Abstract
Competitive inhibition of transcription factors by small proteins is an intriguing component of gene regulatory networks in both animals and plants. The small interfering proteins possess limited sequence homologies to specific transcription factors but lack one or more protein motifs required for transcription factor activities. They interfere with the activities of transcription factors, such as DNA binding and transcriptional activation, by forming nonfunctional heterodimers. A potential example is the Arabidopsis MIF1 (mini zinc finger 1) protein consisting of 101 residues. It has a zinc finger domain but lacks other protein motifs normally present in transcription factors. In this work, we show that MIF1 and its functional homologues physically interact with a group of zinc finger homeodomain (ZHD) transcription factors, such as ZHD5, that regulate floral architecture and leaf development. Gel mobility shift assays revealed that MIF1 blocks the DNA binding activity of ZHD5 homodimers by competitively forming MIF1-ZHD5 heterodimers. Accordingly, the transcriptional activation activity of ZHD5 was significantly suppressed by MIF1 coexpressed transiently in Arabidopsis protoplasts. Notably, MIF1 also prevents ZHD5 from nuclear localization. Although ZHD5 was localized exclusively in the nucleus, it was scattered throughout the cytoplasm when MIF1 was coexpressed. Transgenic plants overexpressing the ZHD5 gene (35S:ZHD5) exhibited accelerated growth with larger leaves. Consistent with the negative regulation of ZHD5 by MIF1, the 35S:ZHD5 phenotypes were diminished by MIF1 coexpression. These observations indicate that MIF1 regulates the ZHD5 activities in a dual step manner: nuclear import and DNA binding.
Keywords: Arabidopsis, DNA-binding Protein, Gene Expression, Plant, Transcription Factors, Zinc Finger
Introduction
Transcription factors are one of the most critical components of gene regulatory networks that determine the overall course of growth and development by modulating diverse cellular and physiological activities in plants. Transcription factor activities are regulated frequently at the transcriptional level. Upon stimulation by internal and external cues, a transcription factor gene is induced, and the newly synthesized transcription factor is transported into the nucleus (1). Recent studies have shown that their activities are also regulated at various steps after gene transcription through RNA and protein metabolism and intermolecular interactions. Mechanisms underlying posttranscriptional control include processing of primary transcripts and transport of mature transcripts into the cytoplasm (2), controlled RNA metabolism by microRNAs and siRNAs (3), and formation of mRNA-ribosome assemblies (4). They are further regulated at the protein level by distinct molecular and biochemical mechanisms, which include posttranslational modifications (5), nucleo-cytoplasmic transport (6), and dynamic formation of homodimers and heterodimers (7).
Controlled activation of dormant transcription factors also plays an important role in gene expression regulation. It has been shown that a small group of NAC (NAM/ATAF1/2/CUC2) and basic leucine zipper transcription factors is stored as dormant forms in association with cellular membranes, including plasma membranes, nuclear membranes, and endoplasmic reticulum membranes (8, 9). When plants are exposed to environmental stresses, the membrane-bound transcription factors are proteolytically activated by either ubiquitin-mediated proteasome activities or by specific membrane-bound proteases (10). It is now perceived that controlled proteolytic activation of membrane-bound transcription factors is a way of quick transcriptional response that ensures plant survival under stressful conditions (11, 12).
An additional, intriguing mechanism controlling transcription factor activities is dynamic formation of nonfunctional heterodimers between transcription factors and competitive peptide inhibitors. The first characterized is the ID1 (inhibitor of DNA binding 1) protein in animals (13). ID1 is a small protein having a helix-loop-helix domain but lacking the basic domain required for DNA binding. It associates with basic helix-loop-helix transcription factors, such as MyoD, E12, and E47, and inhibits their activities. A potential ID1 homologue in plants is the Arabidopsis KDR (kidari) protein. It negatively regulates a basic helix-loop-helix transcription factor HFR1 (long hypocotyl in far RED1) functioning in plant photomorphogenesis (14). It seems that KDR acts as a negative regulator by competitively forming heterodimers with HFR, although it has not yet been explored at the molecular level. Furthermore, it has been recently reported that small leucine zipper-containing proteins ZPRs (little zippers) interact with HD-ZIP III (class III homeodomain-leucine zipper) transcription factors via the ZIP motif and interfere with their DNA binding activities in Arabidopsis (15, 16).
In humans, the SHP (small heterodimer partner) protein, which mediates the HH/Gli (Hedgehog/glioma-associated oncogene homologue) signaling implicated in several malignancies (17), has a structural organization similar to those of the competitive peptide inhibitors in that it has a typical ligand binding domain like the Gli transcription factors but lacks a conventional DNA binding domain (18). However, it is distinct from the ID1 and ZPR competitors in that it suppresses the nuclear transport and transcriptional activation activity of the Gli transcription factors (17), suggesting that competitive peptide inhibitors affect diverse aspects of transcription factor activities.
Recent reexamination of the Arabidopsis genome has revealed that there are more than 3000 ORFs encoding small proteins consisting of less than 100 residues (19–21), which have not been annotated in the original prediction (Arabidopsis Genome Initiative). Notably, at least some of them encode proteins that possess limited sequence similarities to various transcription factors (22). Of particular interest are the MIF (mini zinc finger) proteins (23). The MIF proteins have zinc finger (ZF)2 motifs having sequence similarities to the ZHD transcription factors. However, the homeodomain (HD) domains are missing in the MIF proteins (24). Although it has been suggested that the MIF proteins would be a novel class of ZHD transcription factors, their protein size and distinct domain structure are strikingly similar to those of ID1, KDR, and ZPR3. A MIF homologue, IMA (inhibitor of meristem activity), has been identified in tomatoes (25).
In this work, we demonstrate that the MIF proteins physically interact with a subset of ZHD transcription factors via the ZF motif. In particular, MIF1 negatively regulates the ZHD5 activities by forming nonfunctional heterodimers. The MIF1-ZHD5 heterodimers were excluded from the nucleus. They also showed reduced transcriptional activation activity. Consistent with the negative regulation of ZHD5 by MIF1, the phenotypes of the ZHD5-overexpressing transgenic plants were rescued by MIF1 coexpression. These observations indicate that the MIF1 protein (and other MIF proteins as well) acts as a dominant negative regulator of the ZHD transcription factors, extending the repertoire of competitive peptide inhibitors in plants. We propose that competitive peptide inhibitor-mediated transcriptional control is a genome-wide regulatory scheme.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used were in the Columbia (Col-0) background. Plants were grown in a controlled culture room set at 22 °C with a relative humidity of 50% under long day conditions (16 h light/8 h dark). White light illumination (120 μmol/m2 s) was provided by FLR40D/A fluorescent tubes (Osram, Seoul, Korea). The 35S:MIF1 transgenic plants were produced by expressing the MIF1 gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter in the pK7WG2D Gateway vector (26). The 35S:ZHD5 transgenic plants were similarly produced but using the pB2GW7 Gateway vector. The 35S:MIF1 transgenic plants were genetically crossed with the 35S:ZHD5 transgenic plants, producing the MIF1xZHD5 plants.
