80 buffer. After pre-clear at 90,000 rpm for 10 min, a necessary step in the microtubule co-sedimentation assay, we can hardly get the amount of full-length CENP-U sufficient for the following experiments. Because we established earlier (Fig. 1, F and H) that CENP-U D6 showed a strong interaction with Hec1 in vitro, we used MBP-tagged CENP-U D6 to substitute full-length CENP-U in this case. No pelleting was observed in the absence of microtubules as shown in Fig. 3Aa. Surprisingly, CENP-U D6 can bind directly to taxol-stabilized microtubules on its own (Fig. 3A, b and c, lanes 5 and 6). The result is maybe unexpected; nevertheless, it is reasonable. Previous studies have demonstrated that CENP-O class proteins including CENP-O, -P, -Q, -R, and -U can form a stable complex (31). Most recently, it was shown that CENP-Q binds to microtubules in vitro with an affinity comparable with that of the Ska complex but better than that of the Ndc80 complex (39). Our results not only supported the idea that there are multiple microtubule binding sites at the kinetochore but also contributed to the determination of additional CCAN subunits that can bind microtubules. In addition to the discovery of the direct microtubule binding property of CENP-U D6 in this assay, we also found that a combination of CENP-U D6 with Hec1 resulted in a dramatic increase in microtubule binding affinity relative to the individual components (Fig. 3A). Co-sedimentation along with MBP-CENP-U D6 but not the unrelated protein MBP resulted in an increase of Hec1 portion in the pellets (Fig. 3A, b and d, compare with lane 8, 10, and 12). Similarly, CENP-U D6 binding activity to microtubule was enhanced when Hec1 was present (Fig. 3A, b and c, compare with lane 6 and 12). In the absence of Hec1, the fraction of MBP-CENP-U D6 in the supernatant was 42%, whereas 58% was in the pellet with microtubules. After the addition of Hec1, the portion in the pellet was increased to 82% with only 18% left in the supernatant (Fig. 3Ba). Similarly, the ratios of Hec1 portion in the supernatant versus in the pellet were 38:62, 36:64, and 3:97%, respectively, in the groups MT+Hec1/Nuf2, MT+Hec1/Nuf2+MBP, and MT+Hec1/Nuf2+ MBP-CENP-U D6 (Fig. 3Bb). Mixing fixed amounts of Hec1 (0.2 μm) with varying concentrations of MBP-CENP-U D6 (0–0.5 μm) revealed that a dose-dependent co-pelleting of CENP-U with microtubules (that is, the higher concentration of CENP-U was present), the more robust microtubule binding of Hec1 was observed (supplemental Fig. S3). In summary, we demonstrate that CENP-U cannot only bind directly to microtubules on its own but also displays a cooperative microtubule binding activity with Hec1.
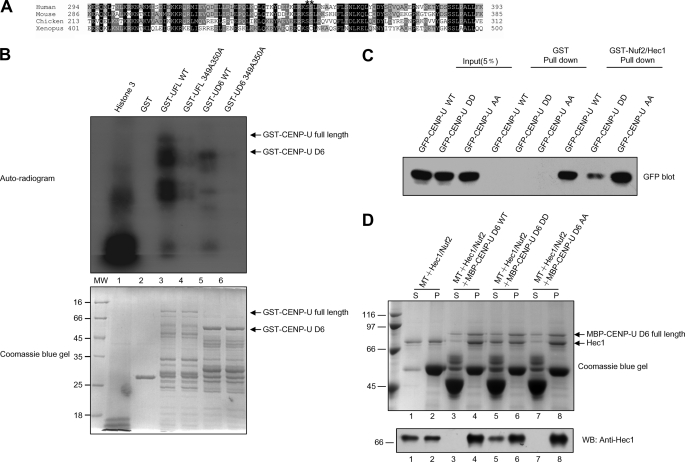
FIGURE 4.
