Abstract
Cytokines produced by islet-infiltrating immune cells induce β-cell apoptosis in type 1 diabetes. The IFN-γ-regulated transcription factors STAT1/IRF-1 have apparently divergent effects on β-cells. Thus, STAT1 promotes apoptosis and inflammation, whereas IRF-1 down-regulates inflammatory mediators. To understand the molecular basis for these differential outcomes within a single signal transduction pathway, we presently characterized the gene networks regulated by STAT1 and IRF-1 in β-cells. This was done by using siRNA approaches coupled to microarray analysis of insulin-producing cells exposed or not to IL-1β and IFN-γ. Relevant microarray findings were further studied in INS-1E cells and primary rat β-cells. STAT1, but not IRF-1, mediates the cytokine-induced loss of the differentiated β-cell phenotype, as indicated by decreased insulin, Pdx1, MafA, and Glut2. Furthermore, STAT1 regulates cytokine-induced apoptosis via up-regulation of the proapoptotic protein DP5. STAT1 and IRF-1 have opposite effects on cytokine-induced chemokine production, with IRF-1 exerting negative feedback inhibition on STAT1 and downstream chemokine expression. The present study elucidates the transcriptional networks through which the IFN-γ/STAT1/IRF-1 axis controls β-cell function/differentiation, demise, and islet inflammation.
Keywords: Apoptosis, Diabetes, Inflammation, Interferon, STAT Transcription Factor, Apoptosis, Inflammation, Interferon-gamma, Pancreatic Beta Cells, Type 1 Diabetes
Introduction
Stressful signals are sensed by cells, which respond via the up- or down-regulation of relevant genes and proteins. This process can be divided in two broad steps; first, cells encode the stress signals internally by modulating the expression, activation, and localization of transcription factors, and second, these transcription factors trigger the expression of key downstream genes that mediate diverse cellular responses to the stress (1, 2).
Signaling events occurring inside the pancreatic β-cells and triggered by diverse stressful mediators are decisive for the survival or death of these cells in type 1 diabetes (T1D)4 (3). T1D is an inflammatory disorder characterized by infiltration of autoreactive immune cells in pancreatic islets (a process termed insulitis) and by selective destruction of insulin-producing β-cells (4). β-cell death in T1D occurs through apoptosis induced by a deadly “dialogue” between β-cells and infiltrating immune cells, where the proinflammatory cytokines IL-1β, IFN-γ, and TNF-α produced by infiltrating macrophages and T cells play a key role (5–8).
We have previously shown that pancreatic β-cells respond to the proinflammatory cytokines IL-1β, TNF-α, and IFN-γ by modifying the expression of complex gene networks under the regulation of at least two master transcription factors, namely NF-κB and STAT1 (3, 5). Inhibition of NF-κB expression in vitro or in vivo prevents cytokine-induced β-cell apoptosis (9, 10), and array analysis of IL-1β and IFN-γ-treated β-cells in the context of NF-κB blockade allowed us to identify several key mediators of β-cell dysfunction and death, including endoplasmic reticulum stress-related factors (11–13). Such information would be of particular interest for STAT1-regulated genes, because the IFN-γ-induced transcription factors STAT1 and downstream IRF-1 have been reported to exert opposite effects in β-cells. Thus, although STAT1 blockade prevents cytokine-induced β-cell death in vitro and diabetes induced by multiple low doses of streptozotocin (14, 15), IRF-1 deficiency does not protect β-cells against cytokines in vitro. On the contrary, it actually exacerbates local production of chemokines, islet graft infiltration, and rejection in non-obese diabetic mice (16). These observations suggest that although STAT1 regulates genes triggering β-cell death and inflammation, IRF-1 may regulate putative anti-inflammatory genes. The nature of these genes, however, remains to be clarified. IRF-1 is induced by IFN-γ through binding of STAT1 to the IRF-1 promoter, but other transcription factors, such as NF-κB, may also induce its expression (17, 18).
To characterize the broad network of genes regulated by STAT1 and IRF-1, we first validated siRNAs targeting each of these transcription factors and then coupled RNA interference to global evaluation of gene expression. This was done by the use of microarray analysis of insulin-producing INS-1E cells exposed or not to proinflammatory cytokines. These microarray findings were subsequently confirmed in INS-1E and primary rat β-cells, and the function of novel STAT1- and IRF-1-dependent genes characterized by RNA interference and additional functional studies. We observed that STAT1, but not IRF-1, up-regulates the proapoptotic protein DP5 (death protein 5/Hrk) and the synthesis of several proinflammatory chemokines, including the CXCR3 ligands CXCL9, -10, and -11, whereas down-regulating genes involved in β-cell differentiation and function. On the other hand, IRF-1 induces many genes independently of STAT1, including IL-6RA, PKIα, or TAP binding protein. Interestingly, our data highlighted a novel role for IRF-1 in providing a negative feedback on STAT1-driven chemokine production through the induction of the regulatory protein SOCS-1. As a whole, these findings allow us to propose a unifying hypothesis explaining the effects of the transcription factors STAT1 and IRF-1 on β-cells.
