Abstract
The lysyl oxidase family is made up of five members: lysyl oxidase (LOX) and lysyl oxidase-like 1–4 (LOXL1-LOXL4). All members share conserved C-terminal catalytic domains that provide for lysyl oxidase or lysyl oxidase-like enzyme activity; and more divergent propeptide regions. LOX family enzyme activities catalyze the final enzymatic conversion required for the formation of normal biosynthetic collagen and elastin cross-links. The importance of lysyl oxidase enzyme activity to normal bone development has long been appreciated, but regulation and roles for specific LOX isoforms in bone formation in vivo is largely unexplored. Fracture healing recapitulates aspects of endochondral bone development. The present study first investigated the expression of all LOX isoforms in fracture healing. A remarkable coincidence of LOXL2 expression with the chondrogenic phase of fracture healing was found, prompting more detailed analyses of LOXL2 expression in normal growth plates, and LOXL2 expression and function in developing ATDC5 chondrogenic cells. Data show that LOXL2 is expressed by pre-hypertrophic and hypertrophic chondrocytes in vivo, and that LOXL2 expression is regulated in vitro as a function of chondrocyte differentiation. Moreover, LOXL2 knockdown studies in vitro show that LOXL2 expression is required for ATDC5 chondrocyte cell line differentiation through regulation of SNAIL and SOX9, important transcription factors that control chondrocyte differentiation. Taken together, data provide evidence that LOXL2, like LOX, is a multifunctional protein. LOXL2 promotes chondrocyte differentiation by mechanisms that are likely to include roles as both a regulator and an effector of chondrocyte differentiation.
Keywords: Bone, Collagen, Connective Tissue, Differentiation, Enzymes, Extracellular Matrix Proteins, Lentivirus, shRNA, Chondrocytes, Lysyl Oxidases
Introduction
The lysyl oxidase family is made up of five members: lysyl oxidase (LOX)3 and lysyl oxidase-like 1–4 (LOXL1-LOXL4) (1). All five members share a conserved C-terminal catalytic domain that provides for lysyl oxidase or lysyl oxidase-like enzyme activity, and more divergent propeptide regions. LOX family enzyme activities catalyze the final enzymatic conversion required for the subsequent formation of normal lysine-derived biosynthetic cross-links found in collagens and elastin (2). The importance of lysyl oxidase enzyme activity to normal bone development has long been known, and is based in part on studies in which enzyme activities are inhibited by lathyrogens in vivo (3), including β-aminopropionitrile, a potent inhibitor of LOX and LOX isoform activity (4). The LOX family can be subdivided into two subgroups. Although all members have similarities in the catalytic C-terminal region, LOX and LOXL1 are more closely related to each other than they are to LOXL2-LOXL4. Moreover, the pro-peptide regions of LOX and LOXL1 have very little similarity to each other and no similarity to LOXL2-LOXL4. The pro-regions of LOXL2-LOXL4 each contain four scavenger receptor cysteine-rich (SRCR) domains whose functions are likely to depend on interactions with other proteins (1). The pro-regions of all LOX family members may have activities that are independent of lysyl oxidase enzyme activity, as has already been shown for the propeptide region of LOX (5–11). Although some information on regulation and functions of LOX in particular in osteoblast development in vitro is known (10, 12), and LOXL4 expression has been previously seen in cartilage and in osteoblast cell lines (13), relatively little information on regulation and roles for other specific LOX isoforms in bone formation in vivo is available.
Fracture healing in mice largely recapitulates endochondral bone development, temporal patterns of chondrocyte and osteoblast differentiation. Expression of corresponding molecular markers are well established (14, 15). An examination of the transcriptome of fracture healing through a large microarray study provided initial data that all five lysyl oxidase-related isoforms were expressed during fracture healing and that some of the isoforms would show specific expression at different developmental stages of healing (16). The present study was initiated to more fully evaluate the expression of all five isoforms in fracture healing. A remarkable coincidence of LOXL2 expression with the chondrogenic phase of fracture healing was found, prompting more detailed analyses of LOXL2 expression in normal growth plates, and LOXL2 expression and function in the ATDC5 cell model of chondrocyte differentiation. Data show that LOXL2 is expressed by pre-hypertrophic and hypertrophic chondrocytes in vivo, and that LOXL2 expression is regulated in vitro as a function of chondrocyte differentiation. Moreover, LOXL2 knockdown studies show that LOXL2 expression plays an essential role in chondrocyte differentiation, regulating levels of SNAIL and SOX9, important transcription factors that control chondrocyte differentiation.
EXPERIMENTAL PROCEDURES
Reagents
Dulbecco's modified Eagle's medium (DMEM), Dulbecco's modified Eagle's medium-F12, penicillin-streptomycin solution, nonessential amino acids, trypsin-EDTA solution, phosphate-buffered saline, sodium pyruvate, NanoOrange® protein quantitation kit, insulin, transferrin, selenite (ITS) were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was from Sigma. Ascorbate 2-phosphate was bought from Fluka Biochemica, Switzerland; cell culture plates were obtained from Corning Inc. (Corning, NY). RNeasy mini-RNA purification kits and Maxi prep kits were purchased from Qiagen (Valencia, CA). TaqMan probes and reverse transcription reagents for real-time PCR were obtained from Applied Biosystems (Foster City, CA). LOXL2 antibody for Western blots was purchased from Abcam (Cambridge, MA). For immunohistochemistry, LOXL2 antibody, biotinylated anti-rabbit IgG, biotinylated anti-goat IgG, non-immune rabbit IgG, and goat IgG was obtained from Santa Cruz Biotechnology. ABC reagents and diaminobenzidine (DAB) substrate, were obtained from Vector Laboratories (Burlingame, CA). Lentivirus constructs were bought from Open Biosystems (Huntsville, AL). Fugene 6 reagent was bought from Roche (Basel, Switzerland) and p24 ELISA kit was purchased from Cell Biolabs, Inc. (San Diego, CA).
Animals
Research was conducted in conformity with all Federal and USDA guidelines under an IACUC approved protocol at the Boston University School of Medicine in Boston, MA. For fracture studies, 8–10 week postbirth C57BL/6J (B6) male mice were obtained from the Jackson Laboratory and housed at the Boston University Medical Center animal housing facility for the duration of each study plus 3 days of acclimation. Growth plate studies were carried out on days 7 and 14 postnatal male mice.
