Abstract
Barth syndrome is an X-linked genetic disorder caused by mutations in the tafazzin (taz) gene and characterized by dilated cardiomyopathy, exercise intolerance, chronic fatigue, delayed growth, and neutropenia. Tafazzin is a mitochondrial transacylase required for cardiolipin remodeling. Although tafazzin function has been studied in non-mammalian model organisms, mammalian genetic loss of function approaches have not been used. We examined the consequences of tafazzin knockdown on sarcomeric mitochondria and cardiac function in mice. Tafazzin knockdown resulted in a dramatic decrease of tetralinoleoyl cardiolipin in cardiac and skeletal muscles and accumulation of monolysocardiolipins and cardiolipin molecular species with aberrant acyl groups. Electron microscopy revealed pathological changes in mitochondria, myofibrils, and mitochondrion-associated membranes in skeletal and cardiac muscles. Echocardiography and magnetic resonance imaging revealed severe cardiac abnormalities, including left ventricular dilation, left ventricular mass reduction, and depression of fractional shortening and ejection fraction in tafazzin-deficient mice. Tafazzin knockdown mice provide the first mammalian model system for Barth syndrome in which the pathophysiological relationships between altered content of mitochondrial phospholipids, ultrastructural abnormalities, myocardial and mitochondrial dysfunction, and clinical outcome can be completely investigated.
Keywords: Cardiolipin, Gene Knockout, Gene Silencing, Mitochondria, Phospholipid, Barth Syndrome, Mitophagy, Mouse Model, Tafazzin, Cardiomyopathy
Introduction
Cardiolipin (CL)2 is a negatively charged mitochondrial polyglycerophospholipid consisting of two phosphatidyl moieties containing four fatty acyl side chains that predominantly resides in the inner mitochondrial membrane at the site of its biosynthesis. Mitochondrial CL content varies in different cell types; it is higher in oxidative tissues. In cardiac mitochondria, CL comprises ∼20% of the total phospholipid amount (1). The fatty acyl chain pattern of CL is tissue-specific. In mammalian cardiac mitochondria, CL species are dominated by tetralinoleoyl cardiolipin with four symmetric linoleic acids 18:2(n-6). In contrast, in neuronal tissues, CL speciation is very diverse as represented by more than 100 different molecular species (2). In mammals, tetralinoleoyl cardiolipin is particularly enriched in highly oxidative tissues, such as heart and skeletal muscle, and composes up to 70–80% of total CL content. In certain marine bivalves, CL contains almost exclusively docosahexaenoic chains, whereas in yeast, the dominant acyl groups are oleic and palmitoleic acid (3). Differences in cardiolipin content and cardiolipin species exist in a variety of tissues, physiological conditions, and diseases (4).
Maintaining the normal level and normal species of CL is crucial for mitochondrial function and structural integrity. CL is involved in the formation of the lamellar crista architecture (5), assembly of electron transport chain complexes (6) and mitochondrial carriers (7), association of enzymes to inner mitochondrial membrane (8, 9), and mitochondrial dynamics (10). CL serves as a proton trap for oxidative phosphorylation, thereby protecting mitochondrial compartments from extreme pH variations (11). Emerging evidence also suggests that CL is essential for activation of caspase-8 and apoptosis (12). Perturbation of CL content and its fatty acyl composition is associated with aging (13, 14), ischemia (13, 15, 16), hypertension (17, 18), diabetes (19), heart failure (20, 21), and tumors (6, 22, 23).
The importance of CL and its fatty acyl composition is emphasized by the recognition that CL is aberrant in patients with Barth syndrome, a rare genetic multisystem disorder (24). In this disorder, total cardiolipin levels are low, monolysocardiolipin (MLCL) levels are greatly increased, and the CL species reveal a mixed acyl group pattern with a significantly lower degree of unsaturation (25). Manifestations in Barth syndrome patients vary by age and severity but include dilated cardiomyopathy, muscle weakness, growth delay, intermittent or chronic neutropenia, 3-methylglutaconic aciduria, chronic fatigue, arrhythmias, and sudden death. Death in infancy and childhood previously occurred in the majority of patients before the advent of improved early cardiac care. Barth syndrome is X-linked, affecting males, and is caused by mutations in tafazzin (taz) gene (26), which is ubiquitously expressed in all human tissues (27). Tafazzin is a mitochondrial transacylase that, after initial synthesis of CL, catalyzes the remodeling of immature CL, resulting in its mature acyl composition with predominance of tetralinoleoyl cardiolipin.
Tafazzin function has been studied in Barth syndrome patients and in non-mammalian model organisms, such as yeast, flies, zebrafish, and in cultured mammalian cells. Although existing models of taz deficiency provide valuable information about the role of taz in CL metabolism and mitochondrial function, these systems cannot be adequately used and manipulated to study the long range effects of gene deletion or mutation in the whole organism and do not allow development and testing of potential therapies.
Therefore, we generated an inducible taz knockdown mouse model by using RNA interference technology (see Fig. 1). We used this newly developed model to investigate the consequences of taz knockdown, which we expected to impact cardiac function and to alter the mitochondria of cardiac and skeletal muscles.
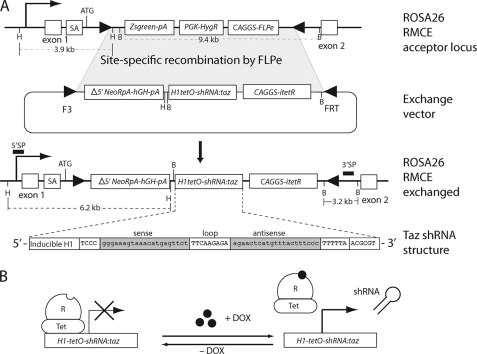
FIGURE 1.
Development of ROSA26-taz shRNATet-On in mouse ES cells. A, mouse ES cells with a pre-engineered RMCE acceptor site were used for the site-directed knock-in of a tetracycline-inducible taz shRNA cassette into the ROSA26 locus. Tafazzin shRNA structure is depicted at the bottom of A. An RMCE acceptor site in the ROSA26 locus consists of the Zsgreen fluorescent protein expression unit (Zsgreen-pA), hygromycin resistance cassette (PGK-HygR), and FLPe recombinase expression module (CAGGS-FLPe). Neomycin selection and the subsequent assay for hygromycin sensitivity were applied to isolate the desired clones (ROSA RMCE exchanged). Selected clones were screened by Southern blot analysis of HindIII and BamHI restriction fragments (not shown). Correctly targeted clones (71% of all neomycin-resistant clones) produce 6.2-kb HindIII and 3.2-kb BamHI fragments on Southern blots with 5′ sense probe (SP) and 3′ sense probe DNA probes (solid black bars), respectively. B, schematic representation of tet repressor (TetR)-mediated control of shRNA expression. Transcription of shRNA is blocked by TetR. Upon induction by dox, TetR is removed from the tet operator sequences (TetO) that is inserted into the promoter, allowing transcription of shRNA. SA, splicing acceptor signal. The technology used in the study to make this tafazzin knockdown mouse is the proprietary property of TaconicArtemis GmbH.