CENP-U-Hec1 interaction is under phospho-regulation by Aurora-B. A, alignment of CENP-U near the Hec1 binding region from human, mouse (NP_082249.1), chicken (NP_001034388.1), and Xenopus (NP_001072545.1). Dark and light shading indicate identical and conserved residues, respectively. Residue numbers at region boundaries are indicated. Asterisks denote the serine residues which are not only conserved but also fit with Aurora-B phosphorylation motif. B, GST-CENP-U proteins, both wild type and mutant, were purified and phosphorylated in vitro using [32P]ATP and active Aurora B as described under “Materials and Methods.” Samples were separated by SDS-PAGE. Lower, a Coomassie Blue-stained gel of the following samples is shown. Markers are histone3 as positive control (lane 1), GST as negative control (lane 2), GST-CENP-U full-length wild type (lane 3), GST-CENP-U full-length349A350A (lane 4), GST-CENP-U D6 wild type (lane 5), and GST-CENP-U D6 349A350A (lane 6). Note that roughly equivalent amounts of wild type and mutant protein were present in the reactions. Upper, the same gel was dried and subsequently incubated with x-ray film. Note that there was dramatic incorporation of 32P into wild type but little into the double mutant of CENP-U proteins. C, GST-Nuf2/Hec1 protein was used to isolate CENP-U wild type and mutants from 293T cell lysates. D, the ability of CENP-U D6 WT, AA, and DD to stimulate microtubule binding of Hec1 was tested in parallel. The final concentration of microtubule was 1 μm. Upper, Coomassie Blue-stained gel. Lower, anti-Hec1 blotting was performed corresponding to the upper stain analysis. S, supernatant; P, pellet.
FIGURE 3.
Synergy in microtubule binding activity after interaction of CENP-U and Hec1. A, shown is a co-sedimentation analysis of mixtures of MBP-CENP-U D6 and Hec1/Nuf2. The final concentration of microtubule was 1 μm. a, no pelleting was observed in the absence of microtubules. b, increased co-sedimentation was observed for both MBP-CENP-U D6 full-length (compare with lane 6 and lane 12, upper arrow) and Hec1 (compare with lane 8, 10, and 12, lower arrow) by Coomassie Blue staining. c and d, anti-MBP and anti-Hec1 blotting were performed corresponding to the abovementioned stain analysis. B, shown is quantification of protein in the indicated reactions. S, supernatant; P, pellet. WB, Western blot.
Aurora-B Phosphorylation Modulates the Interaction of CENP-U and Hec1
Because we demonstrated above that the minimal region of CENP-U responsible for Hec1 binding was aa 230–356 (Fig. 1F), we next sought to test if any mitotic kinases can phosphorylate this region, thus, regulating CENP-U behavior in mitosis. As shown in Fig. 4A, our computational analysis suggested that Ser349 and Ser350 were potential sites for Aurora-B phosphorylation, which are not only in accord with the consensus phosphorylation motif (R/K)X(T/S) for Aurora-B kinase (40) but also conserved among vertebrate. To test whether Ser349 and Ser350 are authentic phosphorylation sites of Aurora-B, we performed an in vitro kinase assay using recombinant GST-tagged CENP-U full-length or D6 truncation (GST-UFL WT, GST-UFL 349A350A, GST-UD6 WT, and GST-UD6 349A350A). As shown in Fig. 4B, both the wild type CENP-U FL and D6 were phosphorylated strongly. Although there is weak incorporation of 32P into UFL 349A350A, indicating that other phosphorylation sites may exist on CENP-U full-length, no detectable incorporation of 32P was observed into UD6 349A350A, which demonstrates that Ser349 and Ser350 are the only two Aurora-B phosphorylation sites within the region of CENP-U-Hec1 interaction. To determine whether phosphorylation of CENP-U by Aurora-B modulates the CENP-U-Hec1 interaction, we conducted a GST pulldown assay. For this experiment, aliquots of 293T cell lysates containing GFP-tagged CENP-U wild type (GFP-CENP-U WT), phospho-mimicking mutant (GFP-CENP-U DD), and non-phosphorylating mutant (GFP-CENP-U AA) was individually incubated with GST-Nuf2/Hec1. As shown in Fig. 4C, a significant reduction in Hec1 binding was observed for the phospho-mimicking mutant compared with that of wild type and non-phosphorylating mutant, indicating that phosphorylation of Ser349 and Ser350 on CENP-U by Aurora-B can down-regulate the binding affinity between CENP-U and Hec1. Because we established above that CENP-U and Hec1 exhibit cooperative binding to microtubules, we next tested the effects of CENP-U mutants on Hec1 microtubule binding affinity. Compared with CENP-U D6 WT and AA mutant, DD mutant is a weaker stimulator for microtubule binding of Hec1 (Fig. 4D), in agreement with their Hec1 binding ability as shown in Fig. 4C.