Analysis of Transcript Levels
Total RNA extraction, reverse transcription, and quantitative real-time RT-PCR (qRT-PCR) were carried out according to the rules that have recently been proposed to ensure reproducible and accurate measurements (27, 28). RNA samples were pretreated extensively with RNase-free DNAse I to get rid of contaminating genomic DNA before use (29). The qRT-PCR primers used are listed in supplemental Table S1.
qRT-PCR was carried out in 96-well blocks with the Applied Biosystems 7500 real-time PCR system using SYBR Green I master mix (Kapa Biosystems, Woburn, MA) in a volume of 20 μl. The primers were designed using the Primer Express software installed into the system. The two-step thermal cycling profile used was 15 s at 94 °C and 1 min at 68 °C. An eIF4A gene (At3g13920) was included in the reactions as an internal control to normalize variations in cDNA amounts used. The qRT-PCRs were carried out in biological triplicates using total RNA samples extracted from three independent plant materials grown under identical growth conditions in individual assays. The comparative ΔΔCT method was used to evaluate relative quantities of each amplified product. The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. PCR specificity was determined by melt curve analysis of amplified products using the standard method installed into the system.
Yeast Two-hybrid Assays
Yeast two-hybrid assays were carried out using the GAL4-based Match Maker two-hybrid system (Clontech, Palo Alto, CA). The yeast strain used was AH109 (MATα trp1 leu2). The vectors used were pGADT7 (activation domain fusion, prey) and pGBKT7 (binding domain fusion, bait). The MIF and ZHD cDNAs were PCR-amplified using gene-specific primer sets (supplemental Table S2) and subcloned into the GAL4 activation domain and binding domain vectors. The bait-prey pairs were cotransformed into yeast cells. To eliminate false positives, the colonies obtained from the cotransformation were confirmed by separate dropping onto SD dropout medium and analyzing the activation of the second reporter gene (lacZ) by β-galactosidase filter lift assays (30). β-Galactosidase activities were also measured by a quantitative liquid culture method (31).
Production of Recombinant Proteins
The pET41a (+) vector containing a glutathione S-transferase (GST) gene fusion and the modified pMAL-c2X Gateway vector containing a maltose-binding protein (MBP) gene fusion were transformed into Escherichia coli Rosetta 2 (D3) cells (Novagen, Darmstadt, Germany). Gene expression was induced by 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 20 °C overnight. Processing of bacterial cells and purification of recombinant proteins were carried out according to the manufacturer's procedure. The purity and quantity of recombinant proteins were determined before use by analyzing on a 12% SDS-PAGE and staining with Coomassie Brilliant Blue R-250.
In Vitro Pull-down Assays
The [35S]methionine-labeled ZHD5 and MIF1 proteins were produced by in vitro translation using the TNT SP6 wheat germ extract-coupled system (Promega, Madison, WI). For in vitro protein-protein interaction assays, 4 μl of the 35S-labeled proteins was incubated with 3 μg of purified GST fusion proteins bound to glutathione-Sepharose beads in 1 ml of buffer A (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, protease inhibitor mixture (Roche Applied Science), and 1 mm PMSF) overnight at 4 °C. The beads were washed five times, each time with 1 ml of ice-cold buffer A containing 0.1% Triton X-100. Bound proteins were eluted with an equal volume of SDS-PAGE loading buffer, boiled for 3 min, resolved on a 12% SDS-PAGE, and subject to autoradiography.
Electrophoretic Mobility Shift Assay (EMSA), EMSA-Western, and Western Blot Analysis
EMSA was performed as described previously (32) using 10 μg of protein and 32P-labeled double-stranded oligonucleotides (33) (see supplemental Table S1).
EMSA-Western analyses were carried out essentially as described previously (34). A “5×” EMSA was run on an 8% nondenaturing polyacrylamide gel. The bands of interest were excised, and proteins were eluted overnight in 200 μl of 2× SDS-PAGE loading buffer at 37 °C. The protein samples were boiled for 5 min before loading on a 12% SDS-PAGE for Western blot analysis.
For Western blot analysis following EMSA, which was performed with an unlabeled oligonucleotide probe, the gel was blotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). A monoclonal anti-MBP antibody conjugated with horseradish peroxidase (HRP; ab49923, Abcam, Cambridge, UK) was used at a 1:10,000 dilution for detection of the ZHD5-MBP protein. For detection of the MIF1-GST protein, a polyclonal anti-GST antibody was used at a 1:10,000 dilution. The secondary antibody used was HRP-conjugated goat anti-rabbit IgG (NA934VS, Amersham Biosciences).
Transient Expression in Arabidopsis Protoplasts
A canonical binding sequence (NNATTA) of the HD domain-containing proteins (35) was fused to the minimal 35S promoter-GUS (β-glucuronidase) reporter gene cassette, in which four tandem repeats of the ATTA sequences were fused to the GUS-coding sequence (36). The effector vector contained a full-size MIF1 or ZHD5 gene driven by the CaMV 35S promoter. Preparation of Arabidopsis mesophyll protoplasts and polyethylene glycol-mediated transformation were carried out as described previously (37). For each transformation, an Arabidopsis protoplast suspension (2 × 106 cells/ml) was transformed with 5 μg of the reporter construct alone or together with the effector construct or a vector control (pCAMBIA 1304). Transformed protoplasts were incubated in the dark for 16 h. GUS activities were measured fluorometrically using 4-methylumbelliferyl-β-d-glucuronide as substrate (36).
Luciferase activity assays were carried out using the Promega luciferase assay system. To normalize transformation efficiencies, the control vector pJD300 containing the CaMV 35S promoter-luciferase cassette was cotransformed with the reporter vector, as described previously (38).
Subcellular Localization Assays
Full-size ZHD5 and MIF1 cDNAs were fused in frame to the 3′-end of the green fluorescent protein (GFP)-coding sequence in the p2FGW7 vector (26). Similarly, a full-size MIF1 cDNA was fused in frame to the 5′-end of the red fluorescent protein (RFP)-coding sequence in the 326-RFP vector (39).
