Abstract
When dietary choline is restricted, most men and postmenopausal women develop multiorgan dysfunction marked by hepatic steatosis (choline deficiency syndrome (CDS)). However, a significant subset of premenopausal women is protected from CDS. Because hepatic PEMT (phosphatidylethanolamine N-methyltransferase) catalyzes de novo biosynthesis of choline and this gene is under estrogenic control, we hypothesized that there are SNPs in PEMT that disrupt the hormonal regulation of PEMT and thereby put women at risk for CDS. In this study, we performed transcript-specific gene expression analysis, which revealed that estrogen regulates PEMT in an isoform-specific fashion. Locus-wide SNP analysis identified a risk-associated haplotype that was selectively associated with loss of hormonal activation. Chromatin immunoprecipitation, analyzed by locus-wide microarray studies, comprehensively identified regions of estrogen receptor binding in PEMT. The polymorphism (rs12325817) most highly linked with the development of CDS (p < 0.00006) was located within 1 kb of the critical estrogen response element. The risk allele failed to bind either the estrogen receptor or the pioneer factor FOXA1. These data demonstrate that allele-specific ablation of estrogen receptor-DNA interaction in the PEMT locus prevents hormone-inducible PEMT expression, conferring risk of CDS in women.
Keywords: Gene Regulation, General Transcription Factors, Genetic Polymorphism, Liver, Phosphatidylcholine, PEMT, Choline, Estrogen
Introduction
In humans, dietary deficiency of choline results in liver and muscle damage characterized by hepatosteatosis and elevated serum levels of hepatic transaminases and creatine phosphokinase, subsequently referred to as choline deficiency-induced syndrome (CDS)5 (1–4). Although CDS occurs in most men and postmenopausal women deprived of dietary choline, the majority (56%) of premenopausal women seem to be protected against CDS (1).
The only de novo source of choline derives from hepatic synthesis catalyzed by PEMT (phosphatidylethanolamine N-methyltransferase) (5, 6). PEMT is regulated by estrogen in human and mouse hepatocytes, but the mechanisms for this regulation are unknown (7). Because endogenous choline production could compensate for insufficient choline intake, we suggest that many premenopausal women may be protected from CDS by induction of PEMT by estrogen. This is important because augmented production of choline may be particularly important during pregnancy and lactation, when demand for choline is especially high due to transport of choline from mother to fetus (8, 9). The National Academy of Sciences set an adequate intake level for choline (10), but in the United States, less than 15% of pregnant women eat the recommended amount (11). In fact, women vary enough in dietary choline intake (from <300 to >500 mg/day) to influence the risk that they will have a baby with a birth defect; at least 25% of women eat so little choline that their pregnancies are at risk (12–14). A 2009 study of 130,000 women in California (14) reported elevated neural tube defect risk associated with lower concentrations of choline in blood and reduced risk with higher levels of choline. Specifically, they observed an odds ratio of 2.4 (95% confidence interval = 1.3–4.7) associated with the lowest decile and an odds ratio of 0.14 (0.02–1.0) associated with the highest decile, both relative to the 25th to 74th percentiles of the control distribution (14).
Although most young women are resistant to CDS, more than 40% of them do require a source of dietary choline, or they develop fatty liver (1). We hypothesize that these women are insensitive to estrogenic activation of PEMT because of a functional polymorphism that alters the interaction of the gene with estrogen. Therefore, these women are dependent on dietary choline. This hypothesis is supported by existing data; Pemt disruption predisposes to hepatic steatosis in mice (15–17), and a functional polymorphism in PEMT (V175M) was significantly associated with nonalcoholic hepatic steatosis in populations in the United States and Japan (18, 19). Indeed, we identified a single nucleotide polymorphism (SNP) in the promoter region of PEMT (rs12325817) that was associated with a greatly increased risk for developing organ dysfunction in premenopausal female carriers when fed a low choline diet (20). The mechanism of action for the effect of this SNP on risk for CDS has not been identified.
To further elucidate the mechanism for allelic differences in PEMT regulation, we identified the CDS risk-associated haploblock. Using allele- and transcript-specific gene expression analyses, we identified the PEMT transcripts that were induced by estrogen and demonstrated that, in primary human hepatocytes, estrogen responsiveness was abrogated in the risk haploblock. We comprehensively analyzed ER-chromatin interaction to identify the location of estrogen regulatory regions in the PEMT locus. We found that the risk haploblock failed to bind the estrogen receptor or the cooperative transcriptional regulator FOXA1 and was unable to mediate hormonal transcriptional regulation.
Our results suggest that a risk haplotype located in the PEMT gene directly abrogates estrogenic regulation of PEMT, explaining how genetic variation influences gender-specific dietary choline requirements, placing young women at risk of CDS.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary human liver cells were provided as a gift by Admet Technologies (Durham, NC). These studies were approved by the institutional review board of the University of North Carolina at Chapel Hill; informed consent was not obtained because the data were analyzed anonymously. Hepatocytes were isolated as described previously (7). Cells were plated at a density of 1.8 × 106 cells/9.6 cm2 on collagen-coated culture plates (BD Biosciences) and incubated for 6 h at 37 °C in humidified air containing 5% CO2 in Williams' complete medium E (Invitrogen) containing 10% dextran-treated charcoal-stripped fetal bovine serum (Biomeda, Foster City, CA) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin; Sigma-Aldrich). Medium was replaced at 6 h with serum-free Williams' complete medium E. All experimentation involving human cells was carried out following a 48-h “recovery period” in serum-free Williams' complete medium E.
ChIP-chip and ChIP-qPCR
Chromatin prepared from formaldehyde-fixed primary hepatocytes after a 45-min incubation with 100 nmol/liter 17-α-estradiol (E2) (Sigma-Aldrich) was immunoprecipitated with anti-ERα (Ab-10, Neomarkers; HC-20, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) or FoxA1 antibodies (Ab5089, Abcam (Cambridge, MA)) prebound to Ultralink protein A/G beads (Pierce) for a minimum of 6 h at 4 °C as described previously (21, 22). Immunoprecipitated chromatin as well as unfractionated input control chromatin were fluorescently labeled and hybridized to a custom designed array (Roche-Nimblegen, Madison, WI) that tiled 430 kb of Chr 17 harboring the PEMT locus and several other genomic locations corresponding to previously identified ER binding sites (21, 23–25) with 60-nucleotide overlapping isothermal probes after eliminating repetitive regions. Chromatin was analyzed by microarray hybridization as described previously (26). Peaks with an average enrichment ratio (log2) >2 and width >150 bp that were independently identified by Mpeak (27) were selected for further study. Primers for quantitative PCR were designed using Primer Express (Primer 3 (28)). Purified DNA was subjected to PCR using the Applied Biosystems SYBR Green Mastermix. Relative DNA quantities were measured using a NanoDrop UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Sequences of qPCR primers are available upon request.