Phosphorylation of CENP-U by Aurora-B Weakens Kinetochore-Microtubule Interaction
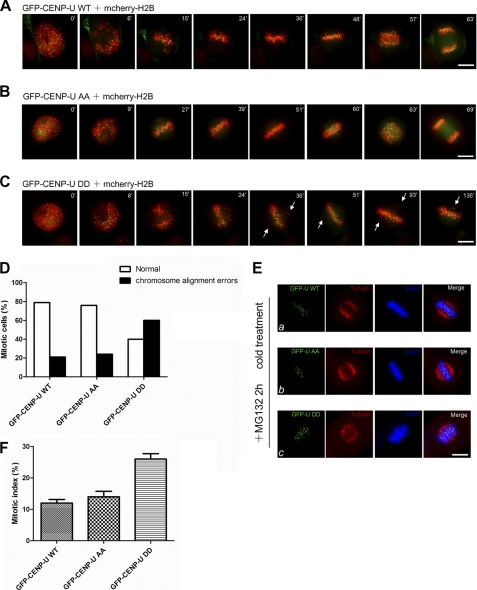
To delineate the functional specificity of the two Aurora-B phosphorylation sites on CENP-U, we examined precisely the chromosome movements in cells expressing GFP-CENP-U WT, AA, and DD. To this end, mcherry-H2B was co-transfected with GFP-CENP-U plasmids to visualize in real-time chromosomes upon nuclear envelope breakdown (NEB). As shown in Fig. 5A, in GFP-CENP-U WT-expressing cells, all chromosomes achieve metaphase alignment 48 min after NEB. The anaphase was initiated 57 min after NEB. The cell underwent complete segregation of sister chromatids 63 min after NEB. Similar to the aforementioned case, in GFP-CENP-U AA-expressing cells all chromosomes achieve metaphase alignment 51 min after NEB. The anaphase was initiated 60 min after NEB. The cell underwent complete segregation of sister chromatids 69 min after NEB (Fig. 5B). Unlike GFP-CENP-U WT- and AA-expressing cells, however, GFP-CENP-U DD-expressing cells were unable to properly align their chromosomes. As shown in Fig. 5C, the cell failed to achieve chromosome alignment even at 135 min after NEB. Besides the severe delay we showed here, we also observed DD-expressing cells exhibiting alignment difficulties eventually progress into anaphase with chromosome bridge (supplemental Fig. S4). We surveyed ∼30 cells in each group. As summarized in Fig. 5D, 79% GFP-CENP-U WT-expressing cells (n = 24) and 76% GFP-CENP-U AA-expressing cells (n = 29) underwent normal mitotic progression. In contrast, 60% GFP-CENP-U DD-expressing cells (n = 30) exhibited chromosome alignment errors (chromosomes failed to achieve metaphase alignment more than 70 min after NEB). Thus, our live cell imaging results demonstrate that overexpressing CENP-U phospho-mimicking mutant perturbs chromosome alignment function.
FIGURE 5.