The pSAT vectors, which were used for bimolecular fluorescence complementation (BiFc) assays (40), were kindly provided by Stanton Gelvin (Purdue University). The ZHD5 and MIF1 cDNAs were fused in frame to the 5′-end of a DNA sequence encoding the N-terminal half of EYFP in the pSATN-nEYFP-C1 vector (E3081) and to the 3′-end of a DNA sequence encoding the C-terminal half of EYFP in the pSATN-cEYFP-C1 vector (E3082), respectively.
The expression constructs were transformed into Arabidopsis protoplasts. Expression of the fusion constructs was examined 24 h after transformation by fluorescence microscopy using a Zeiss LSM510 confocal microscope (Carl Zeiss, Yena, Germany). GFP, RFP, and YFP samples were excited with 488-, 543-, and 514-nm argon laser lines, respectively, with an emission band of 500–530 nm for GFP detection, 565–615 nm for RFP detection, 535–590 nm for YFP detection, and 690–730 nm for chlorophyll autofluorescence.
For transient expression in onion epidermal cells, the p2FGW7 vector having a ZHD5-GFP fusion or a MIF1-GFP fusion and the pSATN-ZHD5-nEYFP-C1 (ZHD5-E3081) and pSATN-MIF1-cEYFP-C1 (MIF1-E3082) vectors were introduced into onion epidermal cells by particle bombardment as described previously (41). After incubation for 20 h at 23 °C, the cells were mounted in phosphate-buffered saline containing 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). The DAPI-stained cells were visualized by bright field and fluorescence microscopy.
Scanning Electron Microscopy
Appropriate plant materials were fixed in fixation solution (2% paraformaldehyde in 25 mm phosphate buffer, pH 7.0) at 4 °C for 24 h. The samples were subsequently incubated in 1% osmium tetroxide in 25 mm phosphate buffer, pH 7.0, at 4 °C for 2 h. They were washed with a series of ethanol dilutions (30, 50, 70, 80, 90, 95, and 100%), each for 10 min. After soaking in 100% isoamyl acetate for 15 min, the samples were subjected to critical point drying, coating with gold, and scanning electron microscopy (SEM) using the JSM 5410LV model (JEOL, Tokyo, Japan).
To examine leaf surfaces, the medial regions in the nonmarginal part of leaves were photographed using the computer image analysis software AnalySIS 2.1 (Soft-Imaging Software GmbH). Cell sizes were measured using the Labwork Image Acquisition and Analysis software (Media Cybernetics, San Diego, CA).
RESULTS
Interaction of MIF Proteins with ZHD Transcription Factors
The MIF proteins have limited sequence similarities to the ZHD transcription factors (23). However, they are structurally distinct from the ZHD proteins. Whereas both have the ZF motifs that mediate protein-protein interactions (42), the MIF proteins lack the HD domains that are required for DNA-protein interactions in many DNA-binding proteins (43). These structural characteristics are analogous to those of the ID1, KDR, and ZPR proteins, which act as competitive inhibitors of the basic leucine zipper and HD-ZIP III transcription factors (13, 15, 16). It was therefore hypothesized that the MIF proteins would be functionally similar to known competitive peptide inhibitors.
We first examined whether the MIF proteins interact with the ZHD proteins by yeast two-hybrid assays using the MIF proteins as baits and the ZHD proteins as preys. We determined the relative strengths of the interactions by measuring cell densities. As inferred from the protein structural analysis, the MIF proteins interacted with a subset of the ZHD proteins. Notably, the interaction patterns were quite diverse. The MIF proteins interacted strongly with ZHD5, ZHD8, ZHD10, and ZHD13 (Fig. 1). In contrast, they did not exhibit any discernible interactions with ZHD4, ZHD11, ZHD12, and ZHD14. Although MIF2 and MIF3 interacted with ZHD3, MIF1 did not. In addition, only MIF3 showed measurable interaction with ZHD9.
FIGURE 1.
Yeast two-hybrid assays on the MIF-ZHD interactions. The MIF-ZHD interactions were examined by yeast two-hybrid assays. Relative strengths of the interactions are indicated. +++, very strong; ++, moderate; +, weak; −, no interaction.
The MIF-ZHD interactions were also examined by measuring β-galactosidase activities in yeast cells expressing different combinations of MIF and ZHD genes. The overall patterns of interactions were quite similar to those obtained by measuring cell growth on selective medium (supplemental Fig. S1). One distinction was the high β-galactosidase activity in yeast cells expressing the MIF1 and ZHD3 genes, which was in contrast to the absence of interaction in the measurements of cell growth on selective medium (Fig. 1). This may be because the MIF1-ZHD3 dimers somehow repress yeast cell growth.
Interaction of MIF1 with ZHD5 via the ZIF Motif
The MIF1 protein has been molecular genetically characterized, and its role in growth hormone signaling has been examined through transgenic approaches and genome-wide expression studies (23). Our data indicated that ZHD5 strongly interacts with MIF2 and MIF3 as well as MIF1 (Fig. 1 and supplemental Fig. S1). Therefore, the MIF1 and ZHD5 proteins were chosen for further analysis.
The MIF1-ZHD5 interactions were further examined by in vitro pull-down assays using a recombinant MIF1-GST fusion protein prepared in E. coli cells and a [35S]methionine-labeled in vitro translation product of the ZHD5 gene. The results showed that ZHD5 bound strongly to MIF1 (Fig. 2A). In contrast, it did not bind to GST, supporting the specific interaction between MIF1 and ZHD5. To verify the MIF1-ZHD5 interaction, a [35S]methionine-labeled MIF1 peptide was produced by in vitro translation, and the ZHD5 protein was produced as a recombinant GST fusion in E. coli cells. In vitro pull-down assays showed that the MIF1 protein bound strongly to the ZHD5-GST fusion (Fig. 2B). However, no visible binding was observed between MIF1 and GST. Together, these observations demonstrate that the MIF1 protein interacts physically with the ZHD5 transcription factor.
FIGURE 2.