Fracture Model
Unilateral fractures were produced in the right femur of 8–10-week-old male mice as previously described (17). The location and quality of fractures was assessed by x-ray analysis while animals were still anesthetized after surgery. Fracture configurations that were comminuted or were not localized to the mid-diaphyseal region were excluded from the study. Days 10 and 14 time points during fracture healing were chosen for the histological studies because the B6 mouse strain has a corresponding peak period of cartilage formation and hypertrophic chondrocyte differentiation (17). For molecular biological studies of fracture healing, mRNA was extracted from callus tissues on day 0 (no fracture) and at intervals up to day 21 of healing.
Histomorphometry Analyses
For histological assessment of fractures, femora with surrounding muscle and soft tissues were fixed, decalcified, sectioned, and stained as previously described (18). For growth plate assessments, femora from either 7- or 14-day-old mice were used.
Cell Culture
ATDC5 cells were cultured in DMEM-F12 Dulbecco's modified Eagle's medium-F12 supplemented with 10% fetal bovine serum and 50 units/ml penicillin and 50 μg/ml streptomycin, in a humidified atmosphere of 37 °C and 5% CO2. For differentiation studies, ATDC5 cells in DMEM-F12 supplemented with 10% FBS were seeded in 6-well tissue culture plates until they reached full confluence. Seven days after reaching visual confluence, culture media were replaced with medium containing in addition 10 μg/ml bovine insulin, 10 μg/ml human transferrin, 3 × 10−8 m sodium selenite (GIBCO®; ITS) and 37.5 μg/ml of ascorbate 2-phosphate. Media were changed every other day throughout the length of the experiment.
RNA Isolation and Real-time qPCR
Total RNA from ATDC5 cells was extracted using the RNeasy® Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Briefly, culture media were aspirated completely from culture plates, cells were disrupted by addition of buffer RLT, and the cell layer was collected. The cell lysate was transferred directly into a Qiashredder spin column, placed in a collection tube and centrifuged at full speed for 2 min. One volume of 70% ethanol was added to the homogenized lysate and mixed by pipetting. Then the sample was transferred into an RNeasy spin column for RNA binding, followed by washing with buffer RW1 and buffer RPE. Bound RNA was eluted in RNase-free water. RNA samples were then stored at −80 °C. RNA samples from fractured femurs, including early fracture callus were harvested at different time points as described (19). RNA obtained was analyzed on 1% agarose gels to ensure RNA quality by visualizing 18 S and 28 S rRNA in proper relative proportions after ethidium bromide staining. RNA was quantified via spectrophotometry (NanoDrop, Thermo Fisher Scientific, Pittsburgh, PA), and 1 μg of RNA per treatment condition was added to 30 μl of reverse transcription reactions (1× RT buffer, 5.5 mm MgCl2, 500 μm per dNTP, 2.5 μm random hexamers, 0.4 units/μl RNase inhibitor, and 3.125 units/μl MultiScribe reverse transcriptase) using the Applied Biosystems Reverse Transcription kit. The reverse transcription thermal cycling conditions are: 25 °C for 10 min, 37 °C for 60 min and 95 °C for 5 min, followed by 4 °C on hold. cDNA was stored at −20 °C prior to use in real-time PCR. 2 μl of each reverse transcription reaction was used for 25 μl of real-time PCR reaction (12.5 μl of Taqman Universal PCR master mix, 1.25 μl specific primer and 2 μl of template) using the 96-well format. TaqMan probe sets (Applied Biosystems, Foster City, CA) employed were for LOX (Mm00495386), LOXL1 (Mm01145738), LOXL2 (Mm00804740), LOXL3 (Mm00442953), LOXL4 (Mm00446385), SOX9 (mm00448840_m), COLII (Mm00491889), COLX (Mm00487041), and Aggrecan (Mm00545794). GAPDH (Mm99999915) was used as an endogenous control. The fold change in the target gene, normalized to endogenous control and relative to the expression of the calibrator sample, was calculated by the 2−ΔΔct method (20).
Western Blot
Total cell extracts from ATDC5 cells at different time intervals were obtained by lysing cells in SDS-PAGE sample buffer (62.5 mm Tris, 10% glycerol, 2% SDS, and 5% β-mercaptoethanol). Protein concentrations were determined with the Nano-Orange protein quantitation kit using bovine serum albumin as standard following the manufacturer's instructions. Protein samples (20 μg) were subjected to 8% SDS-PAGE, and were transferred onto polyvinylidene difluoride membranes (PVDF Perkin Elmer, Boston, MA) overnight in blotting buffer (0.025 m Tris, 0.192 m glycine, and 20% methanol). Membranes were blocked in 5% dry milk dissolved in TBST (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% Tween), for 1 h. Then, the membranes were incubated with blocking solution containing goat anti-LOXL2 antibody (Abcam, Cambridge, MA) (1:400) and kept overnight at 4 °C with mild shaking. Membranes were washed three times with TBS-T for 10 min each and then incubated with horseradish peroxidase-coupled secondary antibodies (1:2500) for 1 h in blocking solution at room temperature with light shaking, and washed with TBS-T three times for 10 min each. Chemiluminescent detection of bound horseradish peroxidase-conjugated secondary antibodies was determined using the ECL Western blotting Detection Reagents (Denville Scientific, Metuchen, NJ) and exposed to film. Membranes were subsequently stripped using Restore Western Stripping Solution (Thermo Scientific/Pierce, Pittsburgh, PA) and re-probed with β-actin antibody (Cell Signaling) for loading control as required. Densitometry was carried out only on films with non-saturating exposures utilizing a Bio-Rad Versadoc Photodocumentation System and Quantity One software.
Immunohistochemistry
Femurs of 9-week-old mice were fractured as described, and tissues collected at intervals of healing (19). Legs from uninjured mice were obtained from 7- and 14-day-old mice, for analysis of epiphyseal growth plates. Samples were fixed and then embedded in paraffin. Paraffin sections (5 μm) were deparaffinized in xylene, and rehydrated through graded alcohols. Microwave heating in 10 mm sodium citrate buffer at pH 6, was used for endogenous antigen retrieval, and then sections were allowed to cool for 20 min. Immunohistochemistry was next carried out as we have previously described in detail (21), but with the following antibodies: primary antibodies were affinity-purified rabbit anti LOX-PP antibody (22), (1:250) and goat-anti LOXL2 antibody (1:25) (Santa Cruz Biotechnology). Secondary antibodies utilized were biotinylated anti-rabbit IgG for LOX-PP and biotinylated anti-goat IgG for LOXL2. Non-immune rabbit IgG and goat IgG (Santa Cruz Biotechnology) served as negative controls for corresponding primary antibodies. Slides were incubated overnight with primary antibodies at 4 °C, and were then processed and photographed as previously described (21).