Transcript-specific Gene Expression
Primary human hepatocytes cultured for 48 h in hormone-free medium in 6-well collagen-coated plates (BD Biosciences) were treated with 100 nm moxesterol. RNA was isolated (RNeasy, Qiagen (Gaithersburg, MD)) and analyzed by quantitative PCR (Taqman, Applied Biosystems (Foster City, CA)) with probes specific for each PEMT transcript (Transcript A, NM_148172, assay ID Hs01002998_m1; Transcript B, NM_007169, assay ID Hs00200354_m1; Transcript C, NM_148173, custom assay ID 4331348). To determine the absolute copy number of the target transcripts, we cloned a cDNA fragment unique to each PEMT transcript isolated from human liver RNA into a TOPOII TA (Invitrogen). The copy number of unknown samples (RNA extracted at various time points from human hepatocytes) was determined by linear regression analysis utilizing transcript-specific standard curves. To correct for differences in both RNA quality and quantity between samples, the expression levels of transcripts of interest were normalized to GAPDH and expressed as a ratio of gene expression change.
Transcripts in Human Liver
Human male liver biopsy samples (Liver Tissue Procurement and Distribution System (Minneapolis, MN); n = 6) were snap-frozen in liquid nitrogen immediately following harvest. RNA was isolated (TRIzol, Invitrogen) and analyzed by quantitative PCR (Taqman, Applied Biosystems) with probes specific for each PEMT transcript as described previously. To correct for differences in both RNA quality and quantity between samples, the expression levels of transcripts of interest were normalized to GAPDH and expressed as a ratio of gene expression change. We also calculated results by normalizing to the amount of cDNA; both calculations yielded similar results.
Identification of the Choline Deficiency Syndrome-associated Haplotype in the PEMT Locus
As part of a University of North Carolina institutional review board-approved study, 57 human subjects were fed a defined diet containing 550 mg of choline/70 kg of body weight/day for 10 days and then were fed a defined low choline diet (<50 mg of choline/70 kg of body weight/day) either until the development of organ dysfunction or for a period of up to 42 days. The design of this study has been described elsewhere (1, 20, 29). Organ dysfunction associated with choline deficiency was defined as a greater than 5-fold increase of serum CPK activity; a 1.5-fold increase in serum activity of aspartate transaminase, alanine transaminase, γ-glutamyl transpeptidase, or lactic dehydrogenase; or an increase in liver fat content of more than 28% (assessed by magnetic resonance imaging) that was resolved upon reintroduction of choline to the diet. DNA isolated from women (n = 64) and men (n = 26) who participated in the study was genotyped for a selection of SNPs in the PEMT locus by sequencing, using primers described previously (20) or by a Taqman® SNP genotyping assay (catalog no. C_9246129_10, Applied Biosystems).
Allele-specific Gene Expression
For allele imbalance experiments, primary hepatocytes were genotyped for the exonic (rs897453) and CDS-associated (rs4646343) SNPs by qPCR using TaqMan primer-probe assays (Assays on Demand SNP genotyping assay; Applied Biosystems): SNP rs897453 (assay ID C___7443062_1_); SNP rs4646343 (assay ID C___9246129_10). To quantify expression differences between the risk and protective allele, a standard curve was generated by mixing genomic DNA from homozygous risk individuals (defined as rs4646343 (A/A) and homozygous protective individuals (rs4646343 (C/C)) at five ratios followed by qPCR with a TaqMan assay specific for each rs897453 allele. cDNA was generated (SSIII First Strand synthesis kit, Invitrogen) from hormone-treated and -untreated hepatocytes heterozygous for SNPs rs897453 and rs4646343. After allele-specific TaqMan, we deduced the ratios of gene expression between the two alleles as described previously (30). Allele-specific detection of PEMT in genomic DNA (predicted to be 50%) served to normalize cDNA ratios. Results are presented as a percentage of risk allele expression.
Transcript-specific allele imbalance experiments were performed with primer-probe sets unique for each transcript (as described above) in hepatocytes homozygous for the risk allele (R) (n = 5), homozygous for the protective allele (P) (n = 6), and heterozygous for the protective/risk alleles (P/R) (n = 5).
Haplotype-specific Estrogen-induced Changes in PEMT Enzyme Activity
Primary human hepatocytes were obtained fresh from CellzDirect (Durham, NC) and cryopreserved from Zen-Bio (RTP). They were genotyped for rs12325817 and rs4646343 as described previously. Cells were plated at a density of 1 × 106 cells/well on collagen-coated 6-well culture plates (BD Biosciences) and incubated for 6 h at 37 °C in humidified air containing 5% CO2 in Hepatocyte Plating Medium (HM-1, Zen-Bio). Medium was replaced at 6 h with serum-free phenol red-free Hepatocyte Maintenance Medium (HM-2, Zen-Bio) for 15 h to recover. The primary cells were then incubated for 24 h in HM-2 with 0 or 100 nmol/liter moxestrol (Steraloids, Newport, RI). Cells were washed twice with 1× PBS, scraped into medium A (250 mmol/liter sucrose, 100 mmol/liter Tris, 0.1 mmol/liter EGTA, and 5 mmol/liter dithiothreitol, pH 8) and homogenized. Homogenate containing 50 μg of protein was used for PEMT activity assays as described previously using phosphatidyldimethylethanolamine (Avanti Polar Lipids, Alabaster, AL) as the methyl acceptor and S-adenosyl-l-[methyl-3H]methionine (American Radiolabeled Chemicals Inc., St. Louis, MO) as the methyl group donor (6). C57BL/6 mouse primary hepatocytes were used as a positive control in these experiments.
Construction of Plasmids and Luciferase Reporter Assay
Genomic DNA was extracted from human hepatocytes and used to amplify a segment of the 5′-flanking region of the transcription start site for PEMT transcript B (NM_007169). The 1.3-kb PCR product was directionally subcloned (BglII/HindIII) into the promoterless expression vector, pGL4-Basic (Promega, Madison, WI), Plasmid was designated as pGL4-PEMT B (supplemental Table 1). ERα binding sites identified by ChIP (peaks II–IV, III/IV, and IV) were amplified from human liver genomic DNA by PCR and subcloned (KpnI/Xho; Xho/Xho) into the pGL4-SV40 vector (Promega; Madison, Wisconsin) and pGL4-PEMT B. Site-directed mutagenesis (Stratagene, La Jolla, CA) of the peak IV pGL4-SV40 construct was performed using a QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Clones were verified by sequencing (University of North Carolina Genome Analysis Facility). MCF7 cells cultured for 24 h in phenol red-free DMEM with 1% charcoal/dextran-treated FBS (Hyclone) were transfected with ERα pGL4-PEMT B and pGL4-SV40 constructs (FuGENE® HD transfection reagent, Roche Applied Science). Twenty-four hours after transfection, cells were treated with 100 nmol/liter moxestrol.