Interaction between MIF1 and ZHD5 via the ZF motif. For the in vitro pull-down assays, the MIF1 and ZHD5 proteins were prepared either as recombinant GST fusions in E. coli cells or as [35S]methionine-labeled in vitro translation products. The GST protein was also included in the assays. − lanes, no addition of [35S]methionine-labeled proteins. The bound proteins were eluted using SDS-PAGE loading buffer and resolved on a 12% SDS-PAGE. One μl of the in vitro translation reaction mixture (50 μl) was loaded (Input). In A, B, and D, part of the Coomassie Blue-stained gel is displayed as a loading control (lower panels). A, in vitro pull-down assays using recombinant MIF1-GST fusion protein and [35S]methionine-labeled ZHD5. B, in vitro pull-down assays using recombinant ZHD5-GST fusion protein and [35S]methionine-labeled MIF1. C, mapping of the interacting domain. A full-size (5-F) and a series of truncated (5-1 to 5-4) ZHD5 forms (top) were produced by in vitro translation. Ten percent of each reaction was loaded on the SDS-PAGE (bottom). The plus and minus symbols indicate either positive interaction (+) or no visible interaction (−), as assayed in D. D, in vitro pull-down assays using a MIF1-GST fusion protein and [35S]methionine-labeled ZHD5 proteins. SM, size marker.
The ZHD proteins form homodimers and/or heterodimers (44). Our data showed that the MIF1 protein bound to the ZHD5 protein, suggesting that the MIF1 protein would also form homodimers. To examine this possibility, we performed in vitro pull-down assays using a [35S]methionine-labeled, in vitro translation product of the MIF1 gene and a recombinant MIF1-GST fusion. As expected, the MIF1 protein bound to the MIF1-GST fusion (supplemental Fig. S2), demonstrating that the MIF1 protein forms homodimers as well as heterodimers with the ZHD proteins. We also examined whether the ZHD5 protein forms homodimers by in vitro pull-down assays using a [35S]methionine-labeled ZHD5 and a recombinant ZHD5-GST fusion. We found that the ZHD5 protein also forms homodimers (supplemental Fig. S3), as has been suggested previously (44). No interaction was detected between ZHD5 and GST.
It was observed that the MIF1 and ZHD5 proteins form heterodimers via the ZF motifs. To map the interacting domain, we produced a series of truncated ZHD5 proteins by in vitro translation (Fig. 2C) and carried out in vitro pull-down assays using a recombinant MIF1-GST fusion protein. The MIF1-GST protein bound specifically to the full-size ZHD5 form (5-F) and a truncated form (5-2) having the ZF motif (Fig. 2D). In contrast, the GST protein did not interact with any of the ZHD5 forms (supplemental Fig. S4). These observations indicate that the ZF motif is responsible for the MIF1-ZHD5 interaction.
Inhibition of DNA Binding of ZHD5 by MIF1
It has been recently reported that the ZHD5 protein (44), and perhaps other ZHD proteins as well (45, 46), binds to a 20-nucleotide sequence (AGTGTCTTGTAATTAAAA). The ATTA core sequence is a canonical binding site for many HD domain-containing proteins (47).
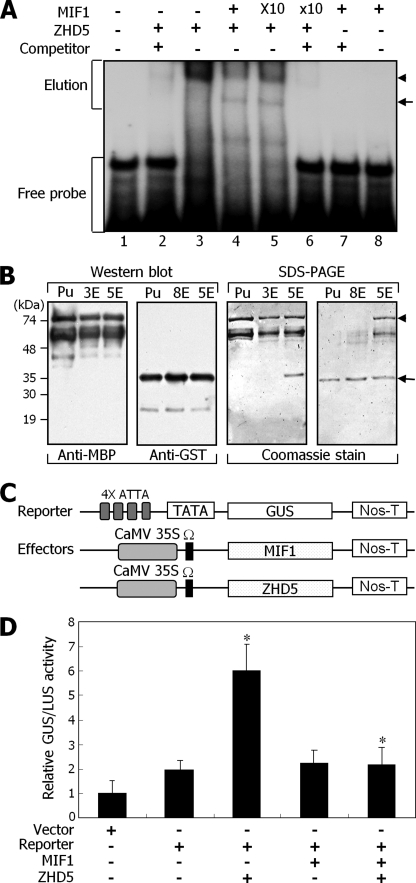
To examine whether the MIF1 protein influences the DNA binding activity of ZHD5, EMSAs were conducted using recombinant MIF1-GST and ZHD5-MBP fusion proteins and a 32P-labeled DNA fragment containing the ATTA core sequence. The ZHD5-MBP fusion protein bound to the DNA fragment possibly as dimers, but the binding disappeared in the presence of an excess amount (100-fold) of competitor DNA (Fig. 3A, lanes 2 and 3, and supplemental Fig. S6, left). Western blot analysis following EMSA (48) revealed that the ZHD5 protein bound to the DNA fragment primarily as dimers, although ZHD5 monomers also bound to the DNA fragment (supplemental Fig. S6).
FIGURE 3.
Inhibition of DNA binding and transcriptional activation activities of ZHD5 by MIF1. A, EMSA. Recombinant MIF1 and ZHD5 proteins were prepared as GST and MBP fusions, respectively, in E. coli cells (see supplemental Fig. S5). Increasing amounts of MIF1 were added to the assays. Excess amounts of competitor DNA (×100) were included in the assays. The arrowhead indicates ZHD5 homodimer (lane 3) or ZHD5-MIF1 heterodimer (lanes 4 and 5). The arrow indicates ZHD5 monomer. B, Western blot and SDS-PAGE analyses of proteins eluted from EMSA-shifted bands. Pu, purified ZHD5-MBP or MIF1-GST fusions from E. coli cells; 3E, 5E, and 8E, proteins eluted from parts, as marked by Elution, of lanes 3, 5, and 8, respectively, of the EMSA gel in A. Western blot analyses were carried out using a monoclonal anti-MBP or a polyclonal anti-GST antibody. The arrowhead indicates ZHD5-MBP, and the arrow indicates MIF1-GST. C, schematic representation of the effector and reporter constructs used. Ω, translation enhancer. Nos-T, Nos terminator. D, transient expression assays in Arabidopsis protoplasts. The empty vector without cDNA insert was included as control. For normalization of transformation efficiencies, the CaMV 35S promoter-luciferase vector was cotransformed in each assay. Three independent measurements were averaged and statistically treated using Student's t test (*, p < 0.01). Error bars, S.E.
However, the MBP protein alone did not bind to the DNA fragment under identical assay conditions (data not shown), confirming that the ZHD5 protein binds specifically to the ATTA-containing DNA fragment. The MIF1-GST fusion protein did not show any detectable binding to the DNA fragment, which is consistent with the lack of DNA binding domain in the MIF1 protein (Fig. 3A, lanes 7 and 8, respectively).
We next included the MIF1-GST fusion protein in the DNA binding assays of the ZHD5-MBP fusion protein. The results showed that binding of the ZHD5-MBP homodimers to the DNA fragment decreased in the presence of the MIF1-GST fusion protein in a dose-dependent manner (Fig. 3A, lanes 4 and 5), indicating that the MIF1 protein prevents the ZHD5 homodimers from DNA binding.