Lentivirus Preparation
All work with lentivirus was performed under BL2 conditions. Viral constructs were bought from Open Biosystems (Huntsville, AL) except for the non-target construct (23) that was obtained via Addgene (Cambridge, MA) and plasmids were isolated using Qiagen Maxi prep kit according to the manufacturer's protocol (Qiagen). The shRNA sequences were: Virus 8 (TRCN0000076708) CCGGGCTGAGAAGAAAGGTGCTCATCTCGAGATGAGCACCTTTCTTCTCAGCTTTTTG; Virus 9 (TRCN0000076709) CCGGGCATGGAAATATCTTCGCCAACTCGAGTTGGCGAAGATATTTCCATGCTTTTTG; Virus 10 (TRCN0000076710) CCGGCCTGGTGCTTAATGCTGAGATCTCGAGATCTCAGCATTAAGCACCAGGTTTTTG; Virus 11 (TRCN0000076711) CCGGCCAAATAGAGAGCCTAAATATCTCGAGA-TATTTAGGCTCTCTATTTGGTTTTTG; Virus 12 (TRCN0000076712) CCGGCAACCAAATAGAGAGCCTAAACTCGAGTTTAGGCTCTCTATTTGGTTGTTTTTG; non-target control virus (plasmid 1864) CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTT-AACCTTAGG.
Lentiviruses were produced by plating 293-T cells at a density of 2 × 106 cells per 10 cm2 at 37 °C under 5% CO2 in DMEM high glucose media, supplemented with 10% FBS and 100 units/ml of penicillin, 100 μg/ml of streptomycin. Cells were co-transfected using Fugene 6 reagent (150 μl) (Roche), and 3 plasmids: lentivirus plasmid, pcMV-VSV-G and pCMV-dR8.2 dvpr at the ratio of 8:1:8. Non-Target lentivirus plasmid served as a negative control (24). The supernatant fluid was collected at 24-h intervals for three consecutive days and was replaced with fresh differentiation media (35 ml). Virus particles were concentrated by ultracentrifugation at 16,500 × g for 90 min and were resuspended in PBS and stored at −70 °C. Virus titer was determined by using a p24 ELISA kit (Cell Biolabs, Inc.) according to the manufacturer's protocol. ATDC5 cells were plated in 6-well plates. Seven days after confluence, the lentivirus transduction was carried out in differentiation media containing 8 μg/ml hexadimethrine bromide. The media were changed the next day and then every other day thereafter.
Alcian blue staining of ATDC5 cultures was performed after fixation of ATDC5 cells with 4% paraformaldehyde for 30 min and washed with water three times and air dried. The staining was performed with the commercially available 1% Alcian Blue solution, pH 2.5 (American Mastertech), for 5 h and washed several times with distilled water before being photographed (25). Alizarin red staining was carried out as described previously (10).
Statistics
Two way ANOVA with Bonferroni post-tests were performed using GraphPad Prism (San Diego) software version 5.02 for Windows; data were considered significant at p < 0.05.
RESULTS
Expression Pattern of LOX Isoforms during Fracture Healing
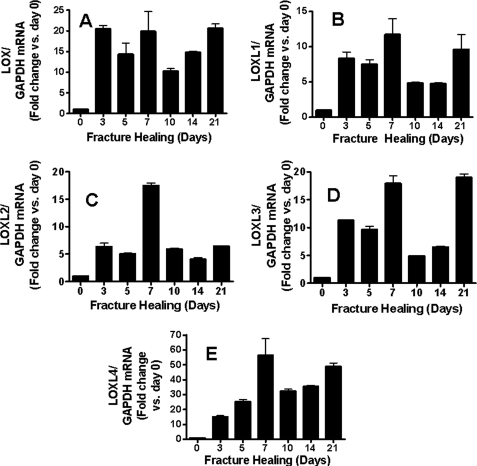
The expression pattern of lysyl oxidase, and its isoforms during fracture repair was first determined. Total RNA samples from mouse fractured femur calluses were extracted at different time points after fracture, and samples were subjected to qPCR. Results show that mRNA levels of LOX, LOXL1, LOXL3, and LOXL4 all had similar bi-phasic expression patterns throughout the healing process with peaks of expression on days 7 and 21 (Fig. 1, A, B, D, and E). The expression pattern for LOXL2, however, was found to be different from the other isoforms with a peak of expression seen on day 7 after fracture (Fig. 1C). This time point corresponds to the onset of the chondrogenic phase of fracture healing (26). We, therefore, developed the hypothesis that LOXL2 could have unique importance in chondrogenesis.
FIGURE 1.
LOXL2 expression as a function of fracture healing is unique among LOX isoforms. Expression of LOX mRNA (A) and its isoforms LOXL1-LOXL4 (B–E, respectively) in normal fracture healing was determined by real-time qPCR. RNA was collected from healing fractures from 3 mice per time point and RNAs pooled; LOX and LOXL1–4 mRNA expressions were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA levels. Data are expressed as fold change relative to day 0.
LOXL2 Protein Is Expressed Predominantly by Chondrocytes in Healing Fractures and in Epiphyseal Growth Plates
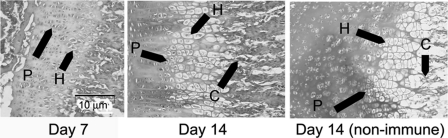
To investigate directly whether LOXL2 is expressed by chondrocytes in vivo, immunohistochemistry studies were carried out on healing fractures introduced into 10-week-old mice and were analyzed on post-fracture days 10 and 14, and on normal epiphyseal growth plates on postnatal days 7 and 14. In healing fractures, LOXL2 protein was strongly detected in chondrocytes abundant on day 10 of healing (supplemental Fig. S1). LOX staining was found to be strongest in osteoblasts lining bone that were highly abundant on day 14, while LOX expression is weaker in chondrocytes. Weaker LOXL2 staining is seen in cells lining bone (osteoblasts) at all time points (supplemental Fig. S2). Similarly, in epiphyseal growth plates, strong staining for LOXL2 was detected in chondrocytes present within the hypertrophic and proliferating zones, and in calcified cartilage (Fig. 2). These findings suggest that LOXL2 is a predominant lysyl oxidase isoform made by chondrocytes in vivo.
FIGURE 2.
LOXL2 expression in epiphyseal growth plates. Paraffin-embedded sections of the growth plates from 7- and 14-day-old mice were immunostained for LOXL2 as described under “Experimental Procedures.” Positive staining is seen in proliferating (P), hypertrophic (H), and calcifying (C) chondrocytes. A day 14 non-immune IgG control counterstained with hematoxylin is shown; scale bar, 10 μm, photographed at 20× magnification.