Human primary hepatocytes grown in serum-free/hormone-depleted medium for at least 2 days in 24-well collagen-coated plates (BD Biosciences) were transfected with JETPEI hepatocyte transfection reagent (Genesee Scientific, San Diego, CA). Twenty-four hours after transfection, cells were treated with 100 mmol/liter moxestrol. Cell lysates were assessed using the Dual Luciferase Reporter Assay system (Promega).
Statistical Analysis
For the luciferase reporter assay and the allele-specific gene expression studies, statistical differences in treatment were determined using Student's t test (31). For the choline depletion study, a Fisher's exact test was used to determine statistical significance of SNP association with organ dysfunction (31). For chromatin studies, a mixed model was applied with signal ratio (immunoprecipitated chromatin/input control) as the response, treatment group (estrogen-treated and -untreated) and genotype (P, P/R, and R) as fixed effects, and genomic region (peaks II, III, IV, and V and negative control) as a random effect (31). For qPCR, a repeated measures model was applied with the gene expression ratio (ln(PEMT/GAPDH)) as the response and treatment group (moxestrol-treated and -untreated), time (4, 8, and 24 h), and genotype as predictors (31). (Analysis was rerun for each transcript.) Terms testing for interactions between the fixed effects predictors were included. Interaction terms with a p value greater than 0.10 were removed, and the models were repeated. An unstructured correlation matrix to account for the correlation of the observations within each subject at each time/region was included in the model (31). For the transcript-specific gene expression analysis in human liver biopsies and the PEMT activity studies, statistical differences were assessed using an analysis of variance and Tukey-Kramer test.
RESULTS
Identification of a CDS-associated Haploblock
We had previously performed a clinical study that assessed human dietary choline requirements (1). In that study, we examined a targeted set of SNPs in genes involved in choline metabolism (five SNPs, including two in PEMT) and identified one SNP in PEMT (rs12325817) that was highly associated with CDS in women (20). We now completed an analysis of an additional 33 women subjected to the same clinical protocol (bringing the total to 64 women) and confirmed the association between risk of CDS and the presence of at least one variant allele marked by rs12325817 (p = 9 × 10−6). In addition, we examined another SNP in PEMT (rs4646343; located within intron IA) in these 64 women and found that rs4646343 (present in the HapMap data base (32, 33)) was highly associated with risk for CDS (p = 6.2 × 10−5). Ninety-two percent of the 64 women with a variant allele for rs12325817 also had a variant allele for rs4646343.
Estrogen Regulation of PEMT Is Transcript-specific
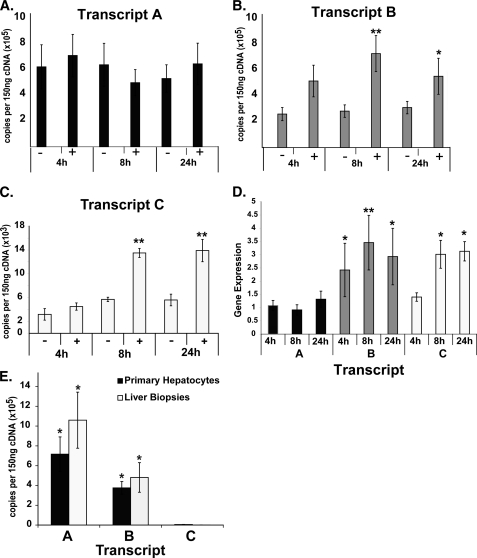
To begin to understand the mechanism for estrogenic control of PEMT, we examined the regulation of the three PEMT transcripts. Each transcript is regulated by a distinct promoter and contains a unique first exon (Fig. 1). Using transcript-specific primer-probe sets and cDNA standards, we quantified the absolute level of expression of each transcript with and without treatment with the estrogen analog moxestrol. This synthetic estrogen was used because it is not subject to cytochrome P450 metabolism expected in primary hepatocytes (34, 35). Although transcript A was most abundant in unstimulated cells (∼7.5 × 105 copies/150 ng of cDNA), it was not induced at any time point tested (Fig. 2, A and D). In contrast, transcripts B and C were significantly induced (Fig. 2, B–D). Upon moxestrol treatment, transcript B became the predominant transcript, exceeding levels of transcript A at 8 h. Transcript C was the least abundant. These data suggest that estrogen selectively activates transcripts B and C.
FIGURE 1.
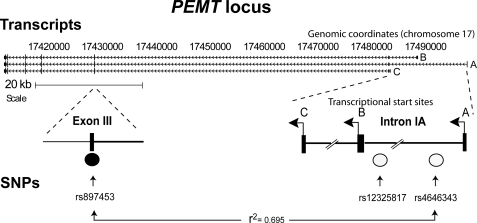
PEMT SNPs are associated with CDS risk in women. Shown is a schematic representation of the PEMT locus, including alternative transcriptional start sites (A, B, and C), and location of SNPs (rs12325817 and rs4646343) highly linked to the CDS phenotype in humans fed a low choline diet. Using HapMap data, other SNPs in this region were found to be in high linkage disequilibrium with rs4646343, including an SNP located in common exon 3 (rs897453) (Centre d'Etude du Polymorphisme Humain (CEPH); r2 = 0.695).
FIGURE 2.
Estrogen regulation of PEMT is transcript-specific. Using RNA isolated from primary hepatocytes cultured in the absence (−) or presence (+) of 100 nmol/liter moxestrol for the lengths of time noted, PEMT transcripts were quantified using transcript-specific primers by quantitative PCR. A, transcript A, initially the most abundant, was not responsive to hormone treatment. B, transcript B was significantly induced in response to hormone treatment at both 8 and 24 h after treatment. C, transcript C was the least abundant but was significantly induced in response to hormone treatment at 8 and 24 h. D, PEMT expression levels, presented as a ratio of treated/untreated for all transcripts (normalized to GAPDH), demonstrated significant induction of B (all time points tested) and transcript C (at the 8 and 24 h time points). E, RNA isolated from male human liver biopsies and primary human hepatocytes was subjected to transcript-specific quantitative PCR. For human biopsy samples, results are presented as an average for all subjects (n = 6), and for human hepatocytes, results are shown as an average of all untreated samples, across all time points. Transcripts A and B were significantly more abundant than transcript C. Transcript A was significantly more abundant than transcript B (n = 5–6/time point; *, p < 0.05; **, p < 0.01). Error bars, S.E.
PEMT A is the major transcript in human liver. In six male human liver biopsy samples, PEMT transcript A mRNA was >6 times more abundant than was transcript B and transcript C mRNA (p < 0.01; Fig. 2E).