To examine whether the ZHD5-MIF1 heterodimers are formed in the EMSA, proteins were eluted from the shifted bands (Fig. 3A, lane 5) and subjected to Western blot analysis using an anti-MBP or an anti-GST antibody. The results showed that the protein complexes bound to DNA contained both the ZHD5 and MIF1 proteins (Fig. 3B), indicating that the ZHD5-MIF1 heterodimers bind to the DNA fragment but with a significantly reduced signal intensity of the DNA-protein complex. This may be related to the previous observation (44), in which it has been found that the ZHD proteins form various heterodimers more readily than homodimers (see “Discussion”).
A question was whether the MIF1 protein influences the transcriptional activation activity of the ZHD5 homodimers and whether the ZHD5-MIF1 heterodimers possess the transcriptional activation activity. To answer the question, leaf mesophyll protoplasts were prepared from Arabidopsis plants harboring a GUS reporter vector, in which a fusion of the GUS gene with four tandem repeats of the ATTA sequence was subcloned upstream of the CaMV 35S minimal promoter (36). A couple of effector vectors were constructed by subcloning the MIF1 or ZHD5 cDNA sequence under the control of the CaMV 35S promoter (Fig. 3C) and transformed into the mesophyll protoplasts. GUS activity assays revealed that expression of the ZHD5 protein increased the GUS activity ∼6-fold (Fig. 3D). In contrast, expression of the MIF1 protein did not have any discernible effects on the GUS activity. When the ZHD5 and MIF1 proteins were coexpressed, the GUS activity decreased to a basal level. These observations indicate that the MIF1 protein inhibits the transcriptional activation activity of the ZHD5 protein. It is also clear that the ZHD5-MIF1 heterodimers are not transcriptionally active.
Modulation of Subcellular Localization of ZHD5 by MIF1
Our data showed that ZHD5 is a transcriptional activator, suggesting that it is a nuclear protein. It was therefore hypothesized that the MIF1 protein may also affect the subcellular localization of ZHD5.
We examined the subcellular localization of ZHD5 using a ZHD5-GFP fusion protein in Arabidopsis protoplasts. The ZHD5-GFP protein was localized predominantly into the nucleus (Fig. 4A). Nuclear localization of the ZHD5-GFP protein was also confirmed by transient expression in onion epidermal cells and DAPI staining (supplemental Fig. S7). We next determined the subcellular localization of MIF1. A MIF1-RFP gene fusion was expressed transiently in Arabidopsis protoplasts. The RFP signal was detected in small vesicle-like structures in the cytoplasm (Fig. 4B), although the identity of the small vesicle-like structures is unknown (49). Weak GFP signal was also detected in the nuclei of a few cells examined (supplemental Fig. S7).
FIGURE 4.
Modulation of the subcellular localization of ZHD5 by MIF1 in Arabidopsis protoplasts. Arabidopsis mesophyll protoplasts were transformed with the indicated constructs, such as ZHD5-GFP (A), MIF1-RFP (B), GFP vector only (C), ZHD5-GFP + MIF1-RFP (D–F), and ZHD5-nYFP + MIF1-cYFP (G–I), and visualized by fluorescence microscopy 24 h after transformation. D and E show the GFP and RFP signals of coexpressed ZHD5-GFP and MIF1-RFP proteins, respectively. G, image of the reconstructed YFP signals (BiFc signals). H, autofluorescence image. F and I, superimposed images.
We next examined whether the MIF1 protein influences the subcellular localization of the ZHD5 protein by employing two different approaches; one is coexpression of the ZHD5-GFP and MIF1-RFP fusions, and the other is reconstitution of YFP signals by BiFc. We first coexpressed the ZHD5-GFP and MIF1-RFP fusions transiently in Arabidopsis protoplasts and visualized both fluorescence markers simultaneously by confocal fluorescence microscopy. Both the GFP and RFP signals were distributed in small vesicle-like structures in the cytoplasm (Fig. 4, D and E), a pattern similar to that of the MIF1-RFP distribution, showing that the subcellular localization of the ZHD5 protein is altered by the MIF1 protein (Fig. 4F).
Image analysis of reconstituted YFP signals (BiFc signals) was also carried out to examine the effects of the MIF1 protein on the subcellular localization of the ZHD5 protein. The BiFc assay involves the reconstitution of YFP fluorescence upon association of the nonfluorescent N-terminal and C-terminal fragments of YFP (50). The ZHD5 protein was fused to the N-terminal fragment of YFP (ZHD5-nYFP), and the MIF1 protein was fused to the C-terminal fragment of YFP (MIF1-cYFP). We observed that ZHD5 forms a BiFc complex with MIF1 (Fig. 4, G–I, and supplemental Fig. S7), indicating that the ZHD5-MIF1 complexes readily form in vivo. Furthermore, the YFP fluorescence was detected in small vesicle-like structures, as observed in the coexpression assays. These observations further confirm the interaction of the MIF1 protein with the ZHD5 protein and demonstrate that the MIF1 protein inhibits the nuclear import of the ZHD5 protein.
Inhibition of ZHD5 Functions by MIF1 in Planta
Our data indicated that the MIF1 protein inhibits DNA binding and nuclear import of the ZHD5 protein. We therefore investigated whether the MIF1 protein inhibits the ZHD5 function in planta. We produced Arabidopsis transgenic plants overexpressing the MIF1 and ZHD5 genes driven by the CaMV 35S promoter.
The 35S:MIF1 transgenic plants exhibited dwarfed growth with small, dark green leaves (Fig. 5A). In addition, the leaf surface was shiny, as has been observed previously (23). Transgenic plants overexpressing the ZHD5 gene (35S:ZHD5) exhibited accelerated growth with larger leaves, which is in inverse to the 35S:MIF1 phenotypes. The 35S:MIF1 transgenic plants were then genetically crossed with the 35S:ZHD5 transgenic plants, resulting in the MIF1xZHD5 plants. qRT-PCR assays of gene transcript levels revealed that overexpression of the MIF1 gene did not influence the ZHD5 gene transcription and vice versa (Fig. 5B), excluding the possibility of MIF1-ZHD5 interactions at the transcriptional level.
FIGURE 5.