Phosphorylation of CENP-U by Aurora-B leads to reduced kinetochore-microtubule interaction. A–C, shown is real-time imaging of chromosome movements in HeLa cells co-transfected with mcherry-H2B and GFP-CENP-U WT, AA, or DD. Chromosomes were marked by mcherry-H2B. Arrows indicate lagging chromosomes during alignment. Bar, 10 μm. D, shown is a graphic representation of chromosome alignment errors in GFP-CENP-U WT-, AA-, and DD-expressing cells. 24 GFP-CENP-U WT-expressing cells, 29 GFP-CENP-U AA-expressing cells, and 30 GFP-CENP-U DD-expressing cells were analyzed. E, overexpression of CENP-U DD results in destabilization of spindle microtubules. HeLa cells were transfected with GFP-CENP-U wild type, AA, and DD mutant for 36 h followed by a 2-h treatment of MG132 and examined for spindle stability in response to cold treatment. Cold-treated wild type (a), AA (b), and DD (c) mutant-expressing cells were stained with tubulin (red) and DAPI (blue). Bar, 10 μm. F, shown is the mitotic index of HeLa cells transfected with the indicated GFP-CENP-U plasmids and immunostained with DAPI 36 h after transfection. An average of 200 GFP-CENP-U-positive cells from three separate experiments was surveyed. Error bars represent S.E.; n = 3 preparations.
Given the importance of CENP-U in stable kinetochore-microtubule attachment as mentioned in Fig. 2, we next investigated whether the phospho-regulated reduction in CENP-U-Hec1 binding affinity can contribute to this behavior, thus, maybe explaining the phenotype observed in real time imaging of GFP-CENP-U DD-expressing cells. To this end, a cold treatment assay was performed. HeLa cells expressing GFP-CENP-U WT, AA, and DD were analyzed by immunofluorescence after cold treatment. As shown in Fig. 5E, cells expressing GFP-CENP-U DD (c) retained fewer cold stable microtubules compared with cells expressing GFP-CENP-U WT (a) and AA (b), as was the case in the CENP-U-depleted cells (Fig. 2D), albeit to a lesser degree. To determine whether DD mutant overexpression can cause an accumulation of mitotic cells, we transfected GFP-CENP-U plasmids into HeLa cells, and DNA morphology was examined by stain with DAPI. The mitotic index in DD-positive cells was increased (26 ± 3%) compared with that of WT positive cells (12 ± 2%) and AA positive cells (14 ± 3%) (Fig. 5F). All together, our results suggest that the CENP-U phospho-mimicking mutant behaves as a dominant negative, increasing the frequency of unstable kinetochore-microtubule attachment.
DISCUSSION
We have identified that CENP-U physically interacts with Hec1 in vitro and in vivo. Our functional analyses demonstrate that this interaction is required for a stable kinetochore-microtubule attachment. Interestingly, the cooperative microtubule binding activity of Hec1 and CENP-U is negatively regulated by Aurora-B kinase.
The importance of CCAN in outer kinetochore assembly has led to the general hypothesis that it makes an indirect structural contribution (2). However, depletion of CENP-U, a subunit of CCAN, does not affect the kinetochore localization of CENP-A, CENP-B, Hec1, CENP-E, CENP-F, Mis12, Aurora-B, or components of CCAN including CENP-H, CENP-M, and CENP-N (6), arguing that CENP-U may not serve as a structural scaffold. Our investigation revealed that CENP-U is a novel microtubule-binding protein, and depletion of CENP-U perturbs kinetochore-microtubule attachment plasticity. Based on our in vivo and in vitro observations, it is tempting to speculate that CENP-U makes a direct contribution to the kinetochore-microtubule attachment rather than just a structural platform. Consistent with our hypothesis, most recently, CCAN was proposed as a direct regulator of kinetochore-microtubule dynamics based on several observations. First, kinetochores lacking CENP-H, a subunit of CCAN, establish bioriented attachments but fail to generate regular oscillations as a result of uncontrolled rate of microtubule plus-end turnover. Second, CENP-H and CENP-I preferentially accumulate on kinetochores bound to growing microtubules. Last, CENP-Q, another subunit of CCAN, binds microtubules in vitro (39). Because both CENP-Q and CENP-U are in a stable subcomplex named CENP-O class proteins (31), our finding that CENP-U is a direct microtubule-binding protein seems more genuine and contributes to the determination of additional CCAN subunits that directly bind microtubules. Recent high resolution imaging of human kinetochore proteins, which shows that CCAN is located in the immediate vicinity of the kinetochore-microtubule plus ends (27), also supports our hypothesis. As our future plan, it would be of great interest to determine whether CENP-U is a direct regulator of kinetochore-microtubule dynamics and whether additional CCAN components, besides CENP-U and CENP-Q, can directly bind microtubules.