Risk Allele Is Not Estrogen-responsive
Using HapMap data, other PEMT SNPs were found to be in high linkage disequilibrium with rs4646343, including an SNP located in common exon 3 (rs897453) (Centre d'Etude du Polymorphisme Humain (CEPH); r2 = 0.67) (Fig. 1) (note that rs12325817 was not assessed in the HapMap study) (25, 26). This non-synonymous SNP encodes a conservative amino acid substitution (Val → Ile). The availability of a common exonic SNP in linkage disequilibrium with the CDS risk-associated SNP enabled examination of allele-specific expression.
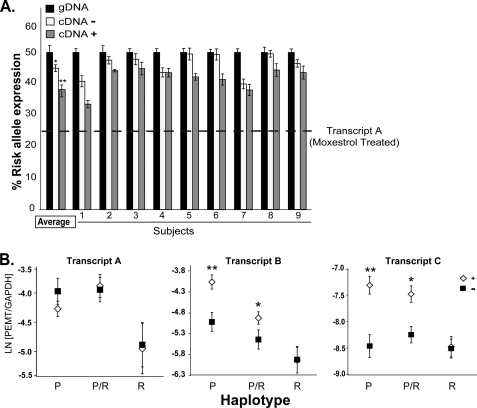
Human liver organ donors were genotyped to determine their SNP status at the CDS-associated allele, rs4646343, and the exonic allele, rs897453. We quantified the relative expression of PEMT from the risk and the non-risk alleles in heterozygous hepatocytes by leveraging the association with the exonic SNP with and without moxestrol treatment. In untreated hepatocytes, the risk allele was underexpressed by ∼4% relative to the protective allele (p = 0.01), and after treatment with moxestrol, the risk allele was underexpressed by ∼8% (p = 0.0002; Fig. 3A).
FIGURE 3.
PEMT risk allele is not estrogen-responsive. A, RNA isolated from primary hepatocytes heterozygous for the risk allele was subjected to allele-specific quantitative PCR. Results were normalized to genomic DNA (black bars). The estimated relative expression of the risk allele cDNA versus the protective allele cDNA was calculated by linear regression analysis. In the absence of 100 nmol/liter moxestrol treatment (−, open bars), the risk allele was underexpressed by ∼4% relative to the protective allele. Upon moxesterol treatment (24 h) (+, gray bars), the risk allele was underexpressed by ∼8%. Based on the transcript-specific expression data, the estimated relative abundance of unregulated transcript A in treated cells is depicted by the dashed line. Results are presented for individual subjects and as an average for all subjects (n = 9). *, p < 0.01; **, p < 0.001. Error bars, S.E. B, RNA was isolated from moxestrol-treated (open diamonds) or untreated hepatocytes (black squares) that were P, R, or P/R. PEMT levels were determined by qPCR. Transcript A was not induced by estrogen in any of the groups. Transcripts B and C were induced by hormone in the P and P/R groups, but in hepatocytes homozygous for the risk allele (R), these transcripts were not induced by hormone treatment. Results are presented as the natural log (LN) of the ratio of PEMT relative to GAPDH gene expression levels in moxestrol-treated versus untreated hepatocytes. (n = 5–6/genotype). *, p < 0.001; **, p < 0.0001. Error bars, S.E.
To explore whether expression of the protective allele may compensate for the underexpression of the risk allele, we compared PEMT transcript levels in human hepatocytes after treatment with moxestrol. In contrast to PEMT transcript A, for which genotype was not associated with differences in expression (p = 0.06), transcripts B and C demonstrated a highly significant interaction between response to treatment and genotype (p = 0.0001 for both). Transcripts B and C were induced less in cells homozygous for the risk allele than in cells heterozygous for the risk allele or homozygous for the protective allele (Fig. 3B).There is also a trend toward decreased basal expression of transcripts A and B in the presence of the risk allele. These allele-specific data further support the association of CDS-linked SNPs with the absence of hormonal PEMT regulation.
Identification of ER Binding Sites in the PEMT Locus in Human Hepatocytes
To identify the mechanism for estrogenic activation of the B and C transcripts, we comprehensively identified sites of ER interaction with the PEMT locus. Chromatin was isolated from primary human hepatocytes before and after treatment with estradiol and immunoprecipitated (ChIP) with estrogen receptor α antibodies (ERα, the major isoform in liver (36)). Immunoprecipitated and total (input control) chromatin were hybridized to a microarray that tiled virtually the entire PEMT locus with densely spaced, overlapping probes (Fig. 4A). Because previous genome-wide studies had identified functional ER binding sites several hundred kb from regulated genes (21), we designed our arrays to include 430 kb surrounding the PEMT gene. As positive controls, we also tiled genomic loci previously shown to bind ER in other cell types. We found that ∼56% of these regions exhibited ER binding in our experiments (data not shown).
FIGURE 4.
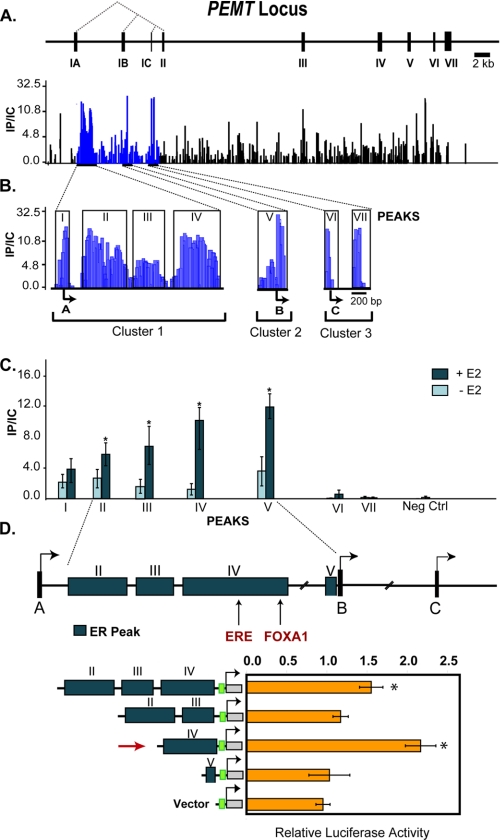
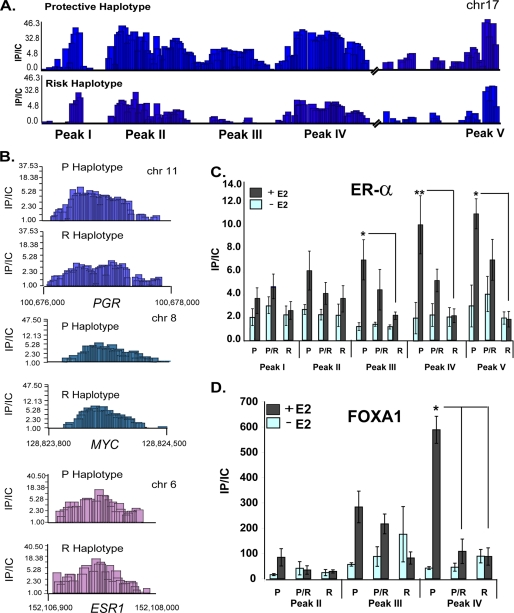
Identification of ER binding sites within PEMT. The PEMT gene encodes three isoforms that are distinguished by unique leading exons, IA, IB, and IC. The dotted lines indicate splicing of exons IA–IC to common exons II–VII. Chromatin isolated from human hepatocytes incubated for 45 min with 100 nmol/liter estradiol (E2) was immunoprecipitated with anti-ERα antibody and hybridized, together with unfractionated chromatin, to a microarray that densely tiled virtually the entire PEMT locus. A, schematic depiction of the PEMT locus. Locus-wide chromatin immunoprecipitation data are expressed as the ratio of immunoprecipitated chromatin to input control (IP/IC). Only positive enrichment ratios of ChIP signal to input control signal are shown. B, seven ER binding regions were mapped within three clusters in close proximity to transcription start sites A, B, and C. The boxed regions indicate identified peaks. C, quantitative PCR was performed on chromatin from cells before and after 45-min exposure to 100 nmol/liter E2 treatment with primers specific to the seven microarray-defined regions (as well as a negative control (neg ctrl)). ER binding to peaks within intron IA and 3′ to the B TSS was significantly increased by E2 (n = 3). *, p < 0.05. D, ER binding regions (peaks II–IV and peak V) from the protective haplotype were cloned upstream of the SV40 promoter and transfected into primary human hepatocytes grown in hormone-depleted media. Cells were either untreated or treated with 100 nmol/liter moxestrol (MOX). Peak IV (indicated by the red arrow) harbors the consensus ERE. The green box represents the SV40 promoter, which drives expression of the luciferase gene. The data represent the average relative luciferase activity normalized for Renilla luciferase and are expressed as -fold induction relative to the activity in the absence of moxestrol. Absolute values for both treated (+) and untreated (−) cells are presented separately in the table. *, p < 0.05 compared with untreated.