Phenotypic recovery of transgenic plants overexpressing ZHD5 by MIF1 coexpression. A, phenotypes of transgenic plants. The MIF1 or ZHD5 gene was overexpressed under the control of the CaMV 35S promoter. Five-week-old transgenic plants grown in soil were photographed. The MIF1xZHD5 plants were produced by genetic cross of the 35S:MIF1 and 35S:ZHD5 transgenic plants. The inlet shows an enlarged view of the 35S:MIF1 transgenic plant. B, expression of the MIF1 and ZHD5 genes in the transgenic plants. Transcript levels were determined by qRT-PCR using total RNA samples extracted from 2-week-old, whole plants grown on MS-agar plates. Biological triplicates were averaged. Error bars, S.E. Statistical significance was determined using Student's t test (*, p < 0.01). C, measurements of plant heights. Thirty plants fully grown in soil were measured and averaged (t test; *, p < 0.01). D, measurements of leaf areas. A series of rosette leaves from representative plants were photographed (left). Leaf areas were measured using the eighth rosette leaves of 20 plants and averaged for each plant group (right). Error bars, S.E. (t test; *, p < 0.01). E, cell sizes. Sizes of the cells from the adaxial sides of leaves were compared by SEM and the Labwork image acquisition and analysis program. Statistical significance of the cell size measurements was determined by Student's t test (p < 0.01). Scale bars, 50 μm. F, floral structures. Floral structures were compared by SEM. Parts of sepals and petals were removed to visualize internal structures. Scale bars, 500 μm.
The overall phenotypes of the MIF1xZHD5 plants resembled those of wild-type (Col-0) plants (Fig. 5A). The height of the MIF1xZHD5 plants was also close to that of wild-type plants (Fig. 5C). The MIF1xZHD5 leaves were somewhat smaller than wild-type and 35S:ZHD5 transgenic leaves but much larger than the 35S:MIF1 transgenic leaves (Fig. 5D). We also examined the leaf epidermal cells by SEM. Although the epidermal cells of the 35S:MIF1 transgenic leaves were smaller than those of the wild-type leaves, they were larger in the 35S:ZHD5 transgenic leaves (Fig. 5E). Notably, the epidermal cells of the MIF1xZHD5 leaves were only slightly larger than those of the wild-type leaves. These observations indicate that the 35S:ZHD5 phenotypes were efficiently rescued by MIF1 expression, further supporting the MIF1 regulation of the ZHD5 activities.
The 35S:MIF1 transgenic plants had distorted floral structures, such as short, crooked filaments and enlarged, twisted gynoecium (Fig. 5F). In contrast, the reproductive organs were essentially normal in the 35S:ZHD5 transgenic plants. The MIF1×ZHD5 plants also had normal floral structures. The morphological recovery of floral structures in the MIF1×ZHD5 plants would be explained by the suppression of the MIF1 effects by the ZHD5 protein through protein-protein interaction.
Taken together, our data demonstrate that the MIF1 protein serves as a negative regulator of the ZHD5 transcription factor by forming nonfunctional heterodimers (Fig. 6). The MIF1 regulation of the ZHD5 activities occurs both in the cytoplasm and nucleus. Whereas the MIF1 protein blocks the nuclear import of the ZHD5 protein in the cytoplasm, it inhibits the DNA binding activity of the ZHD5 homodimers in the nucleus. This view is also consistent with the distribution of the MIF1 protein both in the cytoplasm and nucleus.
FIGURE 6.
Schematic working model of MIF1. The MIF1 protein inhibits nuclear localization of the ZHD5 protein by forming MIF1-ZHD5 heterodimers in the cytoplasm. In the nucleus, the MIF1-ZHD5 heterodimers have a lower DNA binding activity compared with that of the ZHD5 homodimers.
DISCUSSION
We here demonstrated that a small Arabidopsis protein, MIF1, having a ZF motif modulates nuclear import and DNA binding of the ZHD5 transcription factor by forming nonfunctional heterodimers. Biochemical assays and transgenic studies support that the MIF1 protein acts as a dominant negative regulator of ZHD5 and possibly other ZHD transcription factors.
Since the first identification as potential regulators of the C4 phosphoenolpyruvate carboxylase functioning in plant responses to environmental stresses (51), diverse roles of the plant-specific ZHD transcription factors have been proven in growth hormone signaling (52), adaptive responses to environmental stresses (53, 54), pathogen-derived signaling processes (55), and inflorescence stem growth (56). There are 14 ZHD members in Arabidopsis (24, 44). The ZHD proteins form homodimers and a heterodimer (44). We found that MIF1 and its functional homologues MIF2 and MIF3 exhibit a dynamic interaction pattern with a subset of the ZHD transcription factors. This entails that the MIF proteins would be related with a wide array of plant responses to developmental and environmental cues. This view explains the pleiotropic phenotypes of the transgenic plants overexpressing the MIF1 gene in Arabidopsis (23).
We found that the MIF1 protein interacts with the ZHD5 protein via the ZF motif. It has been shown that the ZF motif is necessary and sufficient for the dimer formation of the ZHD proteins (51). It is therefore expected that the MIF1-ZHD5 heterodimer formation competes with the ZHD5-ZHD5 homodimer formation in plant cells, providing a clue as to how the MIF1 protein regulates the activity of the ZHD5 transcription factor. Our observations indicate that the MIF1 protein interferes with the nuclear localization or promotes the nuclear exclusion of the ZHD5 protein in the cytoplasm and suppresses the DNA binding activity of the ZHD5 homodimers in the nucleus. This is also in agreement with the subcellular localization of the MIF1 protein.
Controlled nuclear localization is a well recognized mechanism regulating the activities of various transcription factors and other regulatory proteins in eukaryotes (57, 58). In many cases, nuclear import of transcription factors is regulated by posttranslational modifications, such as protein phosphorylation (59), and degradation of docking proteins (60). It is therefore possible that protein phosphorylation is involved in the subcellular localization of the ZHD5 protein. The MIF1 binding would influence the propensity of the ZHD5 phosphorylation by inducing conformational changes, resulting in disruption of the interaction of the ZHD5 protein with nuclear import and/or export machinery. A recent study has shown that association of a 14-3-3 protein with the basic leucine zipper transcriptional activator RSG (repression of shoot growth) is responsible for the cytoplasmic localization of RSG (61). Similarly, Arabidopsis ovate proteins regulate the subcellular localization of HD-containing transcription factors (62).
The results obtained from EMSAs indicate that the MIF1 protein inhibits the DNA binding of the ZHD5 homodimers. Dimer formation of transcription factors is a way of enhancing specific DNA binding activities (63–65). Interestingly, the MIF1-ZHD5 heterodimers possess DNA binding capacity, unlike what has been observed with the ID1 protein (13). However, the MIF1-ZHD5 heterodimers are transcriptionally inactive, as revealed by the transient expression assays in Arabidopsis protoplasts. A plausible explanation would be that the MIF1 protein inhibits the ZHD5 activity by forming transcriptionally inactive MIF1-ZHD5 heterodimers, which also compete with the ZHD5 homodimers for DNA binding.