EXPERIMENTAL PROCEDURES
Targeting of taz shRNA Allele in ROSA26 Locus in Mouse Embryonic Stem (ES) Cells and Generation of Tafazzin-deficient Mice
Transgenic mice were generated at TaconicArtemis GmbH (Köln, Germany) using proprietary methods and materials. In brief, mouse embryonic stem cells with a pre-engineered recombinase-mediated cassette exchange (RMCE) acceptor site on chromosome 6 were used for targeting insertion of several distinct Tet-On taz shRNA expression cassettes into the ROSA26 locus (28) as depicted Fig. 1. Correctly targeted embryonic stem cell clones were identified by Southern DNA blot analysis and tested for tafazzin mRNA knockdown at the embryonic stem cell stage (not shown). One embryonic stem cell clone that gave acceptable mRNA knockdown (as well as CL deficiency) was injected into tetraploid blastocysts, which eventually resulted in a line of transgenic mice from a single founder (29). Tetraploid blastocysts were prepared from B6D2F1 mice (Harlan).
All animal studies were approved by our Institutional Animal Care and Use Committee. Animals were housed in microisolator cages with temperature-controlled conditions under a 12-h light/dark cycle with free access to drinking water and food. Doxycycline (dox) was administered as part of the food with standard rodent chow containing 3% fat content (RHM1500) formulated by Purina Mills (Gray Summit, MO) and containing 625 mg of dox/kg of chow. Tafazzin silencing in these mice was induced at an early embryonic stage. First, wild type C57BL/6J female mice were placed on a doxycycline-containing diet 4 or 5 days before mating. During the mating period with transgenic males (ROSA26H1/tetO-shRNA:taz) of C57BL/129S6 genetic background, dox was temporarily discontinued to avoid reported potential problems with male fertility (30). After detecting copulatory plugs, females were separated from males and placed back on the dox-containing diet. After 3 weeks, transgenic pups were identified by PCR genotyping of tail genomic DNA. Pups were continuously maintained on the dox-containing diet thereafter. Non-transgenic male littermates fed with dox-containing chow were used as wild type controls in all experiments unless otherwise noted. Some litters of mice that were never induced with dox were also used as non-induced transgenic control mice.
Mice were genotyped by PCR analysis of tail DNA (see supplemental material for primer sequences and PCR conditions). Mice positive for the taz shRNA transgene were identified by the presence of 381-bp DNA product in PCRs.
Mitochondrial Protein Isolation and Western Blot
Mitochondria were isolated from fresh cardiac and skeletal muscles following Dounce tissue homogenization and differential centrifugation using the Mitochondria Isolation kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Mitochondrial proteins were fractionated by SDS-PAGE on a 4–20% gradient gel and semidry blotted on nitrocellulose membrane for 30 min at 200 mA (Bio-Rad). Primary polyclonal antibodies were produced in rabbits against the C-terminal peptide of mouse tafazzin protein (QEEFQRLKMQAEQLHNHFQPGR). The IgG fraction was purified from serum on Protein G-Sepharose (Pierce). Purified antibodies were conjugated with biotin and used at a dilution of 1:500 for Western analysis. Streptavidin-peroxidase conjugate (Roche Applied Science) and ECL-Plus® Western blotting detection reagent (GE Healthcare) were used for band visualization. ImageJ software (National Institutes of Health) was used for quantitative band intensity analyses.
RNA Isolation, cDNA Synthesis, and Quantitative PCR
RNA was isolated from ∼50 mg of frozen tissues using TRIzol® reagent (Invitrogen) according to the manufacturer's protocol, and RNA concentration was determined by UV absorbance at 260 nm. The cDNA was synthesized from RNA using the Omniscript® reverse transcription kit (Qiagen), and quantitative PCR was performed on an Opticon-2 real time thermocycler (Bio-Rad) using SYBR® Green Master Mix (Applied Biosystems). The amount of taz-specific cDNA in different samples was normalized to the content of ribosomal L7 protein mRNA.
Mitochondrial DNA (mtDNA) amounts were determined in whole genomic DNA preparations isolated from different tissues using a DNeasy kit (Qiagen). The mtDNA and nuclear DNA amounts were measured by quantitative PCR as described above for mRNA. Nuclear and mtDNA-specific primer sequences are provided in the supplemental material. All samples were analyzed in triplicate.
Phospholipid Analysis
Phospholipids were extracted from frozen tissues. Tissue samples were homogenized in phosphate-buffered saline followed by the addition of 3 ml of chloroform-methanol (2:1, v/v) and internal standards (IS) (0.4 nmol of tetramyristoyl CL, 0.32 nmol of dimyristoylphosphatidylglycerol, 3 nmol of dimyristoylphosphatidylethanolamine, and 1.5 nmol of dimyristoylphosphatidylcholine; Avanti Polar Lipids, Alabaster, AL). After 20 min of sonication, the mixture was cooled on ice for 15 min and centrifuged for 10 min at 1000 × g. The lower, organic phase was removed, and the upper phase was re-extracted with 3 ml of chloroform-methanol (2:1, v/v). The combined organic phases were dried and dissolved in 150 μl of chloroform/methanol/water (50:45:5, by volume) containing 0.01% ammonia. An aliquot of 10 μl was used for high-performance liquid chromatography (HPLC) mass spectrometry as described previously (31). Mass spectra of CL and MLCL molecular species were obtained by continuous scanning from 400 to 1000 m/z. Semiquantitative determination of selected phospholipid species was performed by determining the area of each phospholipid peak (APL) and that of the added IS (AIS) using Xcalibur 2.0 (Thermo Scientific). If appropriate, the peak areas were corrected for the contribution of naturally occurring isotopes. To compare individual samples, the ratio APL/AIS was multiplied by the amount of added IS. If less than 1 mg of protein was used, the result was also corrected for the protein content of the cell homogenate.
Electron Microscopy
After euthanasia, mice were perfused with 4% formaldehyde and 1% glutaraldehyde in 0.1 m pH 7.5 Pipes buffer (PB). Small tissue pieces were incubated in 4% glutaraldehyde and 2% formaldehyde in PB for 2 h at room temperature and overnight at 4 °C. Solutions were exchanged with 4% glutaraldehyde and 0.4% tannic acid in PB for 30 min and with 1% osmium tetroxide in PB for 1 h. The samples were counterstained with 1% uranyl acetate for 2 h before exchanging the solution with ethanol at progressively lower temperatures (32). Samples were infused with epoxy resin and polymerized with heat. The sample blocks were sectioned to a thickness of 50–70 μm, collected on electron microscope grids, and stained with uranyl acetate and Sato lead stain. The samples were then imaged using a Philips CM12 microscope operated at 120 kV.
Cardiac Imaging
Two-dimensional and motion-mode (M-mode) transthoracic echocardiography was performed as described previously using a Visual Sonics Vevo 770 imaging system and a 30-MHz transducer (33). Off-line analyses included measurement in diastole of interventricular septum (IVSd) and left ventricular free wall (LVFWd) thickness and the left ventricular (LV) end diastolic and end systolic internal dimensions (LVIDd and LVIDs, respectively). LV shortening fraction (SF) was calculated as SF = ((LVIDd − LVIDs)/LVIDd) × 100. Cardiac mass was determined by magnetic resonance imaging using the proprietary software application.