Using the positive control regions to establish peak detection parameters, seven putative ER binding sites were identified in the PEMT gene using chromatin from primary human hepatocytes harboring the protective haplotype (Fig. 4B). Replicate data from a second independent experiment demonstrated identical binding at six of seven sites (data not shown). These seven putative ER binding sites, comprising three clusters, were located between the PEMT A transcriptional start site (TSS) and the shared second intron 3′ to the PEMT C TSS. The major cluster (cluster 1) consisted of four separate regions located ∼6.9 kb 5′ from the TSS for PEMT B.
To quantify differential ER binding to these regions, ChIP-qPCR was performed on chromatin isolated from primary human hepatocytes before and after treatment with estradiol. Each of the regions within intron 1A was examined separately. Compared with untreated chromatin, estrogen significantly increased binding to regions II, III, IV, and V (p < 0.05) (Fig. 4C) but did not induce binding when probed with a non-specific antibody (IgE) (supplemental Fig. 3). Sites I, VI, and VII did not exhibit estrogen-induced binding, similar to a negative control site in the region. Region IV contains a consensus estrogen response element (ERE; GTCA-xxx-TGACC) as well as a FOXA1 site, both of which are evolutionarily conserved between humans and mice. Although the PEMT locus contains 29 predicted imperfect ERE motifs (TRANSFAC (37)), none of these sites demonstrated enrichment.
To examine whether sites of ER binding can act as hormone-responsive transcriptional enhancers in human hepatocytes, the intron IA (containing peaks II–IV) and promoter B (peak V) genomic regions were isolated from human hepatocytes harboring the protective haplotype and cloned upstream of an SV40 minimal promoter (Fig. 4D). Because site IV contains the consensus ERE, this region was also cloned separately from sites I–III. These constructs were transiently transfected into hormone-depleted primary human hepatocytes. The region containing sites II–IV was capable of mediating estrogenic activation (Fig. 4D). When separated, site IV but not sites II/III mediated hormonal activation. Although site V binds ER and induces basal transcription (data not shown), it failed to confer estrogen responsiveness. As further confirmation, these regions were placed upstream of the PEMT B proximal promoter and assayed in MCF7 or liver-derived HepG2 cells. The peaks II–IV and peak IV alone were again able to function as an estrogen-responsive enhancer (p < 0.01; supplemental Figs. 1 and 2). The estrogen receptor is necessary for hormone activity (supplemental Fig. 2).
ER Fails to Bind the CDS Risk Allele
To determine if SNP status affected ER binding to the PEMT locus, we performed microarray-based analysis of chromatin isolated from estrogen-treated hepatocytes homozygous for the risk allele and homozygous for the protective allele. Decreased estrogen binding was evident at sites II, III, and IV in hepatocytes homozygous for the risk allele (Fig. 5A). ER binding to several non-PEMT control sites was unaffected by genotype (Fig. 5B).
FIGURE 5.
Choline deficiency syndrome-associated risk allele fails to bind ER. A, chromatin isolated from 100 nmol/liter estradiol-treated hepatocytes homozygous for the protective or risk alleles was immunoprecipitated with anti-ERα antibodies and hybridized to densely tiled oligonucleotide microarrays. Estrogen receptor complex (ER) binding was decreased for the risk haplotype at peaks II–IV. Chromatin from estradiol-treated (+) or untreated (−) hepatocytes homozygous (P and R) or heterozygous (P/R) for the protective or risk alleles was immunoprecipitated with anti-ERα (B) and anti-FOXA1 (C) antibodies followed by amplification with primers specific for ER-associated regions. Whereas ER binding to peaks II–IV was significantly induced by estrogen for the protective allele, estrogen failed to significantly augment ER binding to the risk allele (n = 5–6/genotype). *, p = 0.0001. Data are expressed as the ratio of immunoprecipitated chromatin to input control (IP/IC). Error bars, S.E. D, ER binding to representative sites on control genes (PGR, ESR1, and MYC) was unaffected by haplotype.
We validated the influence of SNP status on ER binding to the PEMT locus by quantitatively analyzing ER-associated chromatin isolated from R, P, and P/R primary hepatocytes. Chromatin from hepatocytes heterozygous and homozygous for the protective allele exhibited significant differences in ER binding between treated and untreated hepatocytes for sites II–IV (p < 0.0001; Fig. 5C). Chromatin isolated from hepatocytes homozygous for the protective allele exhibited significantly greater ER binding compared with chromatin isolated from homozygous risk hepatocytes for regions III and IV (p < 0.05). In contrast, estrogen treatment failed to induce ER binding in chromatin isolated from subjects homozygous for the risk allele at all sites tested (I–V) (p = 0.12). Because FOXA1 has been shown to act to create a favorable chromatin environment for ER binding, we then examined FOXA1 occupancy at the site 52 bp from the ERE (Fig. 5D). As with ER, FOXA1 chromatin interaction was also greatly influenced by haplotype. Chromatin isolated from hepatocytes homozygous for the protective allele exhibited significantly greater FOXA1 binding compared with chromatin isolated from homozygous risk and heterozygous hepatocytes at region IV (p < 0.05).