LOXL2 Expression Is Regulated as a Function of Chondrocyte Differentiation
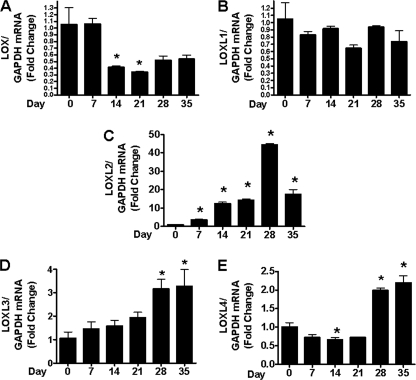
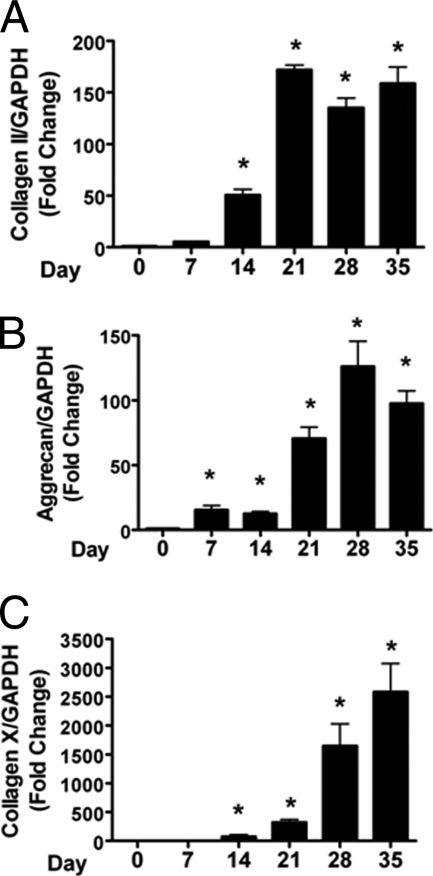
We next wished to determine whether LOXL2 expression is regulated as a function of chondrocyte differentiation. The ATDC5 cell line undergoes a well characterized program of chondrocyte differentiation (27). Cells were plated in 6-well plates in DMEM-F12 growth medium. Seven days after visual confluence, the growth medium was supplemented with ascorbate and insulin-transferrin-selenite solution. Total RNA was isolated at different time points and subjected to quantitative real time PCR. LOX, LOXL1, LOXL3, and LOXL4 mRNA levels showed small changes in expression compared with the robust changes in LOXL2 (Fig. 3). Total mRNA expression of LOXL2 in differentiating ATDC5 cells showed an approximate 4.5-fold increase by day 7, increasing to 12-and 14-fold by day 14–21, and then dramatically reaching its highest level of 43-fold on day 28, decreasing to 18-fold on day 35 (Fig. 3C). To investigate LOXL2 expression in relation to well characterized chondrogenic markers, we examined the expression of collagen type II, X, and aggrecan. Expression of chondrogenic markers was dramatically increased with time as expected (Fig. 4). The sequence of expression of these chondrogenic markers followed the normal pattern of type II collagen expression preceding aggrecan, followed by expression of type X collagen (Fig. 4). Interestingly, LOXL2 expression was significantly elevated as early as day 7 of differentiation, but the peak of LOXL2 mRNA expression was later than the peak of type II collagen expression, but earlier than that of type X collagen.
FIGURE 3.
mRNA expression of LOXL2 is highly increased compared with LOX and other isoforms in differentiating ATDC5 cell line. Quantitative real-time PCR analyses of LOX, LOXL1, 2, 3, and 4, in differentiating ATDC5 cells. RNA was extracted at different time points. The mRNA expression relative to GAPDH was determined and the fold changes were calculated relative to day 0 (*, p < 0.05; ANOVA; n = 3).
FIGURE 4.
Quantitative real-time PCR analyses of (A) type II collagen, (B) aggrecan, and (C) type X collagen in differentiating ATDC5 cells. RNA was extracted at different time points. The mRNA expression relative to GAPDH was determined, and the fold changes were calculated relative to day 0. (*, p < 0.05; ANOVA; n = 3).
LOX and LOXL2 Protein Expression in Differentiating ATDC5 Cells
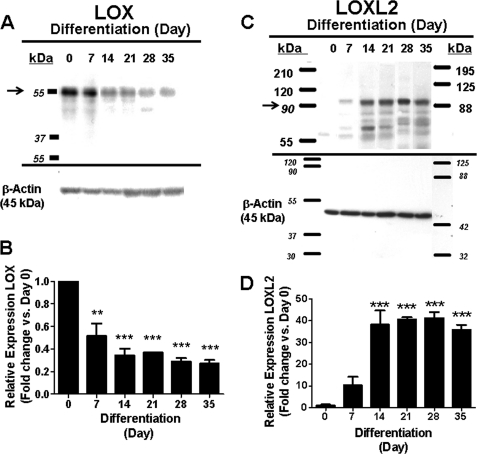
We next wished to determine LOX and LOXL2 protein expression in developing ATDC5 cells. Cells were grown to full confluence and then induced to differentiate as described above. Cells layers were extracted into SDS-PAGE sample buffer at intervals, and equal amounts of proteins were subjected to SDS-PAGE followed by immunoblotting for LOX and LOXL2. Data show that LOX proenzyme levels decrease with differentiation, whereas LOXL2 pro-protein increases ∼40-fold during the first 14 days of differentiation, and remains high through day 35 (Fig. 5). These data are consistent with mRNA regulation seen in Fig. 3.
FIGURE 5.
LOXL2 and LOX protein levels in differentiating ATDC5 cell layers. ATDC5 cells were grown, and protein was extracted from cell layers at different time points, and equal protein samples subjected to Western blotting and, respectively, probed for (A) LOX, and (C) LOXL2; and after stripping for β-actin for normalization. LOX was expressed more in early phases of differentiation. Representative Western blots of three independent experiments are shown. Quantitative densitometry normalized to β-actin is shown for LOX (B), and for LOXL2 (D). (n = 3; **, p < 0.01; ***, p < 0.001; two way ANOVA).