EXPERIMENTAL PROCEDURES
RNA Interference
The siRNAs used in this study are BLOCK-iT StealthTM Select siRNA (Invitrogen) and had the following sequences: siSTAT1 (primer 1), 5′-CCCUAGAAGACUUACAAGAUGAAUA-3′; siSTAT1 (primer 2), 5′-CCAGGCUUGGUGAUUGACCUUGAGA-3′; siIRF-1 (primer 1), 5′-GCCCUCCAUUCAGGCUAUUCCUUGU-3′; siIRF-1 (primer 2), 5′-CCCUGGCUAGAGAUGCAGAUUAAUU-3′; siSOCS-1 (primer 1), 5′-CCGGUACUCCGUGACUACCUGAGUU-3′; and siSOCS-1 (primer 2), 5′-GAGAACCUGGCACGCAUCCCUCUUA-3′. Allstars Negative Control siRNA was used for control-transfected conditions (Qiagen, Venlo, The Netherlands; sequence not provided). The concentration of siRNA used for cell transfection (30 nm) was selected based on dose-response studies (supplemental Fig. 2) (19). DharmaFECT 1 (Thermo Scientific) and Lipofectamine 2000 (Invitrogen) lipid reagents were used for siRNA transfection (19). The efficiency of transfection and the results are similar for both lipid reagents. After transfection, cells were cultured for a 24-h recovery period and subsequently exposed to cytokines.
Cell Treatment and NO Measurement
The following cytokine concentrations were used, based on previous dose-response experiments (6, 20): recombinant human IL-1β (specific activity, 1.8 × 107 units/mg; a kind gift from C. W. Reinolds, National Cancer Institute, Bethesda, MD) at 10 units/ml; recombinant rat IFN-γ (specific activity, 2 × 107 units/mg; R&D Systems, Abingdon, UK) at 100 units/ml; recombinant murine TNF-α (specific activity, 2 × 108 units/mg; Innogenetics, Gent, Belgium) at 1.000 units/ml. Culture supernatants were collected for nitrite determination at an A540 nm using the Griess method. (Nitrite is a stable product of NO oxidation).
Microarray Data Analysis
Total RNA was isolated from INS-1E using the RNeasy mini kit (Qiagen). Aminoallyl antisense-cRNA was obtained, coupled to either fluorophore indocarbocyanine (Cy3) or indodicarbocyanine (Cy5) (Amersham Biosciences) and prepared for hybridization (21). The gene expression profiling study was performed using the GeneChip® Rat Genome 230 2.0 arrays (Affymetrix) containing >31,000 probe sets and covering >28,000 well defined rat genes. Gene expression values were calculated from the Affymetrix Cel files using the robust multichip average algorithm (22) implemented in the ArrayAssist® Expression software (Stratagene). Only probe sets detected as “present” by the Affymetrix MAS5 normalization algorithm in at least two of eight conditions for at least one time point were considered as present and used for further analysis. The robust multichip average algorithm normalized intensities corresponding to the 21,569 filtered probe sets were then imported to the Bioconductor free software where a robust Welch's t test (based on robust estimators of central tendency and dispersion, namely the median and the interquartile range) was applied to compare each experimental condition at different time points (2, 12, or 24 h) to the corresponding control. Probe sets were considered as differentially expressed between two conditions if they had a mean fold change (up or down) >1.5 and a p value <0.02. Lists of cytokine-modified probe sets at each time point included those that were significantly modified when comparing both cytokine-treated to untreated untransfected INS-1E cells and cytokine-treated si-control-transfected to untreated siControl-transfected INS-1E cells. Cytokine-modified probe sets were considered as STAT1-dependent when they were significantly modified when comparing cytokine-treated siSTAT1-transfected and cytokine-treated siControl-transfected samples. Cytokine-regulated and IRF1-dependent probe sets were defined in the same way, by comparing cytokine-treated siIRF1-transfected cells with cytokine-treated siControl-transfected INS1-E cells at each time point. The complete array data will be deposited after publication of this article in the EuroDia database.