Statistical Analysis
Differences between groups were assessed for significance by unpaired Student's t test with assumption of equal variances unless otherwise noted. Results were considered statistically significant if the p value was <0.05. Results are expressed as arithmetic means ± S.E. Statistical calculations were performed using the GraphPad Prism program (GraphPad Software, San Diego, CA).
RESULTS
Generation of taz shRNA Transgenic (TG) Mice
An RMCE strategy with enhanced flippase recombinase (FLPe) and mutant flippase recognition target (FRT) sequences was used for targeted insertion of six distinct taz short hairpin RNA expression modules into the pre-engineered acceptor site at the ROSA26 locus (28, 34) in mouse ES cells (Fig. 1). Correctly targeted clones identified by Southern blot analysis were tested for tafazzin mRNA knockdown and for CL analysis after doxycycline induction (not shown). One targeted ES cell line was acceptable and was used for the creation of FØ generation of chimeric mice by tetraploid-diploid supplementation (29). One founder male generated the tafazzin knockdown line. TG germ line transmission was confirmed in the FØ1 generation pups by PCR analysis of tail DNA.
Induction of shRNA-mediated taz Knockdown
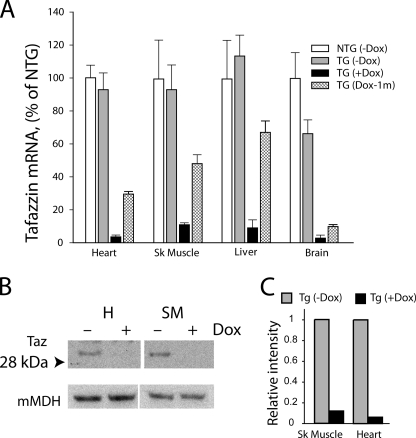
To provide a model for human Barth syndrome consistent with a defective tafazzin gene at conception, tafazzin silencing in mice was induced at an early embryonic stage. The efficiency of shRNA-mediated taz gene silencing was evaluated by quantitative real time RT-PCR analysis of taz mRNA in heart, skeletal muscle, liver, and brain. Silencing of the taz gene was highly efficient in all organs analyzed. Induction of taz shRNA upon administration of dox greatly reduced taz mRNA in heart, skeletal muscle, liver, and brain (Fig. 2A). In non-induced TG mice not treated with the dox-containing diet, no effect of taz shRNA present in the genome was observed in heart, liver, and skeletal muscle because taz mRNA expression was comparable with non-transgenic (NTG) littermates. However, in the brain of non-induced mice, taz mRNA was reduced by 35% as compared with NTG controls. These findings support earlier observations that the regulation of the H1-tet promoter, which drives the tafazzin-shRNA minigene, is tight in mouse cardiac and skeletal muscles (28). Upon induction with dox, taz mRNA levels were reduced in heart to 3.7 ± 1.0%, in skeletal muscle to 11.2 ± 1.2%, in liver to 8.9 ± 3.8%, and in brain to 3.4 ± 1.2% compared with normalized levels of taz mRNA in the same tissues of NTG mice. Interestingly, following withdrawal of the dox-containing diet at 4 weeks, there was partial restoration of taz mRNA in heart, skeletal muscle, and liver to 48, 58, and 64%, respectively, of control levels in NTG mice as assessed 1 month later in 2-month-old animals (Fig. 2A). This observation demonstrates that dox-induced shRNA-mediated knockdown of taz mRNA is at least partially reversible in these tissues.
FIGURE 2.
Dox-inducible shRNA-mediated tafazzin knockdown. A, quantitative RT-PCR analysis of taz mRNA levels in heart, skeletal (Sk) muscle (EDL), liver, and brain of 2-month-old NTG control males, non-induced taz shRNA TG males, dox-induced taz shRNA TG males, and taz shRNA TG males induced only for the first 4 weeks and then withdrawn from the dox-containing diet. Tafazzin mRNA abundance as a percentage of NTG controls is plotted. Data are expressed as the means ± S.E. from three or more experiments. B, representative Western blot of tafazzin protein in mitochondrial extracts of skeletal muscle (SM) and heart (H) probed with anti-tafazzin polyclonal antibodies. Anti-mitochondrial malate dehydrogenase (mMDH) antibodies were used to control for protein loading. Uncropped image of blot is shown in supplementary Fig. 1. C, densitometric analysis of Western blot showing an 86 and 97% reduction of tafazzin protein in skeletal and cardiac muscle-derived mitochondria, respectively.
Tafazzin protein knockdown in cardiac and skeletal muscles was ascertained by Western blot analysis. All existing anti-tafazzin antibodies suffer from poor specificity and cross-reactivity with other polypeptides. However, our affinity-purified antibodies are specific enough to detect tafazzin protein on the Western blots when protein extracts from purified mitochondria are analyzed. As expected, induction with dox reduced tafazzin protein levels in cardiac and skeletal muscles of ROSA26H1/tetO-shRNA:taz males by 97 and 86%, respectively (Fig. 2, B and C), demonstrating that shRNA-mediated tafazzin knockdown is efficient in reducing the tafazzin gene product in mouse striated muscle.
During the first 2 months of observation, tafazzin-deficient mice developed normally without any apparent deviation in body weight compared with NTG littermate controls. After 8 months, TG dox-treated mice weighed 17% less than NTG dox-treated littermates (p = 0.034; Table 1). This is reminiscent of the reduced growth present in surviving individuals with Barth syndrome during childhood and early adolescence (35).
TABLE 1.
Physiological and echocardiographic indices in 2- and 8-month-old taz-deficient (tazdef) and NTG mice
All values are mean ± S.E. Significant differences between NTG and taz-deficient groups are indicated. Hw/Bw, heart weight (milligrams)/body weight (grams) ratio; HR, heart rate (beats/min (bpm)); IVSd, diastolic interventricular septal wall thickness; LVPWd, diastolic left ventricular posterior wall thickness; LVIDd, diastolic left ventricular internal dimension; LVIDs, systolic left ventricular internal dimension; LV Vold, left ventricular diastolic volume; LV Vols, left ventricular systolic volume; FS, left ventricular fractional shortening; EF, left ventricular ejection fraction.