LOXL2 Knockdown
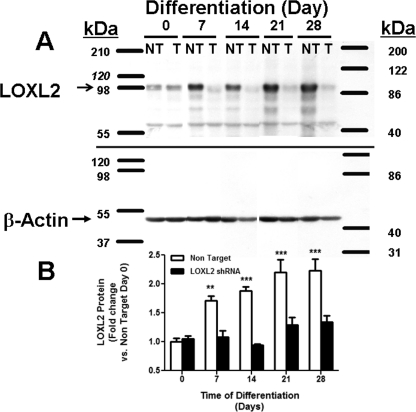
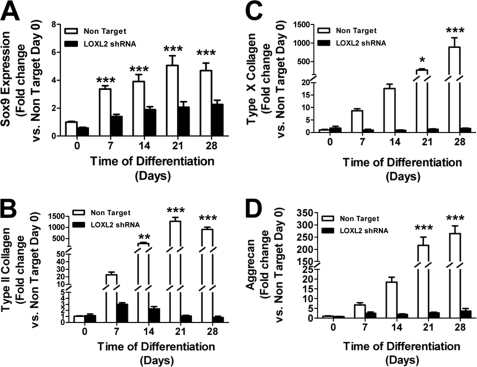
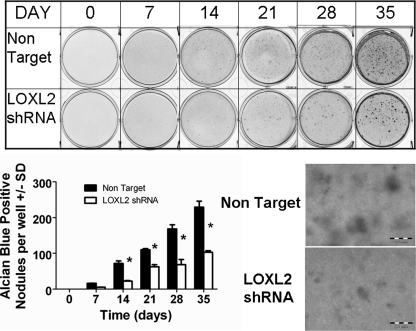
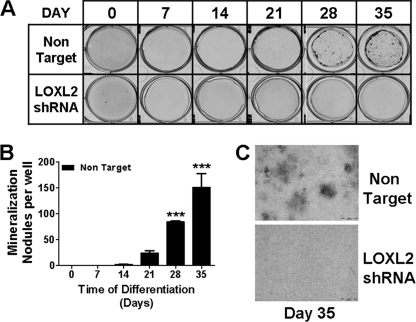
We next wished to determine if LOXL2 plays any role in chondrocyte differentiation, or whether the regulated expression of LOXL2 is simply a consequence of chondrocyte differentiation. Five commercially available lentiviral LOXL2 shRNA constructs were screened for their ability to prevent LOXL2 mRNA increases on day 7 of differentiation. Lentiviruses were produced by co-transfection of 293-T cells with 3 plasmid vectors (lentivirus, pcMV-VSV-G, and pCMV-dR8.2 dvpr) at a ratio of 8:1:8. A Non-Target Virus served as a control (23). As virus 11 gave the most effective knockdown (data not shown), all subsequent studies were performed with this virus, and the non-target virus control. This shRNA clone has no potential to directly knockdown other LOX isoforms, as confirmed by BLAST analyses (supplemental Fig. S3) that shows no common sequence with any other lysyl oxidase isoform, or sufficient similarity to any murine mRNA or gene coding sequence (28, 29). Data in Fig. 6 demonstrate that LOXL2 protein levels are diminished in differentiating ATDC5 cells compared with the non-target virus control, confirming that the shRNA lentivirus successfully diminished LOXL2 production. We next determined the effect of LOXL2 knockdown on the expression of chondrocyte-specific differentiation marker mRNAs. Data in Fig. 7 show remarkable inhibition of expression of the chondrocyte differentiation markers: SOX9, type II collagen, type X collagen, and aggrecan. Alcian blue (Fig. 8) and alizarin red (Fig. 9) staining of differentiating LOXL2 knockdown and non-target lentivirus transduced ATDC5 cells show nearly complete inhibition of chondrocyte-like extracellular matrix, and mineralized nodule formation in LOXL2 knockdown cultures. Interestingly, though LOXL2 shRNA clone 11 is specific LOXL1, LOXL3, and LOXL4, but not LOX mRNAs, were also significantly down-regulated compared with non-target shRNA (supplemental Fig. S4, two way ANOVA). Taken together, these data indicate that regulated expression of LOXL2 helps to control differentiation of ATDC5 chondrocytes, and that LOXL2 expression is not simply a consequence of ATDC5 chondrocyte differentiation. Interestingly, LOXL2 down-regulation appears to have secondary effects on regulation of other LOX isoforms.
FIGURE 6.
LOXL2 protein expression is inhibited by LOXL2 shRNA in differentiating ATDC5 cells. Cell layers were isolated and subjected to Western blotting using a LOXL2 antibody (A); NT, non target virus; T, LOXL2 shRNA virus. Expression of β-actin was measured for normalization. A representative blot of three independent experiments is shown. Data in B are combined from all three experiments (**, p < 0.005; ***, p < 0.0005, two way ANOVA).
FIGURE 7.
Chondrocyte differentiation markers gene expression in ATDC5 cells infected with LOXL2 shRNA lentivirus. A lentivirus knock down of LOXL2 was introduced in differentiating ATDC5 cells. RNA was extracted at day 0, 7, 14, 21, and 28, and then subjected to qPCR for expression of differentiation markers (A) SOX9, (B) type II collagen, (C) type X collagen, (D) aggrecan. All differentiation markers were significantly decreased in the ATDC5 cell line transduced with virus when compared with the non target control virus. The values shown are the average of two independent experiments done with three independent samples each (*, p < 0.05; **, p < 0.005; ***, p < 0.0005, two way ANOVA).
FIGURE 8.
Cartilage-like extracellular matrix accumulation is inhibited by LOXL2 shRNA. Top, LOXL2 and non-target lentivirus transduced ATDC5 cells were cultured, and at intervals fixed and stained with Alcian Blue as described under “Experimental Procedures.” Representative images are shown. Lower left, all stained nodules in each well were counted under a low power microscope. The values shown are the average of three independent samples (*, p < 0.05; **, p < 0.005; ***, p < 0.0005, two way ANOVA) and demonstrate that LOXL2 shRNA blocks nodule formation. Lower right, higher power image of nodules comparing the Non Target cells to the LOXL2 shRNA cells on day 35 showing mineralization nodules only in the Non Target panel. The scale bar is 0.1 mm long.
FIGURE 9.
Mineralization of ATDC5 cells is blocked by LOXL2 shRNA. A, LOXL2 and non-target lentivirus transduced ATDC5 cells were cultured, and at intervals fixed with 10% buffered formalin and stained with Alizarin Red. Representative images are shown. B, all mineralization nodules in each well were counted under a low power microscope. The values shown are the average of three independent samples (*, p < 0.05; **, p < 0.005; ***, p < 0.0005, two way ANOVA) and demonstrate that LOXL2 shRNA blocks nodule formation. C, a higher power image of nodules comparing the Non Target cells to the LOXL2 shRNA cells on day 35 showing mineralization nodules only in the Non Target panel. The scale bar is 0.1 mm long.