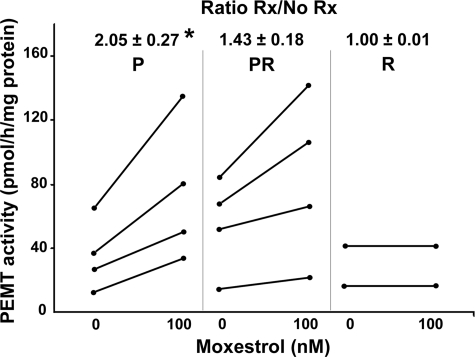
Hepatocytes with the CDS risk allele have decreased PEMT activity. To determine whether the observed decrease in PEMT gene expression resulted in a reduction in functional PEMT enzyme levels, activity was measured in human hepatocytes that were R, P, or P/R after treatment with moxestrol. Untreated fresh hepatocytes had similar activity (16 ± 7 pmol/h/mg protein) as those kept in culture for 48 h (14 ± 4 pmol/h/mg protein; p = 0.67). Hepatocytes homozygous for the protective allele (P) had 2 ± 0.1-fold more activity after moxestrol than did untreated cells, whereas hepatocytes with the risk allele (P/R and R) had 1.3 ± 0.1-fold more activity after moxestrol than did untreated cells (p = 0.0018) (Fig. 6). Only two samples of hepatocytes homozygous harboring the risk allele could be obtained. In both samples, PEMT activity did not increase after moxestrol (estrogen) treatment (Fig. 6). In comparison, hepatocytes from C57BL/6 mice had 2.7 ± 0.1 times more PEMT activity after moxestrol treatment (p < 0.01).
FIGURE 6.
Less PEMT activity is induced by estrogen in human hepatocytes with the risk allele. Primary human hepatocytes that were P, R, or P/R were treated with 0 or 100 nmol/liter moxestrol for 24 h. PEMT activity was measured and reported as absolute values as well as the -fold increase in activity as compared with the untreated cells (mean ± S.D.). The P group (n = 4) had significantly more activity than the P/R (n = 4) and R (n = 2) groups separately or combined (*, p < 0.05). Rx, treatment with moxestrol.
DISCUSSION
For mammals, choline is an essential nutrient. Restricted consumption leads to significant organ dysfunction (1, 20) and increases the risk of giving birth to a baby with a neural tube and/or orofacial defect (12, 13). De novo choline production through PEMT-catalyzed conversion of phosphatidylethanolamine to phosphatidylcholine can compensate for reduced dietary choline intake (38). In a clinical study, we had identified a subset of young women who were protected against CDS (relative to men and postmenopausal women) (1), and we suggested that this protection resulted from an enhanced capacity for de novo choline production by estrogen-mediated induction of PEMT (7).
In this study, we more fully identified the haploblock associated with protection from CDS and determined the mechanism for hormonal regulation of PEMT. Using a targeted (rather than a genome-wide) SNP association study, we identified two PEMT SNPs in linkage disequilibrium that characterized a haplotype associated with greatly increased risk of CDS in women (rs12325817, p = 0.000009; rs4646343, p = 0.000062). Based on the genotype determined by these SNPs, we demonstrated allele- and transcript-specific differences in hormonal responsiveness in primary human hepatocytes. Using locus-wide chromatin immunoprecipitation studies, we identified regions of estrogen receptor contact with DNA that could induce gene expression. Finally, we showed that the risk haplotype was associated with decreased ER binding compared with the same region containing the protective haplotype. ER interaction at all sites was abrogated on the risk allele. Specifically, we found that, in hepatocytes homozygous for the risk allele, ER binding to the region containing the ERE was not induced by estrogen. Interestingly, ER binding at peak V (∼6800 bp from peak IV) also was diminished in the context of the risk allele, suggesting that proximal promoter sites of ER interaction are coordinately disrupted by the risk-associated SNPs.
PEMT generates three transcripts (PEMT A, B, and C). Although transcript A constitutes nearly half of the total PEMT message in estrogen-treated hepatocytes, the development of CDS was associated with diminished PEMT B and C. In heterozygous cells, allele-specific transcript detection demonstrated greatly diminished estrogen-induced expression from the CDS risk allele in the absence of allelic compensation. Because the technique to determine allele-specific expression could not discriminate between transcript isoforms, it is likely that the abundance of transcript A (unaffected allele) masks a significantly greater reduction in transcript B and C expression (affected allele). These data highlight the importance of accounting for transcript isoforms when examining allelic imbalance and could explain the findings of a recent genome-wide association study in human liver, which failed to identify significant allelic expression imbalance associated with PEMT SNP rs897453 (27). In addition, these data suggest that either the degree of PEMT B induction is physiologically relevant or the PEMT B isoform may be acting in a fashion distinct from PEMT A. Total PEMT activity was induced in the presence of moxestrol in hepatocytes homozygous for the protective allele and was abrogated by the risk allele.
To understand why estrogen failed to activate the risk allele, we performed detailed, locus-wide chromatin immunoprecipitation. We identified several putative ER binding sites in the PEMT gene, including a cluster of sites ∼6.8 kb upstream of the TSS of the B transcript (in the first intron of the PEMT A isoform). Previous efforts to map ER binding genome-wide had not detected these sites (21, 24, 36). This difference may reflect the tissue- and/or species-specific nature of this interaction.
We resolved ER binding cluster I (Fig. 4B) into three distinct binding sites (annotated as peaks I–IV). Peak IV mediated estrogen responsiveness in a co-transfection assay. Peak IV contains a canonical ERE motif and a FOXA1 site. In addition to the peak IV canonical ERE region, other sites of ER chromatin contact were identified. In isolation, these were unable to mediate estrogen-responsive transcription, although several of these non-ERE-containing sites confer non-hormone-responsive cis-regulatory activity indicative of a functional enhancer element. These data suggest multiple contact sites for a protein complex that includes ER. Despite the proximity of the ERE to the transcriptional start site of the A transcript, this transcript was not regulated by estrogen.
Direct sequencing of the risk and protective alleles failed to demonstrate alterations of the ERE or FOXA1 recognition sites. Neither the SNP linked to CDS in our original study (rs12325817) nor the SNP identified in this study (rs4646343) resides in an ER binding region. Although it is possible that these SNPs mediate the difference in chromatin structure, there are several other SNPs in the region. Differences in inheritance and the relatively small number of subjects in our clinical study make it difficult to confidently narrow down possibilities among these potentially critical SNPs.