We found that the MIF1 protein interacts with ZHD1, ZHD6, ZHD7, ZHD8, ZHD10, and ZHD13 in addition to ZHD5. A question is whether the MIF1 protein inhibits the activities of other ZHD transcription factors in a similar manner. It is possible that whereas the MIF1 protein and it functional homologues MIF2 and MIF3 may inhibit the activity of some ZHD members by blocking their DNA binding activities, they may interfere with the activity of other ZHD members by modulating their nuclear localization. It has not been examined how other competitive peptide inhibitors, such as ID1, KDR, and ZPR, regulate their target transcription factors. It will be interesting to investigate whether other competitive peptide inhibitors act in a similar manner as observed with the MIF1 protein.
We report here that at least three small proteins, MIF1, MIF2, and MIF3, act as dominant negative regulators of the ZHD transcription factors in Arabidopsis. A phylogenetic analysis of the ZHD and MIF proteins has suggested that the MIF genes might be derived from a ZHD gene, possibly by premature termination of the ZHD gene transcription (24). At least three ZPR proteins and six KDR proteins have been shown or proposed to function in a similar manner as do the MIF proteins in plants (14–16, 66). A small group of ID1 proteins have also been reported in animals (67, 68). A genome-scale screening has shown that there are up to 100 small proteins, consisting of 120 or fewer amino acids, which have a structural organization similar to those of the ZPR, KDR, and MIF proteins (22). We therefore propose that regulation of transcription factor activities by small competitive peptides has been adopted as a transcriptional control mechanism functioning broadly in eukaryotic genomes.
Supplementary Material
Acknowledgments
We are grateful to Dr. Inhwan Hwang (POSTECH) for the p326-RFP vector and to Dr. Stanton Gelvin (Purdue University) for the BiFc vectors. We thank Ms. Kyung-A Ryu (Department of Biological Sciences, Seoul National University) for her help with confocal microscopic analysis. We also thank the National Instrumentation Center for Environmental Management (NICEM), College of Agriculture and Life Sciences and Seoul National University for electron microscopic analysis of plant materials.
This work was supported by Brain Korea 21 Biogreen 21 Grant 20080401034001 and National Research Laboratory Programs and by grants from the Plant Signaling Network Research Center, the National Research Foundation of Korea (2009-0087317 and 2007-03415), and the Agricultural R & D Promotion Center (309017-5), Korea Ministry for Food, Agriculture, Forestry, and Fisheries.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S7.
- ZF
- zinc finger
- AD
- activation domain
- BD
- binding domain
- bHLH
- basic helix-loop-helix
- BiFc
- bimolecular fluorescence complementation
- bZIP
- basic leucine zipper
- CaMV
- cauliflower mosaic virus
- HD
- homeodomain
- HD-ZIP III
- class III homeodomain-leucine zipper
- MTF
- membrane-bound transcription factor
- RSG
- Repression of Shoot Growth
- ZHD
- zinc finger-homeodomain.
REFERENCES
- 1. Whiteside S. T., Goodbourn S. (1993) J. Cell Sci. 104, 949–955 [DOI] [PubMed] [Google Scholar]
- 2. Resnekov O., von Gabain A. (eds) (1996) Cell Biology, NATO ASI Series, p. 267, Springer, New York [Google Scholar]
- 3. Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. (2006) Genes Dev. 20, 515–524 [DOI] [PubMed] [Google Scholar]
- 4. Sommer P., Nehrbass U. (2005) Curr. Opin. Cell Biol. 17, 294–301 [DOI] [PubMed] [Google Scholar]
- 5. Tootle T. L., Rebay I. (2005) BioEssays 27, 285–298 [DOI] [PubMed] [Google Scholar]
- 6. Ziegler E. C., Ghosh S. (2005) Sci. STKE 2005, re6. [DOI] [PubMed] [Google Scholar]
- 7. Lamb P., McKnight S. L. (1991) Trends Biochem. Sci. 16, 417–422 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y. N., Slabaugh E., Brandizzi F. (2008) Curr. Opin. Plant Biol. 11, 695–701 [DOI] [PubMed] [Google Scholar]
- 9. Seo P. J., Kim S. G., Park C. M. (2008) Trends Plant Sci. 13, 550–556 [DOI] [PubMed] [Google Scholar]
- 10. Hoppe T., Rape M., Jentsch S. (2001) Curr. Opin. Cell Biol. 13, 344–348 [DOI] [PubMed] [Google Scholar]
- 11. Kim Y. S., Kim S. G., Park J. E., Park H. Y., Lim M. H., Chua N. H., Park C. M. (2006) Plant Cell 18, 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seo P. J., Kim M. J., Song J. S., Kim Y. S., Kim H. J., Park C. M. (2010) Biochem. J. 427, 359–367 [DOI] [PubMed] [Google Scholar]
- 13. Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. (1990) Cell 61, 49–59 [DOI] [PubMed] [Google Scholar]
- 14. Hyun Y., Lee I. (2006) Plant Mol. Biol. 61, 283–296 [DOI] [PubMed] [Google Scholar]
- 15. Wenkel S., Emery J., Hou B. H., Evans M. M., Barton M. K. (2007) Plant Cell 19, 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim Y. S., Kim S. G., Lee M., Lee I., Park H. Y., Seo P. J., Jung J. H., Kwon E. J., Suh S. W., Paek K. H., Park C. M. (2008) Plant Cell 20, 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim K., Kim K. H., Cho H. K., Kim H. Y., Kim H. H., Cheong J. (2010) Biochem. J. 427, 413–422 [DOI] [PubMed] [Google Scholar]
- 18. Seol W., Choi H. S., Moore D. D. (1996) Science 272, 1336–1339 [DOI] [PubMed] [Google Scholar]
- 19. Frith M. C., Wilming L. G., Forrest A., Kawaji H., Tan S. L., Wahlestedt C., Bajic V. B., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bailey T. L., Huminiecki L. (2006) PLoS Genet. 2, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanada K., Zhang X., Borevitz J. O., Li W. H., Shiu S. H. (2007) Genome Res. 17, 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashimoto Y., Kondo T., Kageyama Y. (2008) Dev. Growth Differ. 50, S269–S276 [DOI] [PubMed] [Google Scholar]
- 22. Yun J., Kim S. G., Hong S., Park C. M. (2008) Plant Signal. Behav. 3, 615–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu W., Ma H. (2006) Plant J. 45, 399–422 [DOI] [PubMed] [Google Scholar]