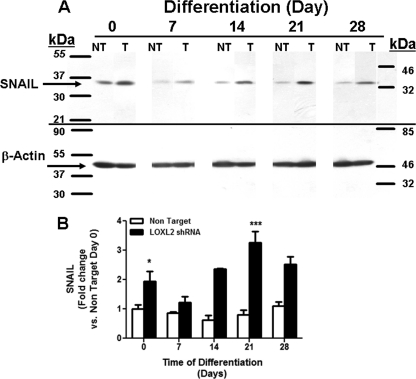
To investigate pathways and potential mechanisms of action by which LOXL2 regulates chondrocyte differentiation, we determined the expression of SNAIL in LOXL2 knockdown cells. SNAIL is a transcription repressor whose diminished expression as a function of chondrocyte differentiation permits chondrocyte differentiation (30, 31). SNAIL has been shown previously to bind to LOXL2, resulting in the stabilization of SNAIL protein and promotion of epithelial to mesenchymal transition in some breast cancer cell lines (32). Interestingly, knockdown of LOXL2 resulted in a potent up-regulation of SNAIL protein levels in ATDC5 cells (Fig. 10), consistent with the effect of LOXL2 on inhibiting chondrocyte differentiation seen here, but in apparent contrast to what occurs in cancer and cancer cell lines (33, 34). This is an interesting finding in light of the observation that LOXL2 also down-regulated SOX9 (Fig. 7A). SOX9 is a transcription factor and upstream master regulator of chondrogenesis. SOX9 is regulated independent of the inhibitory action of the more downstream transcription repressor SNAIL (30). These findings suggest that LOXL2 plays roles in both the positive regulation of transcription factors that promote chondrogensis, and in maintaining correct levels of transcriptional repressors.
FIGURE 10.
SNAIL protein expression is increased by silencing LOXL2 in ATDC5 cells. Cell layers of differentiating non target and LOXL2 shRNA transduced ATDC5 cells were isolated at intervals and subjected to Western blotting for SNAIL. Expression of β-actin served as the internal control. A, representative Western blot; B, densitometric analysis of SNAIL/β-actin. The values shown are the average of three independent samples (*, p < 0.05; ***, p < 0.0005, two way ANOVA).
DISCUSSION
The amine oxidase catalytic activity of LOX is the final enzyme-catalyzed reaction required for the biosynthetic cross-linking of collagen and elastin molecules resulting in stabilization and normal function in the extracellular matrix (35). The importance of LOX activity and LOX-dependent cross-linking has been extensively studied in many tissues including cardiovascular tissues, skin, and bone (12, 36–38). Similarly, the role and importance of LOX dependent cross-links in cartilage is known. The content of type II collagen and of non-reducible collagen cross-links increases during cartilage formation and growth. Moreover, these increases are associated with increased tensile biomechanical properties and the functional integrity of cartilage tissues (39, 40). Mechanical properties of mature cartilage tissue have been attributed to the composition and organization of its collagen network (41, 42). Studies of cartilage explants designed to investigate integrative repair in vitro, show that treatment with BAPN, a specific lysyl oxidase enzyme activity inhibitor (4), results in increased collagen extractablility with a concurrent reduction of dihydroxylysinonorleucine cross-links. BAPN treated explants also display markedly inhibited functional integrative cartilage repair in vitro (43). Cartilage explants pretreated with BAPN, followed by its withdrawal before a period of integrative repair resulted in an ultimate increase in integrative strength of the explant (44). The authors suggest that BAPN pre-treatment resulted in an accumulation of “primed” collagen molecules containing unmodified hydroxylysine residues that were, upon BAPN withdrawal, rapidly oxidized by LOX or a lysyl oxidase isoform(s) (44, 45). Taken together, these studies emphasize that collagen cross-linking and organization resulting from lysyl oxidase activity is required for the correct generation and maintenance of a cartilage-containing extracellular matrix. Hence, studying the expression pattern of lysyl oxidase and its isoforms that are each potentially responsible for generation of these cross-links in cartilage is of considerable importance.
The present study first analyzed the mRNA expression pattern of lysyl oxidase and its isoforms in samples collected from mouse fracture calluses at different time points. Fracture healing entails a chondrogenic phase, producing a template for osteogenesis (46). LOXL2 expression exhibited a striking temporal expression pattern, peaking around day 7 post fracture, a time point that correlates positively with the chondrogenic phase of healing. This finding led to our further analyses of LOXL2 protein expression in healing fractures, in growth plates, and LOXL2 mRNA and protein expression in differentiating chondrogenic ATDC5 cells. Findings of LOXL2 expression primarily in chondrocytes in vivo, and a robust increasing expression of LOXL2 as a function of differentiation in vitro led us to ask whether a deficiency of LOXL2 would alter chondrocyte development in vitro. Indeed, shRNA knockdown of LOXL2 compared with a non-target virus in the ATDC5 cell line completely blocks the expression of chondrocyte differentiation markers and mineralization in vitro.
Chondrocyte differentiation depends upon regulation of the activity of several different transcription factors and transcription repressors that coordinate balances between proliferation and extracellular matrix synthesis. SNAIL is a transcriptional repressor that is expressed at high levels during proliferative phases of chondrocyte development, and its expression decreases as type II collagen expression increases. Overexpression of SNAIL blocks synthesis of type II collagen, and this occurs by transcriptional repression of type II collagen via an E box cis-acting element in its promoter (31). SNAIL does not, however, block the expression of SOX9, a master upstream transcription factor that drives chondrocyte differentiation (30, 31, 47). The finding that LOXL2 knockdown also inhibits upstream SOX9 expression in the ATDC5 chondrogenic cell line indicates that LOXL2 normally promotes chondrocyte differentiation by mechanisms that must include a target upstream of SOX9 expression, perhaps by mechanisms independent of those that regulate SNAIL. For example, FGFR3 signaling up-regulates SNAIL expression, and this pathway is an essential SOX9-independent component of SNAIL regulation in normal chondrogenesis (30). Similarly, because transcription factors are ultimately regulated by growth factor signaling, and the TGF-β superfamily of growth factors is particularly important in the chondrogenic phase of fracture healing (15), it is tempting to speculate that LOXL2 is directly or indirectly involved in TGF-β superfamily growth factor signaling.