The KMN network has received much attention as the core kinetochore-microtubule attachment site, and formation of the entire network can synergistically enhance microtubule binding activity compared with the individual components (2, 9). In our study we also observed that CENP-U and Hec1 display cooperativity in their association with microtubules, thus providing the functional relevance of the CENP-U-Hec1 interaction. Given the fact that the Ndc80 complex plays a key role in the robust kinetochore-microtubule attachment, whereas the perturbation of kinetochore-microtubule attachment in CENP-U-depleted cells is less severe than what was observed in Hec1 depleted cells, we propose that CENP-U may function as a regulatory factor for fine toning the MT binding affinity of Ndc80 complex.
Recent studies support the notion that Aurora B kinase plays a critical role in correcting aberrant kinetochore-microtubule attachments by phosphorylating key substrates at the kinetochore and promoting turnover of kinetochore microtubules (41, 42). Identification of CENP-U as a cognate substrate for Aurora B together with the finding that phosphorylation of CENP-U by Aurora B weakens its binding and cooperativity with Hec1 in stabilizing kinetochore-microtubule interaction extend the list of kinetochore proteins implicated in aberrant microtubule detection and correction.
Aurora-B phosphorylation of downstream substrates, including the Ndc80 complex (9, 24, 40), the Dam1 ring complex (40), and the microtubule-depolymerizing kinesin MCAK (43, 44), appears to result in the destabilization of kinetochore-microtubule attachments (2). In particular, phosphorylation by Aurora-B reduces the binding affinity of the Ndc80 complex for microtubules (9), which provides a potential direct mechanism for eliminating incorrect kinetochore-microtubule attachment (2). We report here that Aurora-B phosphorylates CENP-U in vitro and have mapped two target sites within the Hec1 binding region. Moreover, it has been shown that phospho-mimicking mutation of Aurora-B target sites results in weaker binding activity to Hec1 in vitro and a dominant negative phenotype similar to the CENP-U depletion phenotype in vivo, whereas the effects of expressing the phospho-mimicking mutants in HeLa cells are somewhat less severe than the effects of CENP-U depletion. Combining these data with the observation that CENP-U and Hec1 exhibit a cooperative binding to microtubules, we propose that Aurora-B phosphorylation on CENP-U weakens the CENP-U-Hec1 interaction, thereafter resulting in unstable attachment of the Ndc80 complex with microtubules, which contributes to Aurora-B-dependent elimination of incorrect kinetochore-microtubule attachment in vivo.
Taken together, we established an interrelationship between CENP-U and Hec1 in the stabilization of kinetochore-microtubule attachment, and this interaction is under Aurora-B modulation. We propose that the CENP-U-Hec1 interaction provides a novel link between the CCAN pathway and the KMN-Aurora-B pathway. It is likely that all of the kinetochore outer plate proteins interact to orchestrate the dynamics and plasticity of chromosome segregation in mitosis. The CENP-U-Hec1 interaction established here is a core of the molecular society of mammalian cell kinetochores.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of CENP-U plasmids from Dr. Kyung S. Lee (National Institutes of Health). We thank members of our groups for insightful discussions and technical assistance. The facilities used were supported in part by National Center for Research Resources Grant G12RR03034.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-56292 and CA132389 and National Center for Research Resources Grant UL1 RR025008 from the Clinical and Translational Science Award program. This work was also supported by Chinese Natural Science Foundation Grants 31071184 (to Z. D.), 31000602 (to L. Z.), and 90508002 and 90913016 (to X. Y.), Chinese Academy of Science Grants KSCX1-YW-R-65, KSCX2-YW-H-10, and KSCX2-YW-R-195, Chinese 973 Project Grants 2006CB943603, 2007CB914503, and 2010CB912103, China National Key Projects for Infectious Disease Grant 2008ZX10002-021, a Georgia Cancer Coalition breast cancer research grant, Atlanta Clinical and Translational Science Award Chemical Biology Grant P20RR011104, and Anhui Province Key Project Grant 08040102005.