FOXA1, also known as HNF3α (hepatocyte nuclear factor 3α), together with FOXA2, is critical for liver development in mice (39). FOXA1 functions to open chromatin, increasing the accessibility of other transcription factors (40). Specifically, FOXA1 was recently shown to be critical in determining sites of estrogen receptor activity (41). This cooperative nature of FOXA1 and ER suggests that without FOXA1 binding to PEMT, there may be a loss of ER binding and hormonal regulation of PEMT expression. It is possible that polymorphisms in the risk allele alter the binding of FOXA1 directly or through the recruitment of other chromatin modifiers that could indirectly influence FOXA1 chromatin association.
Based on the results of our study, we predict that women harboring risk-associated SNPs abrogating hormonal regulation of PEMT will have a higher dietary requirement for choline because they cannot augment endogenous production of choline with estrogen. This haplotype could identify young women at risk for liver and muscle damage as well as for abnormal pregnancy outcomes when dietary choline is limited.
Supplementary Material
Acknowledgments
We thank Admet Technologies, who kindly provided human primary hepatocytes, and Zen-Bio, who allowed us to genotype their hepatocyte lots before purchase. We also thank Marie Fogarty, who was instrumental in optimization of allelic imbalance assays, and Chris Miller, who assisted in the development of gene copy number assays. We thank Paul Giresi and Andrew McFadden for technical assistance with ChIP-Chip studies, Wei Sun and Tom Randall for ChIP-Chip and bioinformatic data analysis, and Hye Mee Hwang for assistance with PEMT activity assays. Daniel Menendez kindly provided the pS2 luciferase construct used in control studies.
This work was supported, in whole or in part, by National Institutes of Health Grants DK55865 and AG09525 (to S. Z.) and DK56350 (to the University of North Carolina at Chapel Hill Nutrition Obesity Research Center).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- CDS
- choline deficiency syndrome
- qPCR
- quantitative PCR
- ER
- estrogen receptor
- TSS
- transcriptional start site
- ERE
- estrogen response element
- R
- homozygous for the risk allele
- P
- homozygous for the protective allele
- P/R
- heterozygous for both alleles.
REFERENCES
- 1. Fischer L. M., daCosta K. A., Kwock L., Stewart P. W., Lu T. S., Stabler S. P., Allen R. H., Zeisel S. H. (2007) Am. J. Clin. Nutr. 85, 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Z., Agellon L. B., Vance D. E. (2005) J. Biol. Chem. 280, 37798–37802 [DOI] [PubMed] [Google Scholar]
- 3. Buchman A. L., Ament M. E., Sohel M., Dubin M., Jenden D. J., Roch M., Pownall H., Farley W., Awal M., Ahn C. (2001) JPEN J. Parenter. Enteral Nutr. 25, 260–268 [DOI] [PubMed] [Google Scholar]
- 4. Zeisel S. H., Da Costa K. A., Franklin P. D., Alexander E. A., Lamont J. T., Sheard N. F., Beiser A. (1991) FASEB J. 5, 2093–2098 [PubMed] [Google Scholar]
- 5. Bremer J., Greenberg D. M. (1961) Biochim. Biophys. Acta 46, 205–216 [Google Scholar]
- 6. Ridgway N. D., Vance D. E. (1992) Methods Enzymol. 209, 366–374 [DOI] [PubMed] [Google Scholar]
- 7. Resseguie M., Song J., Niculescu M. D., da Costa K. A., Randall T. A., Zeisel S. H. (2007) FASEB J. 21, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McMahon K. E., Farrell P. M. (1985) Clin. Chim. Acta 149, 1–12 [DOI] [PubMed] [Google Scholar]
- 9. Zeisel S. H., Mar M. H., Zhou Z., da Costa K. A. (1995) J. Nutr. 125, 3049–3054 [DOI] [PubMed] [Google Scholar]
- 10. Institute of Medicine and National Academy of Sciences U.S.A. (1998) Choline: Dietary Reference Intakes for Folate, Thiamin, Riboflavin, Niacin, Vitamin B12, Panthothenic Acid, Biotin, and Choline, Vol. 1, pp. 390–422, National Academy Press, Washington, D. C. [PubMed] [Google Scholar]
- 11. Jensen H. H., Batres-Marquez S. P., Carriquiry A., Schalinske K. L. (2007) FASEB J. 21, lb219 [Google Scholar]
- 12. Shaw G. M., Carmichael S. L., Yang W., Selvin S., Schaffer D. M. (2004) Am. J. Epidemiol. 160, 102–109 [DOI] [PubMed] [Google Scholar]
- 13. Shaw G. M., Carmichael S. L., Laurent C., Rasmussen S. A. (2006) Epidemiology 17, 285–291 [DOI] [PubMed] [Google Scholar]
- 14. Shaw G. M., Finnell R. H., Blom H. J., Carmichael S. L., Vollset S. E., Yang W., Ueland P. M. (2009) Epidemiology 20, 714–719 [DOI] [PubMed] [Google Scholar]
- 15. Zhu X., Song J., Mar M. H., Edwards L. J., Zeisel S. H. (2003) Biochem. J. 370, 987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waite K. A., Cabilio N. R., Vance D. E. (2002) J. Nutr. 132, 68–71 [DOI] [PubMed] [Google Scholar]
- 17. Walkey C. J., Donohue L. R., Bronson R., Agellon L. B., Vance D. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12880–12885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song J., da Costa K. A., Fischer L. M., Kohlmeier M., Kwock L., Wang S., Zeisel S. H. (2005) FASEB J. 19, 1266–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dong H., Wang J., Li C., Hirose A., Nozaki Y., Takahashi M., Ono M., Akisawa N., Iwasaki S., Saibara T., Onishi S. (2007) J. Hepatol. 46, 915–920 [DOI] [PubMed] [Google Scholar]