At this time it is unclear if LOXL2 amine oxidase activity required for collagen cross-linking is most relevant to its role in supporting chondrocyte differentiation, or whether novel LOXL2 functions that are independent of extracellular matrix cross-linking may be involved. For example, a recent study implicates LOX oxidation of a PDGF receptor as a positive regulator of PDGF signaling (48), while LOX oxidation of TGF-β and FGF-2 have been reported and are seen as inhibitory to their respective functions (49, 50). To add to the complexity, it is noted that the released propeptide of LOX itself has important biological activities that are independent of enzyme activity (5–8, 10, 11, 51). Thus, both enzyme activity dependent and independent mechanisms of LOXL2 may be relevant to its effects on chondrocyte development. The LOXL2 N-terminal region, unlike LOX and LOXL1, contains four SRCR domains (1). The SRCR superfamily is comprised of proteins that are either secreted or found on the cell surface and reports suggest that these domains may be involved in protein-protein interactions that can help in cell adhesion and signaling. These SRCR domains could play important roles in some functions of LOXL2. It is now clear that all LOX isoforms including LOXL2 are inhibited by BAPN (52). LOX isoform-specific enzyme activity inhibitors are needed to determine the relative contributions of each LOX isoform enzyme activity to biological effects of members of this multifunctional family of proteins.
It is interesting that studies in tumor cells have found that ectopically expressed LOXL2 can bind to SNAIL, and that this interaction was seen to stabilize SNAIL by repressing E-cadherin expression and promoting epithelial to mesenchymal transition in MDCK cells and in a breast cancer cell line, thereby promoting an invasive phenotype (32). The present study now shows that inhibition of LOXL2 levels in ATDC5 cells up-regulates SNAIL. As noted, this is accompanied by consequent inhibition of all other measured endpoints related to chondrocyte differentiation. The molecular basis for the opposite effects of LOXL2 on SNAIL levels in different cell systems is unknown, but is likely to be related to the invasive properties of cancer cells on the one hand, and the extracellular matrix producing function of fibrogenic cells on the other.
In summary, the present study shows for the first time that LOXL2 expression is regulated in a temporal manner during fracture repair in vivo. The LOXL2 expression pattern both in healing fractures and in developing growth plates indicate that LOXL2 is abundantly expressed in chondrocytes. In vitro differentiation of chondrogenic ATDC5 cells shows that LOXL2 has a strong temporal regulation pattern consistent with a role in type II collagen maturation. Most important, knockdown of LOXL2 results in inhibition of ATDC5 chondrocyte development in vitro, accompanied by increased SNAIL expression and decreased SOX9 expression. Together, these findings indicate that LOXL2 is likely to play a vital role in chondrogenesis, and that a greater understanding of LOXL2 activities and functions will provide potential new avenues to investigate the control of cartilage formation, growth, metabolism, and repair.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NIDCR R01 DE 14066 (to P. C. T.) and NIAMS P01AR049920 and AR044927 (to L. C. G.), and the Dept. of Defense Grant DAMD17-03-1-0576 (to L. C. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- LOX
- lysyl oxidase
- ANOVA
- analysis of variance
- SRCR
- scavenger receptor cysteine-rich.
REFERENCES
- 1. Csiszar K. (2001) Prog. Nucleic Acids Res. Mol. Biol. 70, 1–32 [DOI] [PubMed] [Google Scholar]
- 2. Kagan H. M., Trackman P. C. (1991) Am. J. Respir. Cell Mol. Biol. 5, 206–210 [DOI] [PubMed] [Google Scholar]
- 3. Selye H. (1957) Rev. Can. Biol. 16, 1–82 [PubMed] [Google Scholar]
- 4. Tang S. S., Trackman P. C., Kagan H. M. (1983) J. Biol. Chem. 258, 4331–4338 [PubMed] [Google Scholar]
- 5. Min C., Kirsch K. H., Zhao Y., Jeay S., Palamakumbura A. H., Trackman P. C., Sonenshein G. E. (2007) Cancer Res. 67, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 6. Min C., Yu Z., Kirsch K. H., Zhao Y., Vora S. R., Trackman P. C., Spicer D. B., Rosenberg L., Palmer J. R., Sonenshein G. E. (2009) Cancer Res. 69, 6685–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Palamakumbura A. H., Jeay S., Guo Y., Pischon N., Sommer P., Sonenshein G. E., Trackman P. C. (2004) J. Biol. Chem. 279, 40593–40600 [DOI] [PubMed] [Google Scholar]
- 8. Palamakumbura A. H., Vora S. R., Nugent M. A., Kirsch K. H., Sonenshein G. E., Trackman P. C. (2009) Oncogene 28, 3390–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vora S. R., Palamakumbura A. H., Mitsi M., Guo Y., Pischon N., Nugent M. A., Trackman P. C. (2010) J. Biol. Chem. 285, 7384–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vora S. R., Palamakumbura A. H., Mitsi M., Guo Y., Pischon N., Nugent M. A., Trackman P. C. (2010) J. Biol. Chem. 285, 2384–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu M., Min C., Wang X., Yu Z., Kirsch K. H., Trackman P. C., Sonenshein G. E. (2007) Cancer Res. 67, 6278–6285 [DOI] [PubMed] [Google Scholar]
- 12. Hong H. H., Pischon N., Santana R. B., Palamakumbura A. H., Chase H. B., Gantz D., Guo Y., Uzel M. I., Ma D., Trackman P. C. (2004) J. Cell. Physiol. 200, 53–62 [DOI] [PubMed] [Google Scholar]
- 13. Ito H., Akiyama H., Iguchi H., Iyama K., Miyamoto M., Ohsawa K., Nakamura T. (2001) J. Biol. Chem. 276, 24023–24029 [DOI] [PubMed] [Google Scholar]
- 14. Gerstenfeld L. C., Cullinane D. M., Barnes G. L., Graves D. T., Einhorn T. A. (2003) J. Cell. Biochem. 88, 873–884 [DOI] [PubMed] [Google Scholar]