| Variable | 2 months |
8 months |
||
|---|---|---|---|---|
| NTG (n = 6) | tazdef (n = 4) | NTG (n = 4) | tazdef (n = 4) | |
| Body weight (g) | 22.7 ± 0.737 | 22.2 ± 0.5 | 38.6 ± 2.3 | 32.2 ± 1.7a |
| Hw/Bw (mg/g) | 5.0 ± 0.2 | 5.2 ± 0.7 | 3.9 ± 0.1 | 4.3 ± 0.3a |
| HRb (bpm) | 442 ± 10 | 383 ± 38 | 450 ± 13 | 460 ± 30 |
| IVSd (mm) | 0.65 ± 0.03 | 0.71 ± 0.04 | 0.68 ± 0.03 | 0.50 ± 0.07c |
| LVPWd (mm) | 0.55 ± 0.04 | 0.68 ± 0.03a | 0.60 ± 0.02 | 0.41 ± 0.06c |
| LVIDd (mm) | 4.24 ± 0.13 | 4.13 ± 0.20 | 4.63 ± 0.19 | 4.98 ± 0.10 |
| LVIDs (mm) | 3.08 ± 0.02 | 2.88 ± 0.20 | 2.92 ± 0.16 | 3.80 ± 0.07c |
| LV Vold (μl) | 80.9 ± 5.8 | 77.0 ± 8.6 | 88.2 ± 6.9 | 117.1 ± 5.4c |
| LV Vols (μl) | 38.1 ± 4.8 | 32.9 ± 5.5 | 36.8 ± 3.7 | 61.8 ± 2.6c |
| FS (%) | 28 ± 2 | 31 ± 2 | 31 ± 2 | 24 ± 1c |
| EF (%) | 54 ± 4 | 58 ± 3 | 58 ± 2 | 47 ± 1c |
| LV walld (μl) | 74.1 ± 5.2 | 83.7 ± 8.4 | 85.1 ± 1.2 | 74.6 ± 2.3a |
a p < 0.05.
b Anesthetized heart rate.
c p < 0.001.
d Left ventricular volume determined by cardiac MRI.
Phospholipid Analysis
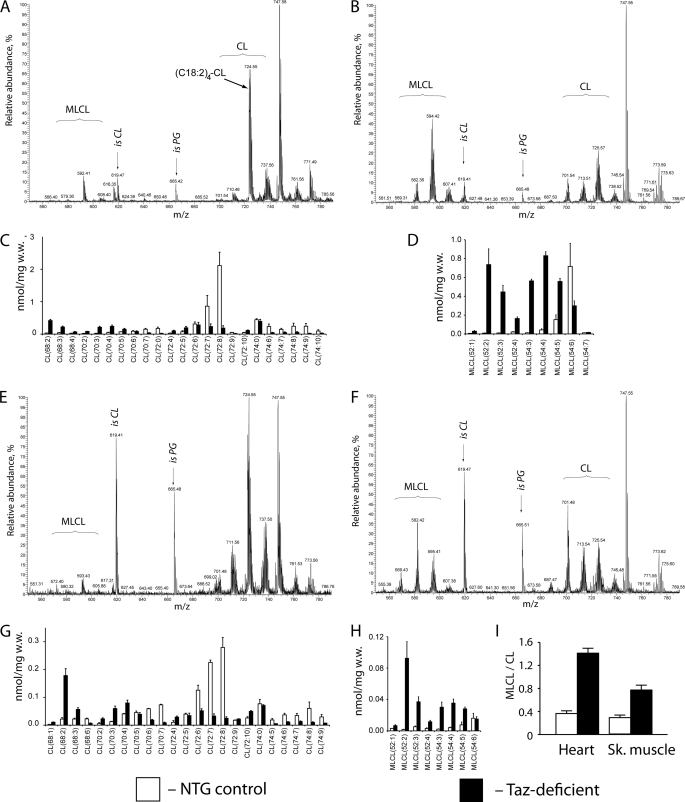
The only well established biochemical consequence of mutations in the taz gene in various organisms is reduction of tetralinoleoyl cardiolipin with concomitant accumulation of MLCL species and other CL species with more saturated acyl groups. To determine whether taz-deficient mice exhibit CL defects similar to those observed in Barth syndrome patients, CL and MLCL spectra were analyzed in cardiac and skeletal muscles of 2-month-old taz-deficient mice and NTG littermate controls. Lipid extracts from cardiac and skeletal muscles (extensor digitorum longus (EDL)) were subjected to LC-MS/MS to measure changes in phospholipid molecular species spectra with acyl moieties that have 66 (CL(66), m/z ∼685–690) to 74 (CL(74), m/z ∼730–740) carbon atoms (Fig. 3, A, C, E, and G). C72:8 and C72:7, representing (C18:2)4-CL and (C18:2)3/(C18:1)1-CL, are the most abundant molecular species of CL in cardiac and skeletal muscles of NTG mice. This observation is consistent with existing data of CL profiles in mammalian cells (36, 37). As expected, taz knockdown induced a dramatic decrease in CL content and a shift toward more saturated CL species in cardiac and skeletal muscles of 2-month-old mice. In NTG control hearts, the major molecular species with C18 acyl side chains (Fig. 2C), CL(72:8) and CL(72:7), constitute 32 and 13% of total CL, respectively. Tafazzin deficiency drastically altered the pattern of CL speciation in the heart and reduced the 72:8 and 72:7 CL molecular species to 5 and 6%, respectively. Alterations in content of C18 cardiolipins were accompanied by accumulation of molecular species with shorter acyl side chains and with higher degrees of saturation (Fig. 3C, CL(68:2)–CL(70:5)).
FIGURE 3.
Cardiolipin and monolysocardiolipin analyses in heart and skeletal muscles of NTG control and taz-deficient mice. Representative MS profiles of phospholipids from heart tissue of NTG (A) and taz-deficient (B) mice are shown. C and D, acyl group profiles of CL (C) and MLCL (D) from hearts of NTG and taz-deficient mice (n = 4 for each group). E and F, representative MS profiles of phospholipids from skeletal muscle of NTG (E) and taz-deficient (F) mice. G and H, acyl group profiles of CL (G) and MLCL (H) from skeletal (Sk.) muscle of NTG and taz-deficient mice. I, MLCL/CL ratios for heart and skeletal muscle from NTG and taz-deficient mice (n = 4 for each group). Data are expressed as the means ± S.E. isCL and isPG denote internal standards of cardiolipin and phosphatidylglycerol (0.4 nmol), respectively.
In the EDL, a fast skeletal muscle, the total cardiolipin level of NTG dox-treated control mice was lower than in the heart, perhaps reflecting the lower mitochondrial content in this highly glycolytic tissue. Similar to the results obtained in the heart, taz knockdown in the EDL muscle resulted in complete alteration of the molecular spectrum of fully acylated cardiolipins. In control EDL muscle, 72:8 and 72:7 CLs constituted 19 and 15%, respectively, of the cardiolipin material, whereas in taz-deficient muscles, these values were diminished to 2 and 3%, respectively. As in the heart, these changes were also accompanied by dramatic increases in content of CL molecular species with shorter and asymmetric side acyl groups (Fig. 3G, species 68:2–72:5).
Tafazzin knockdown also induced robust accumulation of MLCLs both in cardiac and skeletal muscles. As a result of these changes, the MLCL/CL ratio was drastically increased both in cardiac and skeletal muscles (Fig. 3, D, H, and I). This is consistent with biochemical changes demonstrated in tissue and blood samples taken from Barth syndrome patients (25, 38).