Western Blots
Cells were washed, lysed, resolved by 8–10% SDS-PAGE, and transferred to a nitrocellulose membrane as described (19). The antibodies used were as follows: anti-STAT1 (catalog no. sc-346) and anti-IRF-1 (catalog no. sc-640) from Santa Cruz Biotechnology (Santa Cruz, CA); antiphospho-STAT1 (Tyr701; catalog no. 9171) from Cell Signaling (Danvers, MA); anti-PTPN2 (clone 252294) from R&D Systems (Abingdon, UK), and anti-α-tubulin (product no. T9026) from Sigma. HRP-conjugated anti-rabbit or anti-mouse IgG (Lucron Bioproducts, De Pinte, Belgium) were used as secondary antibodies. Immunoreactive bands were revealed using the SuperSignal West Femto chemiluminescent substrate (Thermo Scientific), detected using a LAS-3000 CCD camera and quantified with the Aida Analysis software (Fujifilm).
Culture of Primary FACS-sorted Rat β-Cells and INS-1E Cells
Male Wistar rats (Charles River Laboratories, Brussels, Belgium) were housed and used according to the guidelines of the Belgian Regulations for Animal Care. Islets were isolated by collagenase digestion and hand picked under a stereomicroscope. β-cells were purified by autofluorescence-activated cell sorting (FACSAria, BD Bioscience, San Jose, CA) (50). The preparations contained 90.4 ± 3.2% β-cells (n = 9). β-cells were cultured for 2 days in Ham's F-10 medium containing 10 mm glucose, 2 mm GlutaMAX, 50 μm 3-isobutyl-1-methylxanthine, 5% FBS, 0.5% charcoal-absorbed BSA (Boehringer, Indianapolis, IN), 50 units/ml penicillin, and 50 μg/ml streptomycin (50). During cytokine exposure, cells were cultured in the same medium but without serum. The rat insulin-producing INS-1E cell line (a kind gift from Dr. C. Wollheim, Centre Medical Universitaire, Geneva, Switzerland) was cultured as described previously (20).
Infection with Recombinant Adenoviruses
Cells were left uninfected or infected either with Ad-Luc (luciferase-expressing virus) or Ad-srIκB (a virus expressing an NF-κB super repressor protein) (23). Cells were infected for 2 h at 37 °C with a multiplicity of infection of 10. The multiplicity of infection was selected based on lowest toxicity by viral infection combined with highest blockade of NF-κB activation. After infection (24 h), cells were treated with cytokines. We have shown previously that infection of β-cells with Ad-srIκB at the multiplicity of infection used in the present study does not change its function (23).
mRNA Extraction and Real-time PCR
Poly(A)+ mRNA was isolated from INS-1E cells or rat primary β-cells using the Dynabeads mRNA DIRECTTM kit (Invitrogen) and reverse transcribed as described previously (50). The real-time PCR amplification reaction was done as described (50), using SYBR Green and compared with a standard curve. Expression values were corrected for the housekeeping gene GAPDH; we have previously shown that cytokines do not modify GAPDH expression (19). The primers used in this study are listed below: rat GAPDH Std 5′-ATGACTCTACCCACGGCAAG-3′ (F) and 5′-TGTGAGGGAGATGCTCAGTG-3′ (R) (975 bp); rat GAPDH RT 5′-AGTTCAACGGCACAGTCAAG-3′ (F) and 5′-TACTCAGCACCAGCATCACC-3′ (R) (118 bp); rat STAT1 Std 5′-CCTCTTCCAGCAGCTC-3′ (F) and 5′-ACTGCCAACTCAGCAC-3′ (R) (596 bp); rat STAT1 RT 5′-TGAGTTCCGACACCTGCAACTGAA-3′ (F) and 5′-AGGTGGTCTCAAGGTCAATCACCA-3′ (R) (102 bp); rat IRF-1 Std 5′-CTATTCCCAAAGAACTGCTGCCCT-3′ (F) and 5′-GGTGGCGTTTCCAATGTTCAGAGT-3′ (R) (604 bp); rat IRF-1 RT 5′-AAAGAACTGCTGCCCTTCCCAA-3′ (F) and 5′-GCAAAGTCCACAGAGAAAGTGCCA-3′ (R) (122 bp); rat insulin 1 Std 5′-CATCAGCAAGCAGGTCATTG-3′ (F) and 5′-TGCAGCACTGATCCACAATG-3′ (R) (316 bp); rat insulin 1 RT 5′-ACCTTTGTGGTCCTCACCTG-3′ (F) and 5′-AGCTCCAGTTGTGGCACTTG-3′ (R) (118 bp); rat Glut2 Std 5′-GTCCAGAAAGCCCCAGATAC-3′ (F) and 5′-CCCCAGCAAAAAGGAAGAAC-3′ (R) (699 bp); rat Glut2 RT 5′-TCAGCCAGCCTGTGTATGCA-3′ (F) and 5′-TCCACAAGCAGCACAGAGACA-3′ (R) (89 bp); rat Pdx1 Std 5′-CGCATGAAGTGGAAGAAAGAG-3′ (F) and 5′-TGTTATGGGACCGCTCAAGTT-3′ (R) (601 bp); rat Pdx1 RT 5′-GGTATAGCCAGCGAGATGCT-3′ (F) and 5′-TCAGGTGGGAGCCTGATTCT-3′ (R) (153 bp); rat MafA Std 5′-AGCTGGTGTCCATGTCAGTG-3′ (F) and 5′-CGTATTTCTCCTTGTACAGG-3′ (R) (251 bp); rat MafA RT 5′-AAGGAGGAGGTCATCCGACT-3′ (F) and 5′-TCTGGAGCTGGCACTTCTCG-3′ (R) (120 bp); rat DP5 Std 5′-CATGTCCTGTATGCCACCTG-3′ (F) and 5′-GCTCAGACGTGGAGGTCTTC-3′ (R) (612 bp); rat DP5 RT 5′-TCTGGAAGACACCCTCTGCT-3′ (F) and 5′-CACAGAGTCCCACCATGTTG-3′ (R) (93 bp); rat CXCL10 Std 5′-GAAGCACCATGAACCCAAGT-3′ (F) and 5′-GGGTAAAGGGAGGTGGAGAG-3′ (R) (380 bp); rat CXCL10 RT 5′-GCAAGTCTATCCTGTCCGCAT-3′ (F) and 5′-GGGTAAAGGGAGGTGGAGAGA-3′ (R) (117 bp); rat CXCL9 Std 5′-GGAGTTCGAGGAACCCTAGT-3′ (F) and 5′-CAGAGCGCTTGTTGGTAAA-3′ (R) (354 bp); rat CXCL9 RT 5′-CAAGGCACATTCCACTACAA-3′ (F) and 5′-CCTTGCTGAATCTGGGTCTA-3′ (R) (132 bp); rat CXCL1 Std 5′-TCCAACAGAGCACCATGGTC-3′ (F) and 5′-TCTCCATTACTTGGGGACAC-3′ (R) (310 bp); rat CXCL1 RT 5′-TCCAGAGTTTGAAGGTGATG-3′ (F) and 5′-AGCATCTTTTGGACAATCTTC-3′ (R) (131 bp); rat CCL20 Std 5′-GGGGGTACTGCTGGCTTAC-3′ (F) and 5′-CCAGAAAAGCATCCGTTTTTAC-3′ (R) (274 bp); rat CCL20 RT 5′-GACTGCTGCCTCACGTACAC-3′ (F) and 5′-CGACTTCAGGTGAAAGATGATAG-3′ (R) (120 bp); rat SOCS-1 Std 5′-ACGCCTGCGGCTTCTACT-3′ (F) and 5′-GGAACTCAGGTAGTCACGGAGTAC-3′ (R) (395 bp); and rat SOCS-1 RT 5′-CGAGCTGCTGGAGCACTAC-3′ (F) and 5′-GGAACTCAGGTAGTCACGGAGTA-3′ (R) (89 bp).
Assessment of Cell Viability
The percentage of viable, apoptotic, and necrotic cells was determined following a 15-min incubation with the DNA-binding dyes propidium iodide (5 μg/ml, Sigma) and Hoechst 33342 (5 μg/ml, Sigma) (50). A minimum of 500 cells was counted in each experimental condition. Viability was evaluated by two independent observers, one of them being unaware of sample identity. The agreement between findings obtained by the two observers was >90%. Results are expressed as percent apoptosis, calculated as (number of apoptotic cells/total number of cells) × 100. Apoptosis was confirmed in some experiments by the Cell Death Detection ELISAPLUS kit (Roche Diagnostics), which detects cytoplasmic fragmented DNA.
Promoter Reporter Assays
INS-1E cells were transfected as described previously (10) with pRL-CMV encoding Renilla luciferase (Promega) and either a luciferase promoter-reporter construct containing five NF-κB consensus binding sites (NF-κB reporter) or three GAS consensus sequences (STAT1 reporter) or the fragment (−1568/+81) from the rat DP5 promoter (DP5 promoter experiments) (28). Luciferase activities were assayed after 24 h of cytokine treatment (20).