- 15. Ai-Aql Z. S., Alagl A. S., Graves D. T., Gerstenfeld L. C., Einhorn T. A. (2008) J. Dent. Res. 87, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bais M., McLean J., Sebastiani P., Young M., Wigner N., Smith T., Kotton D. N., Einhorn T. A., Gerstenfeld L. C. (2009) PLoS One 4, e5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jepsen K. J., Price C., Silkman L. J., Nicholls F. H., Nasser P., Hu B., Hadi N., Alapatt M., Stapleton S. N., Kakar S., Einhorn T. A., Gerstenfeld L. C. (2008) J. Bone Miner. Res. 23, 1204–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerstenfeld L. C., Alkhiary Y. M., Krall E. A., Nicholls F. H., Stapleton S. N., Fitch J. L., Bauer M., Kayal R., Graves D. T., Jepsen K. J., Einhorn T. A. (2006) J. Histochem. Cytochem. 54, 1215–1228 [DOI] [PubMed] [Google Scholar]
- 19. Kayal R. A., Tsatsas D., Bauer M. A., Allen B., Al-Sebaei M. O., Kakar S., Leone C. W., Morgan E. F., Gerstenfeld L. C., Einhorn T. A., Graves D. T. (2007) J. Bone Miner Res. 22, 560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huggett J., Dheda K., Bustin S., Zumla A. (2005) Genes Immun. 6, 279–284 [DOI] [PubMed] [Google Scholar]
- 21. Uzel M. I., Kantarci A., Hong H. H., Uygur C., Sheff M. C., Firatli E., Trackman P. C. (2001) J. Periodontol. 72, 921–931 [DOI] [PubMed] [Google Scholar]
- 22. Hurtado P. A., Vora S., Sume S. S., Yang D., St Hilaire C., Guo Y., Palamakumbura A. H., Schreiber B. M., Ravid K., Trackman P. C. (2008) Biochem. Biophys. Res. Commun. 366, 156–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 24. Bais M. V., Wigner N., Young M., Toholka R., Graves D. T., Morgan E. F., Gerstenfeld L. C., Einhorn T. A. (2009) Bone 45, 254–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakatani S., Mano H., Im R., Shimizu J., Wada M. (2007) Biol. Pharm. Bull 30, 433–438 [DOI] [PubMed] [Google Scholar]
- 26. Ferguson C., Alpern E., Miclau T., Helms J. A. (1999) Mech. Dev. 87, 57–66 [DOI] [PubMed] [Google Scholar]
- 27. Atsumi T., Miwa Y., Kimata K., Ikawa Y. (1990) Cell Differ. Dev. 30, 109–116 [DOI] [PubMed] [Google Scholar]
- 28. Du Q., Thonberg H., Wang J., Wahlestedt C., Liang Z. (2005) Nucleic Acids Res. 33, 1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elbashir S. M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. (2001) EMBO J. 20, 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Frutos C. A., Vega S., Manzanares M., Flores J. M., Huertas H., Martínez-Frías M. L., Nieto M. A. (2007) Dev. Cell 13, 872–883 [DOI] [PubMed] [Google Scholar]
- 31. Seki K., Fujimori T., Savagner P., Hata A., Aikawa T., Ogata N., Nabeshima Y., Kaechoong L. (2003) J. Biol. Chem. 278, 41862–41870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peinado H., Del Carmen Iglesias-de la Cruz M., Olmeda D., Csiszar K., Fong K. S., Vega S., Nieto M. A., Cano A., Portillo F. (2005) EMBO J. 24, 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hollosi P., Yakushiji J. K., Fong K. S., Csiszar K., Fong S. F. (2009) Int. J. Cancer 125, 318–327 [DOI] [PubMed] [Google Scholar]
- 34. Peinado H., Moreno-Bueno G., Hardisson D., Pérez-Gómez E., Santos V., Mendiola M., de Diego J. I., Nistal M., Quintanilla M., Portillo F., Cano A. (2008) Cancer Res. 68, 4541–4550 [DOI] [PubMed] [Google Scholar]
- 35. Kagan H. (1986) in Biology and Regulation of Extracellular Matrix: A Series. Regulation of Matrix Accumulation (Mecham R. P. ed), pp. 321–398, Academic Press, Orlando [Google Scholar]
- 36. Mäki J. M., Räsänen J., Tikkanen H., Sormunen R., Mäkikallio K., Kivirikko K. I., Soininen R. (2002) Circulation 106, 2503–2509 [DOI] [PubMed] [Google Scholar]
- 37. Lees S., Eyre D. R., Barnard S. M. (1990) Connect Tissue Res. 24, 95–105 [DOI] [PubMed] [Google Scholar]
- 38. Oxlund H., Barckman M., Ortoft G., Andreassen T. T. (1995) Bone 17, 365S–371S [DOI] [PubMed] [Google Scholar]
- 39. Williamson A. K., Chen A. C., Masuda K., Thonar E. J., Sah R. L. (2003) J. Orthop. Res. 21, 872–880 [DOI] [PubMed] [Google Scholar]
- 40. Williamson A. K., Masuda K., Thonar E. J., Sah R. L. (2003) Tissue Eng. 9, 625–634 [DOI] [PubMed] [Google Scholar]
- 41. Bank R. A., Soudry M., Maroudas A., Mizrahi J., TeKoppele J. M. (2000) Arthritis Rheum. 43, 2202–2210 [DOI] [PubMed] [Google Scholar]
- 42. Basser P. J., Schneiderman R., Bank R. A., Wachtel E., Maroudas A. (1998) Arch. Biochem. Biophys. 351, 207–219 [DOI] [PubMed] [Google Scholar]
- 43. Ahsan T., Lottman L. M., Harwood F., Amiel D., Sah R. L. (1999) J. Orthop. Res. 17, 850–857 [DOI] [PubMed] [Google Scholar]
- 44. McGowan K. B., Sah R. L. (2005) J. Orthop. Res. 23, 594–601 [DOI] [PubMed] [Google Scholar]
- 45. DiMicco M. A., Sah R. L. (2001) J. Orthop. Res. 19, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 46. Ford J. L., Robinson D. E., Scammell B. E. (2004) J. Orthop. Res. 22, 368–375 [DOI] [PubMed] [Google Scholar]
- 47. Akiyama H. (2008) Mod. Rheumatol. 18, 213–219 [DOI] [PubMed] [Google Scholar]
- 48. Lucero H. A., Ravid K., Grimsby J. L., Rich C. B., DiCamillo S. J., Mäki J. M., Myllyharju J., Kagan H. M. (2008) J. Biol. Chem. 283, 24103–24117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Atsawasuwan P., Mochida Y., Katafuchi M., Kaku M., Fong K. S., Csiszar K., Yamauchi M. (2008) J. Biol. Chem. 283, 34229–34240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li W., Nugent M. A., Zhao Y., Chau A. N., Li S. J., Chou I. N., Liu G., Kagan H. M. (2003) J. Cell. Biochem. 88, 152–164 [DOI] [PubMed] [Google Scholar]
- 51. Vora S. R., Guo Y., Stephens D. N., Salih E., Vu E. D., Kirsch K. H., Sonenshein G. E., Trackman P. C. (2010) Biochemistry 49, 2962–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rodriguez H. M., Vaysberg M., Mikels A., McCauley S., Velayo A. C., Garcia C., Smith V. (2010) J. Biol. Chem. 285, 20964–20974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.