Mitochondrial Abnormalities in Cardiac and Skeletal Muscles of taz-deficient Mice
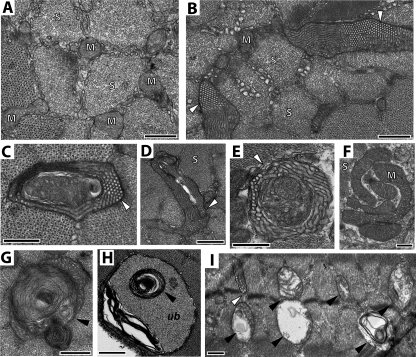
Previous studies demonstrate the gross structural abnormalities in cardiac and skeletal muscle mitochondria from tissue samples of Barth syndrome patients (39). We examined whether our taz-deficient mice exhibit similar abnormalities. Electron micrographs of cardiac muscle of 2-month-old taz-deficient mice did not reveal any significant abnormalities (not shown). However, in 8-month-old mice, abnormalities of both mitochondrial and sarcomeric structure were evident (Fig. 4). Although the majority of mitochondria were normal in size and structure, mitochondria were frequently present in large aggregates between sarcomeres. In longitudinal sections of cardiac ventricles from 8-month-old taz-deficient mice, sarcomeres are poorly arrayed as compared with NTG controls (Fig. 4, A and B). Mitochondria were aggregated and varied in shape and size. Multiple concentric layers of densely packed cristae with onion-shaped morphology were frequent and may represent mitochondria undergoing degeneration or mitophagy (Fig. 4C). Mitochondria in taz-deficient cardiac muscle frequently contained patches of swollen tubular cristae (Fig. 4C). Large vacuoles were often observed inside the mitochondria (Fig. 4D). On the cross-sections of both ventricular and atrial muscles, membrane vesicles, adjacent to the mitochondria, were greatly changed in tafazzin-deficient hearts. These membranous structures (mitochondrion-associated membranes (MAMs)) are formed from endoplasmic reticulum (ER) membrane vesicles (40). MAMs are important for ER-mitochondrion cross-communication and calcium handling by mitochondria. MAMs also facilitate phospholipid transport between the ER and mitochondria. In 2-month-old mice, MAMs appear slightly enlarged. However, in older mice the differences in MAM appearance and morphology were much more pronounced. In taz-deficient hearts, MAMs were abundant, dilated, and often contained multiple membrane layers with a concentric pattern.
FIGURE 4.
Defects of cardiac mitochondria and mitochondrion-associated membranes in taz-deficient mice. A and B, representative electron micrographs of longitudinal sections of cardiac ventricular muscle of 8-month-old NTG (A) and taz-deficient (B) mice. C and D, representative micrographs of deteriorating mitochondria in ventricular (C) and atrial (D) muscle from taz-deficient hearts. White arrows in C and K point to mitochondria undergoing mitophagy. Mitochondria with large central vacuoles are abundantly observed in taz-deficient cardiomyocytes (black arrowhead in D). MAMs, denoted by white arrowheads in panels E–J, are greatly affected in taz-deficient hearts. E and G show representative cross-sections of cardiac muscle of 2-month-old (E) and 8-month-old (G) control mice, respectively. In 2-month-old taz-deficient mice, MAMs appear slightly enlarged (F); however, in older mice, differences in MAM size and morphology become more pronounced (H–J). Scale bars, 500 nm. In all panels, mitochondria are marked with letter “M,” and sarcomeres are marked with the letter “S.”
We also examined the ultrastructural morphology of the highly glycolytic EDL muscle from NTG and tafazzin-deficient mice at both 2 and 8 months of age. In 2-month-old NTG mice, mitochondria varied in size with round or elongated shapes (Fig. 5A). Cristae of the EDL mitochondria were less organized than in cardiac mitochondria and exhibited both laminar and tubular shapes in a single mitochondrion. Significant proportions of mitochondria in tafazzin-deficient mice of both age groups contained multiple layers creating “onion skin-like” concentric cristae and were likely undergoing mitophagy (Fig. 5, G, H, and I). In taz-deficient EDL, we observed giant mitochondria-like structures that were encircled with a well defined outer membrane. These bizarre structures were abundantly present in muscle and had a unique and unusual inner membrane architecture with parallel lamellar cristae extended into the tubular segment with a highly organized honeycomb-like pattern or paracrystalline array (Fig. 4, B–E and I). Paracrystalline inclusions and honeycomb-like formations in mitochondria have been found in bivalve species (41) and in mammalian cells with pathological alterations in lipid and phospholipid metabolism (42, 43) and conceivably are derived from the mitochondrial cristae under stress conditions (43). These structures were specific for the muscles of taz-deficient mice and were not observed in any control tissues.
FIGURE 5.
Mitochondrial defects in skeletal muscle of taz-deficient mice. Representative cross-sectional micrographs of EDL (A–C, E, and G–I) and soleus muscles (F) are shown. A, cross-section of EDL from a 2-month-old NTG mouse. Honeycomb-like formations, denoted by white arrowheads, are abundantly present in EDL of 2-month-old (B–E) and 8-month-old (I) taz-deficient mice. F, aggregated mitochondria in the soleus muscle of a 2-month-old taz-deficient mouse. G, H, and I are representative micrographs of mitophagy (black arrowheads) in EDL muscle of 2-month-old (G) and 8-month-old (I) taz-deficient mice. H shows a grossly abnormal mitochondrion (black arrowhead) surrounded by an autophagic vacuole filled with ubiquitinated material (ub). Scale bars, 500 nm. In all panels, mitochondria are marked with the letter “M,” and sarcomeres are marked with the letter “S.”
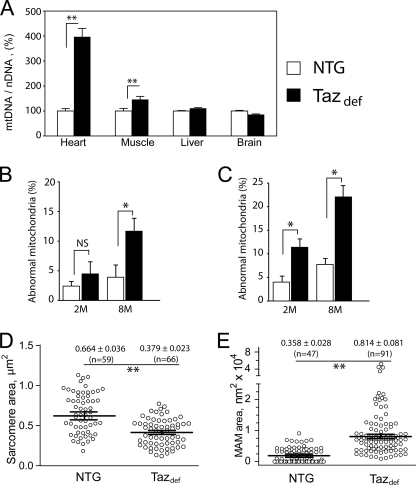
Evaluation of electron micrographs of cardiac and soleus muscles indicated that mitochondria in taz-deficient tissues were more abundant and aggregated in clusters than in NTG mice, which may indicate mitochondrial proliferation (Figs. 4D and 5F). We analyzed mitochondrial areas on randomly chosen cross-sections of 2-month-old hearts. No significant increases in average mitochondrial size were found (not shown). However, quantitative PCR analysis revealed a 4-fold increase of mtDNA content in hearts of 2-month-old tafazzin-deficient mice compared with NTG control mice. In skeletal muscle, the mtDNA amount was modestly elevated (∼30% over controls). Interestingly, these changes of mtDNA content are specific for sarcomeric tissues and were not observed in liver or brain (Fig. 6A).
FIGURE 6.