Overexpression of Rat STAT1 and Rat IRF-1
The expression vectors pCMV-STAT1 (Imagene) and pCMV-IRF-1 (Thermo Scientific) were transfected in INS-1E cells using Lipofectamine 2000 (Invitrogen) as described previously (10). After overnight incubation, the medium was changed, and cells were exposed to cytokines as indicated.
Statistical Analysis
Data are presented as mean ± S.E. Comparisons were performed by two-tailed paired Student's t test or by ANOVA followed by Student's t test with Bonferroni correction. A p value <0.05 was considered statistically significant.
RESULTS
STAT1 Knockdown Protects INS-1E and Primary β-Cells against Cytokine-induced Apoptosis
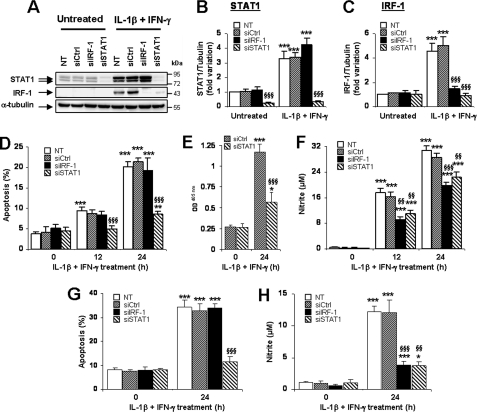
Transfection of siRNAs targeting STAT1 and IRF-1 (siSTAT1 and siIRF-1) respectively inhibited IL-1β and IFN-γ-induced STAT1 and IRF-1 induction by 89 and 72% (Fig. 1, A–C). STAT1 silencing also inhibited cytokine-induced IRF-1 expression, which was expected because IRF-1 is a downstream target of STAT1 (Fig. 1, A and C). An irrelevant siRNA used as control (si-control) did not modify IRF-1 or STAT1 expressions. STAT1 silencing potently inhibited IL-1β and IFN-γ-induced apoptosis after 12 and 24 h, whereas neither si-control nor siIRF-1 protected INS-1E cells (Fig. 1D). This was confirmed by a second method that detects cytoplasmic fragmented DNA (Fig. 1E). Both IRF-1 and STAT1 silencing decreased NO production by 38–48% and 27–35% after 12 and 24 h of treatment, respectively (Fig. 1F). Transfection of siSTAT1 and siIRF-1 in primary β-cell decreased cytokine-induced STAT1 and IRF-1 expressions by 72–87% and 70–89%, respectively (supplemental Fig. 1, A and B). STAT1 silencing did not affect IRF-1 expression in primary β-cells (supplemental Fig. 1B), suggesting that IRF-1 expression is less dependent on STAT1 in primary cells. NF-κB also contributed to cytokine-induced IRF-1 up-regulation in β-cells, as the addition of IL-1β or TNF-α (both NF-κB activators) augmented IFN-γ-induced IRF-1 induction in INS-1E cells (supplemental Fig. 1, C and D), whereas an NF-κB blockade using a super-repressor IκBα (23) inhibited cytokine-induced IRF-1 expression (supplemental Fig. 1E). STAT1 knockdown also protected primary β-cells against IL-1β and IFN-γ-induced apoptosis, whereas both siIRF-1 and siSTAT1 inhibited cytokine-induced NO production (Fig. 1, G and H). This suggests that protection induced by STAT1 knockdown is at least in part independent on NO production. Similar findings were observed in INS-1E cells using additional siRNAs for each target gene (siIRF-1 primer 2 and siSTAT1 primer 2, supplemental Fig. 2), with siSTAT1 primers 1 and 2 protecting INS-1E cells against both IL-1β and IFN-γ and TNF-α and IFN-γ-induced apoptosis (supplemental Fig. 2).
FIGURE 1.