The on-line version of this article (available at http://www.jbc.org) contains supplemental S1–S4.
- KMN
- Knl1-Mis12-Ndc80
- CENP-U
- centromere-associated protein U
- CCAN
- constitutive centromere-associated network
- MT
- microtubule
- NEB
- nuclear envelope breakdown
- aa
- amino acids
- MBP
- maltose binding protein
- ACA
- anti-centromere antibody
- AA
- S349AS350A
- DD
- S349DS350D.
REFERENCES
- 1. Brinkley B. R., Stubblefield E. (1966) Chromosoma 19, 28–43 [DOI] [PubMed] [Google Scholar]
- 2. Cheeseman I. M., Desai A. (2008) Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 3. Cleveland D. W., Mao Y., Sullivan K. F. (2003) Cell 112, 407–421 [DOI] [PubMed] [Google Scholar]
- 4. Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., 3rd, Oegema K., Desai A. (2004) Genes Dev. 18, 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. (2004) Genes Cells 9, 105–120 [DOI] [PubMed] [Google Scholar]
- 6. Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 7. Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. (2006) Nat. Cell Biol. 8, 446–457 [DOI] [PubMed] [Google Scholar]
- 8. Santaguida S., Musacchio A. (2009) EMBO J. 28, 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006) Cell 127, 983–997 [DOI] [PubMed] [Google Scholar]
- 10. McCleland M. L., Kallio M. J., Barrett-Wilt G. A., Kestner C. A., Shabanowitz J., Hunt D. F., Gorbsky G. J., Stukenberg P. T. (2004) Curr. Biol. 14, 131–137 [DOI] [PubMed] [Google Scholar]
- 11. Wigge P. A., Kilmartin J. V. (2001) J. Cell Biol. 152, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCleland M. L., Gardner R. D., Kallio M. J., Daum J. R., Gorbsky G. J., Burke D. J., Stukenberg P. T. (2003) Genes Dev. 17, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin-Lluesma S., Stucke V. M., Nigg E. A. (2002) Science 297, 2267–2270 [DOI] [PubMed] [Google Scholar]
- 14. Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. (2003) J. Cell Sci. 116, 3347–3362 [DOI] [PubMed] [Google Scholar]
- 15. DeLuca J. G., Moree B., Hickey J. M., Kilmartin J. V., Salmon E. D. (2002) J. Cell Biol. 159, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He X., Rines D. R., Espelin C. W., Sorger P. K. (2001) Cell 106, 195–206 [DOI] [PubMed] [Google Scholar]
- 17. Wei R. R., Sorger P. K., Harrison S. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ciferri C., De Luca J., Monzani S., Ferrari K. J., Ristic D., Wyman C., Stark H., Kilmartin J., Salmon E. D., Musacchio A. (2005) J. Biol. Chem. 280, 29088–29095 [DOI] [PubMed] [Google Scholar]
- 19. DeLuca J. G., Dong Y., Hergert P., Strauss J., Hickey J. M., Salmon E. D., McEwen B. F. (2005) Mol. Biol. Cell 16, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei R. R., Al-Bassam J., Harrison S. C. (2007) Nat. Struct. Mol. Biol. 14, 54–59 [DOI] [PubMed] [Google Scholar]
- 21. Wei R. R., Schnell J. R., Larsen N. A., Sorger P. K., Chou J. J., Harrison S. C. (2006) Structure 14, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 22. Miller S. A., Johnson M. L., Stukenberg P. T. (2008) Curr. Biol. 18, 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guimaraes G. J., Dong Y., McEwen B. F., Deluca J. G. (2008) Curr. Biol. 18, 1778–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. (2006) Cell 127, 969–982 [DOI] [PubMed] [Google Scholar]