Mitochondrial proliferation and altered ultrastructure in taz-deficient mice. A compares mtDNA content with nuclear DNA (nDNA) content in various tissues of NTG and taz-deficient mice as a measure of mitochondrial abundance. mtDNA to nuclear DNA ratios as a percentage of NTG controls with S.E. are plotted. B and C, morphometric analyses of mitochondria in cardiac (B) and skeletal (C) muscles. The proportions of cardiac and skeletal muscle mitochondria displaying various structural abnormalities (Figs. 3 and 4) are plotted. Data are means with S.E. of three experiments in each of which we evaluated four or more mitochondrial cross-sections per group. D and E, quantitative morphometric analyses of sarcomere (D) and MAM (E) areas in the cross-sectional micrographs from heart of 8-month-old mice. Numbers are means ± S.E., and the numbers of experimental points are in parentheses. Asterisks denote significant differences between groups (NS, non-significant; *, p < 0.05; **, p < 0.001). The differences in means in D and E were assessed by non-parametric Mann-Whitney U test.
To quantify changes in the morphology of mitochondria, MAMs, and sarcomeres, randomly chosen digital photomicrographs of cardiac and skeletal muscles were subjected to quantitative morphological and statistical analyses. In the skeletal muscles of 2-month-old mice, taz knockdown significantly increased the number of mitochondria with various inner membrane abnormalities (swollen cristae, presence of honeycomb-like structures, concentric layers, and vacuoles), whereas in cardiac muscle, the changes of mitochondrial morphology were not statistically significant (Fig. 5C). In contrast, for 8-month-old mice, taz deficiency induced robust and statistically significant changes of mitochondrial morphology both in cardiac and skeletal muscles. Moreover, sarcomere areas were significantly smaller, and MAMs were more dilated in hearts of 8-month-old taz-deficient mice compared with NTG littermates (Fig. 5, E and F). These results suggest that continuous cardiolipin deficiency has a progressive and cumulative detrimental effect on mitochondrial structures in sarcomeric tissues.
Impaired Cardiac Function in taz-deficient Mice
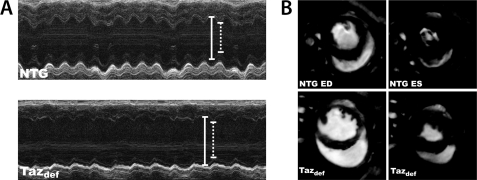
Barth syndrome is characterized by dilated cardiomyopathy often accompanied with left ventricular non-compaction. In severe cases, these cardiac abnormalities lead to LV dysfunction, profound heart failure, and death or cardiac transplantation. To determine whether tafazzin deficiency and a subsequently reduced CL level will result in cardiac disease similar to that of Barth syndrome, cardiac function was examined at 2 and 8 months in tafazzin-deficient and NTG mice by echocardiography and magnetic resonance imaging. Tafazzin deficiency had no overall effect on the measured parameters of cardiac function at 2 months. In contrast, 8-month-old taz-deficient mice exhibited LV dilation (Table 1 and Fig. 7). Echocardiography revealed that taz deficiency led to significant cardiac enlargement with increases of left ventricular diameter and volume both at end diastole and end systole (LVIDd, LVIDs, LV Vold, and LV Vols in Table 1, respectively) and reduced diastolic thickness of left ventricular posterior wall (LVPWd) and interventricular septum (IVSd). In addition, fractional shortening and ejection fraction were significantly reduced in the 8-month-old group (Table 1). Cardiac MRI revealed a 14% reduction of LV mass (LV wall volume) in 8-month-old tafazzin-deficient mice as compared with NTG controls (Table 1 and Fig. 7).
FIGURE 7.
Impaired cardiac function in 8-month-old taz-deficient (Tazdef) mice. A, representative M-mode echocardiographic tracings of NTG and taz-deficient mice. Diastolic dimension is indicated by solid lines, and systolic dimension is indicated by dotted lines. Chamber dilation and thinning of the ventricular walls are evident in the taz-deficient mouse. B, representative short axis cardiac MRI of NTG and taz-deficient mice at end diastole (ED) and end systole (ES).
DISCUSSION
It is increasingly appreciated that CL and its various molecular species play important roles in maintaining optimal activities of oxidative phosphorylation, electron transport chain complex function, and proper mitochondrial crista architecture as well as being intimately involved in apoptosis (5, 12, 32). In 2002, it was discovered that cardiolipin is decreased in skin fibroblasts of patients with Barth syndrome, an X-linked mitochondrial disease caused by mutations in tafazzin gene located on chromosome Xq28 (24). Although tafazzin function has been intensively investigated during the last decade, these studies were limited to non-mammalian systems such as yeast, flies, fish, and cultured cells. Although cardiac anatomy, mitochondrial energetics, thermogenesis, and homeostasis in flies and zebrafish are quite different than in mammals, tafazzin mutants in these two species nonetheless shared some similarities to the human disorder. However, to truly understand a multiorgan system disorder such as Barth syndrome, a mammalian model provides significant advantages for study and is thus highly desirable.
Here, we demonstrate the consequences of tafazzin knockdown in mice using the inducible shRNA-mediated silencing system. These tafazzin mRNA-deficient mice represent the first approximation of human Barth syndrome in a mammalian experimental model. Our data show that shRNA-mediated knockdown of tafazzin is efficient and drastically reduces tafazzin mRNA and tafazzin protein levels in heart and skeletal muscles, the organs prominently affected in Barth syndrome patients. Furthermore, we demonstrated that tafazzin mRNA knockdown could be partially restored postnatally because 4 weeks after withdrawal from doxycycline treatment taz mRNA expression was restored both in cardiac and skeletal muscles to 30 and 50% of their original levels, respectively. This finding with the inducible taz knockdown mice implies the plausibility and potential of corrective human gene therapy in existing Barth syndrome patients. This tafazzin knockdown mouse model may also be able to determine the threshold levels of tafazzin expression/function needed to reverse different aspects of the overall phenotype.
As expected, after 2 months of silencing, taz knockdown caused a dramatic reduction of mature cardiolipins and a very robust increase of MLCLs and hence the MLCL/CL ratio in heart and skeletal muscle. These results are consistent with the existing data from patients with Barth syndrome.
Tafazzin deficiency and subsequent CL deficiency/alteration resulted in a significant increase of mtDNA content in heart and skeletal muscle, reflecting mitochondrial proliferation, which is often observed in cells and tissues obtained from Barth syndrome patients and other disorders of mitochondrial energy production such as fatty acid oxidation deficiency (39, 44). These increases were restricted to the heart and skeletal muscle and were not observed in liver or brain tissues of tafazzin-deficient mice. This tissue-specific effect may reflect the high level of energy demand of these highly oxidative tissues.
Human Barth syndrome is often characterized by an infantile onset of dilated cardiomyopathy. Conversely, the effect of taz knockdown and CL deficiency on mitochondrial crista architecture we observed was rather modest in 2-month-old mice. This surprising observation was supported by the lack of any significant defects in cardiac function in 2-month-old taz knockdown mice under basal conditions. However, this finding is in alignment with our recent discovery of gross mitochondrial structural abnormalities in flying muscle and the lack of any major defects in cardiac mitochondria in a Drosophila model of taz deficiency (5). It is plausible that applying additional stressors, such as exposure to cold, increased exercise, or inotropic drugs, would cause a cardiac phenotype to develop in 2-month-old taz-deficient mice.