- 24. Hu W., dePamphilis C. W., Ma H. (2008) J. Integr. Plant Biol. 50, 1031–1045 [DOI] [PubMed] [Google Scholar]
- 25. Sicard A., Petit J., Mouras A., Chevalier C., Hernould M. (2008) Plant J. 55, 415–427 [DOI] [PubMed] [Google Scholar]
- 26. Karimi M., Inzé D., Depicker A. (2002) Trends Plant Sci. 7, 193–195 [DOI] [PubMed] [Google Scholar]
- 27. Udvardi M. K., Czechowski T., Scheible W. R. (2008) Plant Cell 20, 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ponchel F., Toomes C., Bransfield K., Leong F. T., Douglas S. H., Field S. L., Bell S. M., Combaret V., Puisieux A., Mighell A. J., Robinson P. A., Inglehearn C. F., Isaacs J. D., Markham A. F. (2003) BMC Biotechnol. 3, 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rozen S., Skaletsky H. J. (2000) Bioinformatics Methods and Protocols: Methods in Molecular Biology, pp. 365–386, Humana Press, NJ: [DOI] [PubMed] [Google Scholar]
- 30. Breeden L., Nasmyth K. (1987) Cell 48, 389–397 [DOI] [PubMed] [Google Scholar]
- 31. Miller J. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Hellman L. M., Fried M. G. (2007) Nat. Protoc. 2, 1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duby A. (1987) in Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. eds) Unit 6.4, pp. 1–10, Greene Publishing Associates and John Wiley & Sons, New York [Google Scholar]
- 34. Furlong E. E., Keon N. K., Thornton F. D., Rein T., Martin F. (1996) J. Biol. Chem. 271, 29688–29697 [DOI] [PubMed] [Google Scholar]
- 35. Connolly J. P., Augustine J. G., Francklyn C. (1999) Nucleic Acids Res. 27, 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987) EMBO J. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sheen J. (2001) Plant Physiol. 127, 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- 38. Luehrsen K. R., de Wet J. R., Walbot V. (1992) Methods Enzymol. 216, 397–414 [DOI] [PubMed] [Google Scholar]
- 39. Kim H., Park M., Kim S. J., Hwang I. (2005) Plant Cell 17, 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee L. Y., Gelvin S. B. (2008) Plant Physiol. 146, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klein T. M., Fromm M., Weissinger A., Tomes D., Schaaf S., Sletten M., Sanford J. C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4305–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith G. E., Darling D. S. (2003) Mol. Biol. Rep. 30, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klug A. (2010) Annu. Rev. Biochem. 79, 213–231 [DOI] [PubMed] [Google Scholar]
- 44. Tan Q. K., Irish V. F. (2006) Plant Physiol. 140, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park H. C., Kim M. L., Lee S. M., Bahk J. D., Yun D. J., Lim C. O., Hong J. C., Lee S. Y., Cho M. J., Chung W. S. (2007) Nucleic Acids Res. 35, 3612–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tran L. S., Nakashima K., Sakuma Y., Osakabe Y., Qin F., Simpson S. D., Maruyama K., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. (2007) Plant J. 49, 46–63 [DOI] [PubMed] [Google Scholar]
- 47. Gehring W. J., Affolter M., Bürglin T. (1994) Annu. Rev. Biochem. 63, 487–526 [DOI] [PubMed] [Google Scholar]
- 48. Khoury Christianson A. M., Kafatos F. C. (1993) Nucleic Acids Res. 21, 4416–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Crawford K. M., Zambryski P. C. (2001) Plant Physiol. 125, 1802–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Citovsky V., Lee L. Y., Vyas S., Glick E., Chen M. H., Vainstein A., Gafni Y., Gelvin S. B., Tzfira T. (2006) J. Mol. Biol. 362, 1120–1131 [DOI] [PubMed] [Google Scholar]
- 51. Windhövel A., Hein I., Dabrowa R., Stockhaus J. (2001) Plant Mol. Biol. 45, 201–214 [DOI] [PubMed] [Google Scholar]
- 52. Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002) EMBO J. 21, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Franklin K. A., Praekelt U., Stoddart W. M., Billingham O. E., Halliday K. J., Whitelam G. C. (2003) Plant Physiol. 131, 1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu J., Shi H., Lee B. H., Damsz B., Cheng S., Stirm V., Zhu J. K., Hasegawa P. M., Bressan R. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coego A., Ramirez V., Gil M. J., Flors V., Mauch-Mani B., Vera P. (2005) Plant Cell 17, 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith H. M., Campbell B. C., Hake S. (2004) Curr. Biol. 14, 812–817 [DOI] [PubMed] [Google Scholar]
- 57. Chariot A., Gielen J., Merville M. P., Bours V. (1999) Biochem. Pharmacol. 58, 1851–1857 [DOI] [PubMed] [Google Scholar]
- 58. Zhang J. H., Barr V. A., Mo Y., Rojkova A. M., Liu S., Simonds W. F. (2001) J. Biol. Chem. 276, 10284–10289 [DOI] [PubMed] [Google Scholar]
- 59. Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. (1991) Cell 66, 743–758 [DOI] [PubMed] [Google Scholar]
- 60. Li X., Shou W., Kloc M., Reddy B. A., Etkin L. D. (1994) J. Cell Biol. 124, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Igarashi D., Ishida S., Fukazawa J., Takahashi Y. (2001) Plant Cell 13, 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hackbusch J., Richter K., Müller J., Salamini F., Uhrig J. F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee K. A. (1992) J. Cell Sci. 103, 9–14 [DOI] [PubMed] [Google Scholar]
- 64. Park O. K., Schaefer L. K., Wang W., Schaefer T. S. (2000) J. Biol. Chem. 275, 32244–32249 [DOI] [PubMed] [Google Scholar]
- 65. Reményi A., Tomilin A., Pohl E., Lins K., Philippsen A., Reinbold R., Schöler H. R., Wilmanns M. (2001) Mol. Cell 8, 569–580 [DOI] [PubMed] [Google Scholar]
- 66. Lee S., Lee S., Yang K. Y., Kim Y. M., Park S. Y., Kim S. Y., Soh M. S. (2006) Plant Cell Physiol. 47, 591–600 [DOI] [PubMed] [Google Scholar]
- 67. Ohtani N., Zebedee Z., Huot T. J., Stinson J. A., Sugimoto M., Ohashi Y., Sharrocks A. D., Peters G., Hara E. (2001) Nature 409, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 68. Toledo-Ortiz G., Huq E., Quail P. H. (2003) Plant Cell 15, 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.