- 25. Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J. G., De Wulf P., Salek M., Rappsilber J., Moores C. A., Salmon E. D., Musacchio A. (2008) Cell 133, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Powers A. F., Franck A. D., Gestaut D. R., Cooper J., Gracyzk B., Wei R. R., Wordeman L., Davis T. N., Asbury C. L. (2009) Cell 136, 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wan X., O'Quinn R. P., Pierce H. L., Joglekar A. P., Gall W. E., DeLuca J. G., Carroll C. W., Liu S. T., Yen T. J., McEwen B. F., Stukenberg P. T., Desai A., Salmon E. D. (2009) Cell 137, 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Du J., Cai X., Yao J., Ding X., Wu Q., Pei S., Jiang K., Zhang Y., Wang W., Shi Y., Lai Y., Shen J., Teng M., Huang H., Fei Q., Reddy E. S., Zhu J., Jin C., Yao X. (2008) Oncogene 27, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. (2006) Genes Cells 11, 673–684 [DOI] [PubMed] [Google Scholar]
- 30. Minoshima Y., Hori T., Okada M., Kimura H., Haraguchi T., Hiraoka Y., Bao Y. C., Kawashima T., Kitamura T., Fukagawa T. (2005) Mol. Cell. Biol. 25, 10315–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hori T., Okada M., Maenaka K., Fukagawa T. (2008) Mol. Biol. Cell 19, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kang Y. H., Park J. E., Yu L. R., Soung N. K., Yun S. M., Bang J. K., Seong Y. S., Yu H., Garfield S., Veenstra T. D., Lee K. S. (2006) Mol. Cell 24, 409–422 [DOI] [PubMed] [Google Scholar]
- 33. Cao X., Ding X., Guo Z., Zhou R., Wang F., Long F., Wu F., Bi F., Wang Q., Fan D., Forte J. G., Teng M., Yao X. (2005) J. Biol. Chem. 280, 13584–13592 [DOI] [PubMed] [Google Scholar]
- 34. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) J. Biol. Chem. 279, 20049–20057 [DOI] [PubMed] [Google Scholar]
- 35. Yao X., Anderson K. L., Cleveland D. W. (1997) J. Cell Biol. 139, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu D., Ding X., Du J., Cai X., Huang Y., Ward T., Shaw A., Yang Y., Hu R., Jin C., Yao X. (2007) J. Biol. Chem. 282, 21415–21424 [DOI] [PubMed] [Google Scholar]
- 37. Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. (2000) Nat. Cell Biol. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 38. Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., Cui T., Hu R., Dou Z., Zhu J., Xie W., Rao Z., Ding X., Yao X. (2008) J. Biol. Chem. 283, 26726–26736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amaro A. C., Samora C. P., Holtackers R., Wang E., Kingston I. J., Alonso M., Lampson M., McAinsh A. D., Meraldi P. (2010) Nat. Cell Biol. 12, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. (2002) Cell 111, 163–172 [DOI] [PubMed] [Google Scholar]
- 41. Pinsky B. A., Biggins S. (2005) Trends Cell Biol. 15, 486–493 [DOI] [PubMed] [Google Scholar]
- 42. Ruchaud S., Carmena M., Earnshaw W. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 798–812 [DOI] [PubMed] [Google Scholar]
- 43. Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. (2004) Curr. Biol. 14, 273–286 [DOI] [PubMed] [Google Scholar]
- 44. Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. (2004) Dev. Cell 6, 253–268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.