siRNA-mediated STAT1 knockdown protects INS-1E and primary rat β-cells against cytokine-induced apoptosis. A–F, insulin-producing INS-1E cells were left untransfected (NT), or transfected with 30 nm si-control, siIRF-1, or siSTAT1. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 12 or 24 h as indicated. A, STAT1, IRF-1, and α-tubulin protein expression were evaluated by Western blot. B and C, mean optical density measurements of STAT1 and IRF-1 Western blots corrected for protein loading by α-tubulin. Results are mean fold variation ± S.E. of five independent experiments. D and E, apoptosis was evaluated using Hoechst 33342/propidium iodide staining (D) and a Cell Death Detection ELISAPLUS kit (E). F, nitrite concentrations in supernatants were evaluated as described under “Experimental Procedures.” Results are mean ± S.E. of five independent experiments. G and H, primary FACS-sorted rat β-cells were cultured for 2 days and then left untransfected or transfected with 30 nm of si-control, siIRF-1, or siSTAT1 as indicated. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 24 h as indicated. G, apoptosis was evaluated using Hoechst 33342/propidium iodide staining. H, nitrite concentrations in supernatants. Results are mean ± S.E. of five independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus untreated (i.e. not treated with cytokines) or untreated transfected with the same siRNA. §§, p < 0.01 and §§§, p < 0.001 versus untransfected and si-control treated with cytokines at the same time point, ANOVA followed by Student's t test with Bonferroni correction.
Analysis of Gene Networks and Pathways Regulated by IRF-1 and STAT1 in INS-1E Cells
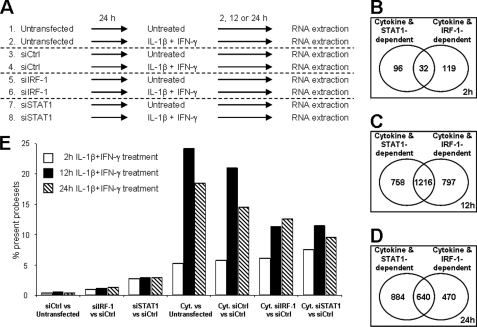
INS-1E cells were left untransfected or transfected with si-control, siIRF-1, or siSTAT1, and subsequently treated with IL-1β and IFN-γ for 2, 12, or 24 h (Fig. 2A). In the microarray analysis, 21.569 probe sets corresponding to 10.874 annotated genes were detected as present. The complete list of probe sets present for each time point is provided as supplemental Tables 2, 3, and 4. Less than 0.5% of the genes were differentially regulated between the untransfected and si-control-transfected conditions, suggesting that the transfection per se had only minor influence on the cells (Fig. 2E). STAT1 silencing affected the expression of 2.6–2.9% of the genes in untreated cells, suggesting a role for STAT1 in the control of basal β-cell function (supplemental Table 5). Cytokine treatment in untransfected and si-control-transfected conditions regulated 21–24% and 14–18% of the genes at 12 and 24 h, respectively (Fig. 2E and supplemental Tables 3 and 4), whereas only ∼5% of the genes were regulated by cytokines at 2 h (Fig. 2E and supplemental Table 2). At 12 h, ∼50% of cytokine-regulated genes were at least partially modified by IRF-1 or STAT1 knockdown (Fig. 2C), whereas at 24 h, 42 and 58% of the cytokine-regulated genes were IRF-1- or STAT1-dependent, respectively (Fig. 2D). The effects of siSTAT1 and siIRF-1 on cytokine-induced genes showed only partial superposition, with 18–33% of the genes regulated by only IRF-1 or STAT1 at 24 h (Fig. 2D). Analysis using Ingenuity Pathways Analysis software revealed that functions associated with cell cycle and development, inflammatory response, and endocrine system disorder were significantly affected by STAT1 silencing (supplemental Fig. 3A). Interestingly, cytokine-regulated functions such as cell death, antimicrobial response, inflammatory response, and antigen presentation were affected by IRF-1 deficiency, suggesting an inhibitory role of IRF-1 in these processes (supplemental Fig. 3A). Canonical pathways regulated by cytokines, including HMGB1, NF-κB, and IL-17 signaling, death receptor, and apoptosis signaling were significantly altered by STAT1 knockdown (supplemental Fig. 3B). Supplemental Table 1 represents selected genes with a putative role in β-cell function/dysfunction and death using a method described previously (24). Based on both the “handmade” and unbiased analysis, we selected three pathways of high relevance in the context of T1D, namely β-cell function/differentiation, apoptosis, and inflammation for additional studies.
FIGURE 2.
Analysis of gene networks regulated by cytokines, IRF-1, and STAT1 in INS-1E cells. INS-1E cells were left untransfected or transfected with 30 nm of si-control, siIRF-1, or siSTAT1 as described in the legend to Fig. 1. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 2, 12, or 24 h as indicated, before being harvested for RNA extraction and array analysis. A, schematic representation of the microarray conditions (three independent experiments). B–D, Venn diagrams showing the number of β-cell genes whose expression was modified by cytokines and that were either STAT1- or IRF-1-dependent after 2 h (B), 12 h (C), or 24 h (D) of cytokine (Cyt.) exposure. E, mean percentage of probe sets considered as present in the three microarray experiments that were differentially regulated by STAT1/IRF-1 silencing and/or cytokine treatment at 2 h (white bars), 12 h (black bars), or 24 h (hatched bars) of cytokine treatment in the different conditions indicated. Results of three independent array experiments were analyzed. mRNA expression was considered as modified by cytokines when p < 0.02 and fold change >1.5 as compared with untransfected cells not treated with cytokines (Untransfected) or si-control-transfected cells not treated with cytokines (siCtrl).