- 20. da Costa K. A., Kozyreva O. G., Song J., Galanko J. A., Fischer L. M., Zeisel S. H. (2006) FASEB J. 20, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., Brown M. (2006) Nat. Genet. 38, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 22. Lee T. I., Johnstone S. E., Young R. A. (2006) Nat. Protoc. 1, 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 24. Kwon Y. S., Garcia-Bassets I., Hutt K. R., Cheng C. S., Jin M., Liu D., Benner C., Wang D., Ye Z., Bibikova M., Fan J. B., Duan L., Glass C. K., Rosenfeld M. G., Fu X. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin C. Y., Vega V. B., Thomsen J. S., Zhang T., Kong S. L., Xie M., Chiu K. P., Lipovich L., Barnett D. H., Stossi F., Yeo A., George J., Kuznetsov V. A., Lee Y. K., Charn T. H., Palanisamy N., Miller L. D., Cheung E., Katzenellenbogen B. S., Ruan Y., Bourque G., Wei C. L., Liu E. T. (2007) PLoS Genet 3, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Geen H., Nicolet C. M., Blahnik K., Green R., Farnham P. J. (2006) BioTechniques 41, 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng M., Barrera L. O., Ren B., Wu Y. N. (2007) Biometrics 63, 787–796 [DOI] [PubMed] [Google Scholar]
- 28. Rozen S., Skaletsky H. (2000) Methods Mol. Biol. 132, 365–386 [DOI] [PubMed] [Google Scholar]
- 29. Busby M. G., Fischer L., da Costa K. A., Thompson D., Mar M. H., Zeisel S. H. (2004) J. Am. Diet Assoc. 104, 1836–1845 [DOI] [PubMed] [Google Scholar]
- 30. Lo H. S., Wang Z., Hu Y., Yang H. H., Gere S., Buetow K. H., Lee M. P. (2003) Genome Res. 13, 1855–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. SAS Institute Inc. (1996) SAS/STATSoftware: Changes and Enhancements through Release 6.11, SAS Institute Inc., Cary, NC [Google Scholar]
- 32. The International HapMap Consortium (2005) Nature 437, 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., Stuve L. L., Gibbs R. A., Belmont J. W., Boudreau A., Hardenbol P., Leal S. M., Pasternak S., Wheeler D. A., Willis T. D., Yu F., Yang H., Zeng C., Gao Y., Hu H., Hu W., Li C., Lin W., Liu S., Pan H., Tang X., Wang J., Wang W., Yu J., Zhang B., Zhang Q., Zhao H., Zhao H., Zhou J., Gabriel S. B., Barry R., Blumenstiel B., Camargo A., Defelice M., Faggart M., Goyette M., Gupta S., Moore J., Nguyen H., Onofrio R. C., Parkin M., Roy J., Stahl E., Winchester E., Ziaugra L., Altshuler D., Shen Y., Yao Z., Huang W., Chu X., He Y., Jin L., Liu Y., Shen Y., Sun W., Wang H., Wang Y., Wang Y., Xiong X., Xu L., Waye M. M., Tsui S. K., Xue H., Wong J. T., Galver L. M., Fan J. B., Gunderson K., Murray S. S., Oliphant A. R., Chee M. S., Montpetit A., Chagnon F., Ferretti V., Leboeuf M., Olivier J. F., Phillips M. S., Roumy S., Sallée C., Verner A., Hudson T. J., Kwok P. Y., Cai D., Koboldt D. C., Miller R. D., Pawlikowska L., Taillon-Miller P., Xiao M., Tsui L. C., Mak W., Song Y. Q., Tam P. K., Nakamura Y., Kawaguchi T., Kitamoto T., Morizono T., Nagashima A., Ohnishi Y., Sekine A., Tanaka T., Tsunoda T., Deloukas P., Bird C. P., Delgado M., Dermitzakis E. T., Gwilliam R., Hunt S., Morrison J., Powell D., Stranger B. E., Whittaker P., Bentley D. R., Daly M. J., de Bakker P. I., Barrett J., Chretien Y. R., Maller J., McCarroll S., Patterson N., Pe'er I., Price A., Purcell S., Richter D. J., Sabeti P., Saxena R., Schaffner S. F., Sham P. C., Varilly P., Altshuler D., Stein L. D., Krishnan L., Smith A. V., Tello-Ruiz M. K., Thorisson G. A., Chakravarti A., Chen P. E., Cutler D. J., Kashuk C. S., Lin S., Abecasis G. R., Guan W., Li Y., Munro H. M., Qin Z. S., Thomas D. J., McVean G., Auton A., Bottolo L., Cardin N., Eyheramendy S., Freeman C., Marchini J., Myers S., Spencer C., Stephens M., Donnelly P., Cardon L. R., Clarke G., Evans D. M., Morris A. P., Weir B. S., Tsunoda T., Mullikin J. C., Sherry S. T., Feolo M., Skol A., Zhang H., Zeng C., Zhao H., Matsuda I., Fukushima Y., Macer D. R., Suda E., Rotimi C. N., Adebamowo C. A., Ajayi I., Aniagwu T., Marshall P. A., Nkwodimmah C., Royal C. D., Leppert M. F., Dixon M., Peiffer A., Qiu R., Kent A., Kato K., Niikawa N., Adewole I. F., Knoppers B. M., Foster M. W., Clayton E. W., Watkin J., Gibbs R. A., Belmont J. W., Muzny D., Nazareth L., Sodergren E., Weinstock G. M., Wheeler D. A., Yakub I., Gabriel S. B., Onofrio R. C., Richter D. J., Ziaugra L., Birren B. W., Daly M. J., Altshuler D., Wilson R. K., Fulton L. L., Rogers J., Burton J., Carter N. P., Clee C. M., Griffiths M., Jones M. C., McLay K., Plumb R. W., Ross M. T., Sims S. K., Willey D. L., Chen Z., Han H., Kang L., Godbout M., Wallenburg J. C., L'Archevêque P., Bellemare G., Saeki K., Wang H., An D., Fu H., Li Q., Wang Z., Wang R., Holden A. L., Brooks L. D., McEwen J. E., Guyer M. S., Wang V. O., Peterson J. L., Shi M., Spiegel J., Sung L. M., Zacharia L. F., Collins F. S., Kennedy K., Jamieson R., Stewart J. (2007) Nature 449, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mattick S., Glenn K., de Haan G., Shapiro D. J. (1997) J. Steroid Biochem. Mol. Biol. 60, 285–294 [DOI] [PubMed] [Google Scholar]
- 35. Barkhem T., Andersson-Ross C., Höglund M., Nilsson S. (1997) J. Steroid Biochem. Mol. Biol. 62, 53–64 [DOI] [PubMed] [Google Scholar]
- 36. Gao H., Fält S., Sandelin A., Gustafsson J. A., Dahlman-Wright K. (2008) Mol. Endocrinol. 22, 10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., Kloos D. U., Land S., Lewicki-Potapov B., Michael H., Münch R., Reuter I., Rotert S., Saxel H., Scheer M., Thiele S., Wingender E. (2003) Nucleic Acids Res. 31, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeisel S. H. (2006) Annu. Rev. Nutr. 26, 229–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee C. S., Friedman J. R., Fulmer J. T., Kaestner K. H. (2005) Nature 435, 944–947 [DOI] [PubMed] [Google Scholar]
- 40. Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., Zaret K. S. (2002) Mol. Cell 9, 279–289 [DOI] [PubMed] [Google Scholar]
- 41. Eeckhoute J., Lupien M., Meyer C. A., Verzi M. P., Shivdasani R. A., Liu X. S., Brown M. (2009) Genome Res. 19, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.