As taz-deficient mice age and reach 8 months, however, morphological defects become more evident in their hearts. Cardiac sarcomeres become significantly thinner and exhibit disarray; mitochondria proliferate and become clustered with more variations in size and morphology. On electron micrographs, we observed overwhelming evidence of mitochondrial deterioration and mitophagy: onion skin-like formations and large voids inside mitochondria were abundantly present on electron micrographs of both ventricular and atrial muscle of 8-month-old taz-deficient hearts.
Impaired mitochondrial fusion and fission are detrimental to mitochondrial survival. Cardiolipin is required for anchoring, assembly, and GTPase activity of dynamin-related Mgm1/OPA1 protein that regulates mitochondrial fusion in the cell (10, 45). Therefore, CL abnormalities may negatively affect mitochondrial dynamics and survival.
Defective mitochondrial protein transport is also detrimental to mitochondrial survival. Cardiolipin participates in the translocation of precursor proteins into the mitochondrial intermembrane space and matrix (46, 47) and is necessary for membrane binding of yeast Tim44, the membrane anchor for the ATP-driven protein import engine. Moreover, mitochondrial protein quality control and removal of misfolded proteins and protein precursors depend upon cardiolipin-mediated assembly of the prohibitin-AAA+ protease complex in the inner mitochondrial membrane (48). Therefore, it is likely that cardiolipin abnormalities, along with reduced mitochondrial membrane potential and ATP production, diminish mitochondrial protein transport and recycling capacities and apply additional stress to mitochondria. These will be addressed in future studies.
We observed severe deterioration of MAMs in taz-deficient cardiomyocytes. These membranous vesicles are derived from ER positioned in close proximity (∼5 nm) to the outer mitochondrial membrane and serve as synapses between ER and mitochondria. MAM function is not well understood but is probably essential for bidirectional Ca2+ transport between mitochondria and ER. Calcium transport between these two organelles is crucial for buffering intracellular Ca2+ and regulating mitochondrial ATP production rate in response to changing energy demands. The nature of tethering of mitochondrion and ER calcium release units is unclear but is likely to involve SNARE complex proteins (49). There is no direct evidence that links CL with ER-mitochondrion interaction, although CL may mediate cross-talk between mitochondria and vacuoles in yeast (50). Defects in calcium handling in cardiomyocytes and cardiac conducting cells caused by the breakdown of ER-mitochondrion cross-communication may contribute to lethal ventricular arrhythmias reported in human Barth syndrome patients.
Unlike the heart, ultrastructural abnormalities in the taz-deficient skeletal muscle were obvious by the age of 2 months. It is remarkable that total cardiolipin content was not significantly affected by taz knockdown in EDL muscle (not shown), but the level of tetralinoleoyl CL was markedly reduced, and MLCL content was dramatically elevated. These findings suggest that acyl composition of the CL and/or MLCL level rather than just the CL content alone are important determinants in the tafazzin-deficient phenotype.
In conclusion, shRNA-mediated knockdown of tafazzin in mice causes a drastic reduction of tetralinoleoyl cardiolipin and accumulation of monolysocardiolipins in striated muscles. Cardiac muscle and skeletal muscle of tafazzin-deficient mice demonstrate ultrastructural defects, including mitochondrial proliferation, myofibrillar disarray, mitophagy, and abnormalities in MAMs. Tafazzin deficiency results in left ventricular dilation, loss of LV muscle mass, and reduction of cardiac function, all features of human Barth syndrome. Our mammalian model system will allow further characterization of the mechanisms underlying Barth syndrome phenotypes as well as testing of potential gene and drug therapies.
Supplementary Material
Acknowledgment
The Barth Syndrome Foundation funded the development the tafazzin knockdown mouse model of Barth syndrome and distributed it to interested researchers.
This work was supported by a grant from the Barth Syndrome Foundation (to Z. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, primer sequences, and PCR conditions.
- CL
- cardiolipin
- taz
- tafazzin
- MLCL
- monolysocardiolipin
- RMCE
- recombinase-mediated cassette exchange
- dox
- doxycycline
- mtDNA
- mitochondrial DNA
- IS
- internal standards
- LV
- left ventricular
- TG
- transgenic
- NTG
- non-transgenic
- EDL
- extensor digitorum longus
- MAM
- mitochondrion-associated membrane
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Schlame M., Ren M., Xu Y., Greenberg M. L., Haller I. (2005) Chem. Phys. Lipids 138, 38–49 [DOI] [PubMed] [Google Scholar]
- 2. Cheng H., Mancuso D. J., Jiang X., Guan S., Yang J., Yang K., Sun G., Gross R. W., Han X. (2008) Biochemistry 47, 5869–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gu Z., Valianpour F., Chen S., Vaz F. M., Hakkaart G. A., Wanders R. J., Greenberg M. L. (2004) Mol. Microbiol. 51, 149–158 [DOI] [PubMed] [Google Scholar]
- 4. Kiebish M. A., Bell R., Yang K., Phan T., Zhao Z., Ames W., Seyfried T. N., Gross R. W., Chuang J. H., Han X. (2010) J. Lipid Res. 51, 2153–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Acehan D., Khuchua Z., Houtkooper R. H., Malhotra A., Kaufman J., Vaz F. M., Ren M., Rockman H. A., Stokes D. L., Schlame M. (2009) Mitochondrion 9, 86–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kiebish M. A., Han X., Cheng H., Chuang J. H., Seyfried T. N. (2008) J. Lipid Res. 49, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klingenberg M. (2009) Biochim. Biophys. Acta 1788, 2048–2058 [DOI] [PubMed] [Google Scholar]
- 8. Lacombe M. L., Tokarska-Schlattner M., Epand R. F., Boissan M., Epand R. M., Schlattner U. (2009) Biochimie 91, 779–783 [DOI] [PubMed] [Google Scholar]
- 9. Schlattner U., Tokarska-Schlattner M., Ramirez S., Brückner A., Kay L., Polge C., Epand R. F., Lee R. M., Lacombe M. L., Epand R. M. (2009) Biochim. Biophys. Acta 1788, 2032–2047 [DOI] [PubMed] [Google Scholar]
- 10. Ban T., Heymann J. A., Song Z., Hinshaw J. E., Chan D. C. (2010) Hum. Mol. Genet. 19, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haines T. H., Dencher N. A. (2002) FEBS Lett. 528, 35–39 [DOI] [PubMed] [Google Scholar]