STAT1 Silencing Partially Prevents Cytokine-induced Down-regulation of Genes Involved in β-Cell Function and Differentiation
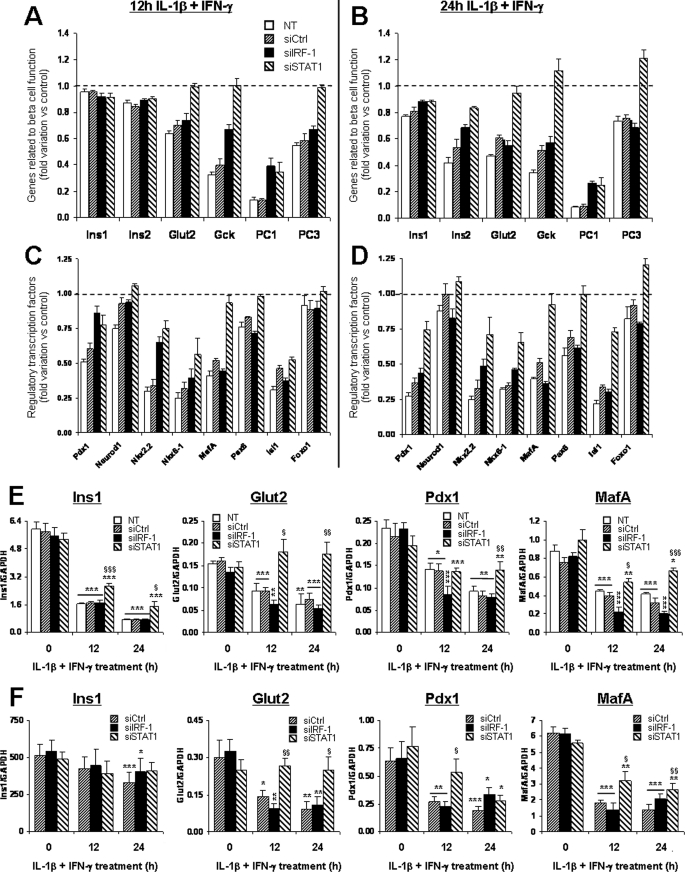
Exposure of the β-cell to cytokines decreases the expression of genes involved in β-cell differentiation and function (24, 25). The present microarray analysis confirmed these findings, since genes such as insulin, Glut2, glucokinase, Pdx1, MafA, and others were significantly down-regulated after 12 and 24 h of IL-1β and IFN-γ exposure (Fig. 3, A–D). Importantly, STAT1 silencing partially prevented the down-regulation of several of these genes, including insulin, Glut2, glucokinase, proconvertase 1 and 3 (Fig. 3, A and B), and all of the transcription factors involved in β-cell differentiation (Fig. 3, C and D). IRF-1 knockdown prevented the down-regulation of proconvertase 1, glucokinase, Pdx1, and Nkx2.2 at 12 h of cytokine (Fig. 3, A and C), but this protective effect was lost at 24 h (Fig. 3, B and D). These results were confirmed by real-time PCR for selected genes (insulin 1, Glut2, Pdx1, and MafA) in INS-1E cells (Fig. 3E) and primary β-cells (Fig. 3F). This suggests that STAT1 mediates at least in part the deleterious effects of cytokines on β-cell differentiation during inflammation.
FIGURE 3.
STAT1 silencing partially prevents cytokine-induced down-regulation of genes involved in β-cell differentiation and function. INS-1E cells (A–E) or primary FACS-purified rat β-cells (F) were left untransfected (NT) or transfected with 30 nm of si-control, siIRF-1, or siSTAT1. After 24 h of recovery post-transfection, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 2, 12, or 24 h as indicated. Expression of genes related to β-cell function (A and B) or regulatory transcription factors (C and D) were analyzed by microarray. Results represent the mean fold variations ± S.E. of the genes as compared with untreated controls after 12 h (A and C) or 24 h (B and D) of cytokine treatment (n = 3). Statistical analyses for the represented genes are described in supplemental Table 1. E, independent confirmation experiments in INS-1E cells Ins1, Glut2, Pdx1, and MafA mRNA expression were assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of four independent experiments. F, confirmation experiments in primary rat β-cells. Ins1, Glut2, Pdx1, and MafA mRNA expression was assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of five independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus untreated (i.e. not treated with cytokines) or untreated transfected with the same siRNA. §, p < 0.05; §§, p < 0.01; and §§§, p < 0.001 versus untransfected and si-control treated with cytokines at the same time point, ANOVA followed by Student's t test with Bonferroni correction.