- 12. Gonzalvez F., Schug Z. T., Houtkooper R. H., MacKenzie E. D., Brooks D. G., Wanders R. J., Petit P. X., Vaz F. M., Gottlieb E. (2008) J. Cell Biol. 183, 681–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lesnefsky E. J., Hoppel C. L. (2008) Biochim. Biophys. Acta 1777, 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee H. J., Mayette J., Rapoport S. I., Bazinet R. P. (2006) Lipids Health Dis. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lesnefsky E. J., Chen Q., Slabe T. J., Stoll M. S., Minkler P. E., Hassan M. O., Tandler B., Hoppel C. L. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H258–H267 [DOI] [PubMed] [Google Scholar]
- 16. Lesnefsky E. J., Minkler P., Hoppel C. L. (2009) J. Mol. Cell. Cardiol. 46, 1008–1015 [DOI] [PubMed] [Google Scholar]
- 17. Sparagna G. C., Johnson C. A., McCune S. A., Moore R. L., Murphy R. C. (2005) J. Lipid Res. 46, 1196–1204 [DOI] [PubMed] [Google Scholar]
- 18. Zachman D. K., Chicco A. J., McCune S. A., Murphy R. C., Moore R. L., Sparagna G. C. (2010) J. Lipid Res. 51, 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han X., Yang J., Cheng H., Yang K., Abendschein D. R., Gross R. W. (2005) Biochemistry 44, 16684–16694 [DOI] [PubMed] [Google Scholar]
- 20. Heerdt P. M., Schlame M., Jehle R., Barbone A., Burkhoff D., Blanck T. J. (2002) Ann. Thorac. Surg. 73, 1216–1221 [DOI] [PubMed] [Google Scholar]
- 21. Sparagna G. C., Chicco A. J., Murphy R. C., Bristow M. R., Johnson C. A., Rees M. L., Maxey M. L., McCune S. A., Moore R. L. (2007) J. Lipid Res. 48, 1559–1570 [DOI] [PubMed] [Google Scholar]
- 22. Kiebish M. A., Han X., Cheng H., Chuang J. H., Seyfried T. N. (2008) Lipids 43, 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandra F., Degli Esposti M., Ndebele K., Gona P., Knight D., Rosenquist M., Khosravi-Far R. (2005) Cancer Res. 65, 8286–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valianpour F., Wanders R. J., Overmars H., Vreken P., Van Gennip A. H., Baas F., Plecko B., Santer R., Becker K., Barth P. G. (2002) J. Pediatr. 141, 729–733 [DOI] [PubMed] [Google Scholar]
- 25. Houtkooper R. H., Rodenburg R. J., Thiels C., van Lenthe H., Stet F., Poll-The B. T., Stone J. E., Steward C. G., Wanders R. J., Smeitink J., Kulik W., Vaz F. M. (2009) Anal. Biochem. 387, 230–237 [DOI] [PubMed] [Google Scholar]
- 26. Bione S., D'Adamo P., Maestrini E., Gedeon A. K., Bolhuis P. A., Toniolo D. (1996) Nat. Genet. 12, 385–389 [DOI] [PubMed] [Google Scholar]
- 27. Houtkooper R. H., Turkenburg M., Poll-The B. T., Karall D., Pérez-Cerdá C., Morrone A., Malvagia S., Wanders R. J., Kulik W., Vaz F. M. (2009) Biochim. Biophys. Acta 1788, 2003–2014 [DOI] [PubMed] [Google Scholar]
- 28. Seibler J., Kleinridders A., Küter-Luks B., Niehaves S., Brüning J. C., Schwenk F. (2007) Nucleic Acids Res. 35, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eggan K., Akutsu H., Loring J., Jackson-Grusby L., Klemm M., Rideout W. M., 3rd, Yanagimachi R., Jaenisch R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malhotra A., Edelman-Novemsky I., Xu Y., Plesken H., Ma J., Schlame M., Ren M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2337–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Houtkooper R. H., Akbari H., van Lenthe H., Kulik W., Wanders R. J., Frentzen M., Vaz F. M. (2006) FEBS Lett. 580, 3059–3064 [DOI] [PubMed] [Google Scholar]
- 32. Acehan D., Xu Y., Stokes D. L., Schlame M. (2007) Lab. Invest. 87, 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hinton R. B., Jr., Alfieri C. M., Witt S. A., Glascock B. J., Khoury P. R., Benson D. W., Yutzey K. E. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H2480–H2488 [DOI] [PubMed] [Google Scholar]
- 34. Seibler J., Küter-Luks B., Kern H., Streu S., Plum L., Mauer J., Kühn R., Brüning J. C., Schwenk F. (2005) Nucleic Acids Res. 33, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kelley R. I., Cheatham J. P., Clark B. J., Nigro M. A., Powell B. R., Sherwood G. W., Sladky J. T., Swisher W. P. (1991) J. Pediatr. 119, 738–747 [DOI] [PubMed] [Google Scholar]
- 36. Schlame M., Beyer K., Hayer-Hartl M., Klingenberg M. (1991) Eur. J. Biochem. 199, 459–466 [DOI] [PubMed] [Google Scholar]
- 37. Schlame M., Brody S., Hostetler K. Y. (1993) Eur. J. Biochem. 212, 727–735 [DOI] [PubMed] [Google Scholar]
- 38. Kulik W., van Lenthe H., Stet F. S., Houtkooper R. H., Kemp H., Stone J. E., Steward C. G., Wanders R. J., Vaz F. M. (2008) Clin. Chem. 54, 371–378 [DOI] [PubMed] [Google Scholar]
- 39. Bissler J. J., Tsoras M., Göring H. H., Hug P., Chuck G., Tombragel E., McGraw C., Schlotman J., Ralston M. A., Hug G. (2002) Lab. Invest. 82, 335–344 [DOI] [PubMed] [Google Scholar]
- 40. Boncompagni S., Rossi A. E., Micaroni M., Beznoussenko G. V., Polishchuk R. S., Dirksen R. T., Protasi F. (2009) Mol. Biol. Cell 20, 1058–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hawkins W. E., Howse H. D., Foster C. A. (1980) Cell Tissue Res. 209, 87–94 [DOI] [PubMed] [Google Scholar]
- 42. Svoboda D. J., Manning R. T. (1964) Am. J. Pathol. 44, 645–662 [PMC free article] [PubMed] [Google Scholar]
- 43. Wada J., Kanwar Y. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Exil V. J., Roberts R. L., Sims H., McLaughlin J. E., Malkin R. A., Gardner C. D., Ni G., Rottman J. N., Strauss A. W. (2003) Circ. Res. 93, 448–455 [DOI] [PubMed] [Google Scholar]
- 45. DeVay R. M., Dominguez-Ramirez L., Lackner L. L., Hoppins S., Stahlberg H., Nunnari J. (2009) J. Cell Biol. 186, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eilers M., Endo T., Schatz G. (1989) J. Biol. Chem. 264, 2945–2950 [PubMed] [Google Scholar]
- 47. Gebert N., Joshi A. S., Kutik S., Becker T., McKenzie M., Guan X. L., Mooga V. P., Stroud D. A., Kulkarni G., Wenk M. R., Rehling P., Meisinger C., Ryan M. T., Wiedemann N., Greenberg M. L., Pfanner N. (2009) Curr. Biol. 19, 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Gestel R. A., Rijken P. J., Surinova S., O'Flaherty M., Heck A. J., Killian J. A., de Kroon A. I., Slijper M. (2010) J. Proteomics 73, 806–814 [DOI] [PubMed] [Google Scholar]
- 49. Jägerström S., Polesie S., Wickström Y., Johansson B. R., Schröder H. D., Højlund K., Boström P. (2009) Cell Biol. Int. 33, 934–940 [DOI] [PubMed] [Google Scholar]
- 50. Chen S., Tarsio M., Kane P. M., Greenberg M. L. (2008) Mol. Biol. Cell 19, 5047–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.