Induction of the Proapoptotic Protein DP5 Is Partially Prevented after STAT1 Knockdown
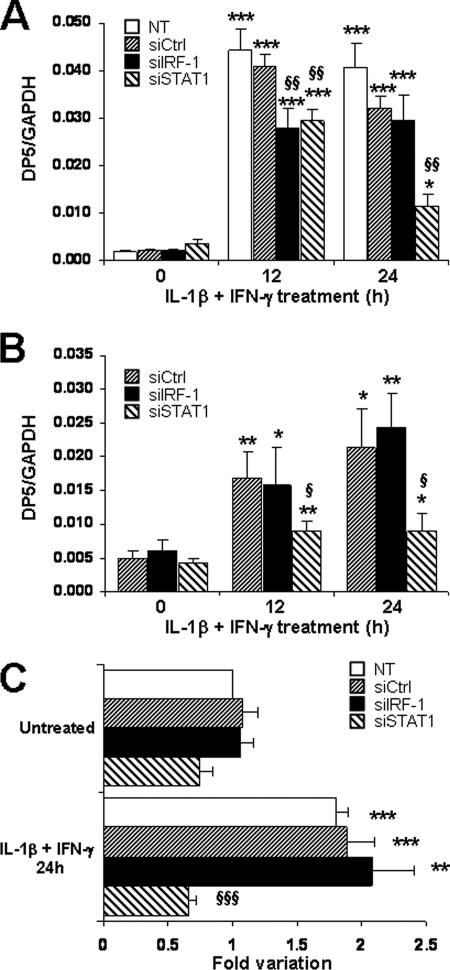
DP5 (Hrk) is a proapoptotic BH3-only member of the Bcl2 family (26), which plays a central role in cytokine-induced β-cell apoptosis (27). The microarray data demonstrated that DP5 induction was hampered by IRF-1 and STAT1 knockdown at 12 h of cytokine treatment (supplemental Table 1). Real-time PCR analysis in INS-1E cells and primary β-cells confirmed that STAT1 silencing prevented DP5 induction at 12 and 24 h of cytokine treatment (Fig. 4, A and B), whereas siIRF-1 effect was only transient. Using a luciferase reporter containing the promoter sequence from the rat DP5 gene (28), we observed that STAT1 silencing completely abolished cytokine-induced DP5 promoter expression, whereas it was induced by 2-fold in untransfected, si-control- or siIRF-1-transfected cells (Fig. 4C). We then tested whether STAT1 overexpression affects cytokine-induced DP5 up-regulation. The transfection of a rat STAT1 expression vector in INS-1E cells resulted in increased STAT1 expression at both mRNA and protein levels (supplemental Fig. 4, A and B) and exacerbated DP5 induction after 16–24 h of IL-1β and IFN-γ treatment (supplemental Fig. 4C). These findings suggest that STAT1 participates in cytokine-dependent DP5 up-regulation, a key mechanism of cytokine-induced β-cell apoptosis (27). Of note, parallel knockdown of STAT1 and DP5 by specific siRNAs induced a more marked inhibition of cytokine-induced apoptosis than DP5 or STAT1 silencing alone (supplemental Fig. 4, D and E). This suggests that STAT1 regulates other proapoptotic signals than DP5.
FIGURE 4.
Induction of the proapoptotic protein DP5 is partially prevented after STAT1 knockdown. INS-1E cells (A) or primary FACS-sorted rat β-cells (B) were transfected and treated as described in Fig. 1. A and B, DP5 mRNA expression was assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of four (A) or five (B) independent experiments. C, INS-1E cells were co-transfected with the DP5 promoter luciferase reporter and control pRL-CMV alone (NT) or in combination with si-control, siIRF-1, or siSTAT1. After 1 day of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 24 h as indicated. Results are mean ± S.E. of five independent experiments and represent fold variation as compared with untreated control condition. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus untreated (i.e. not treated with cytokines) or untreated transfected with the same siRNA. §, p < 0.05; §§, p < 0.01; and §§§ p < 0.001 versus NT & si-control treated with cytokines at the same time point, ANOVA followed by Student's t test with Bonferroni correction.
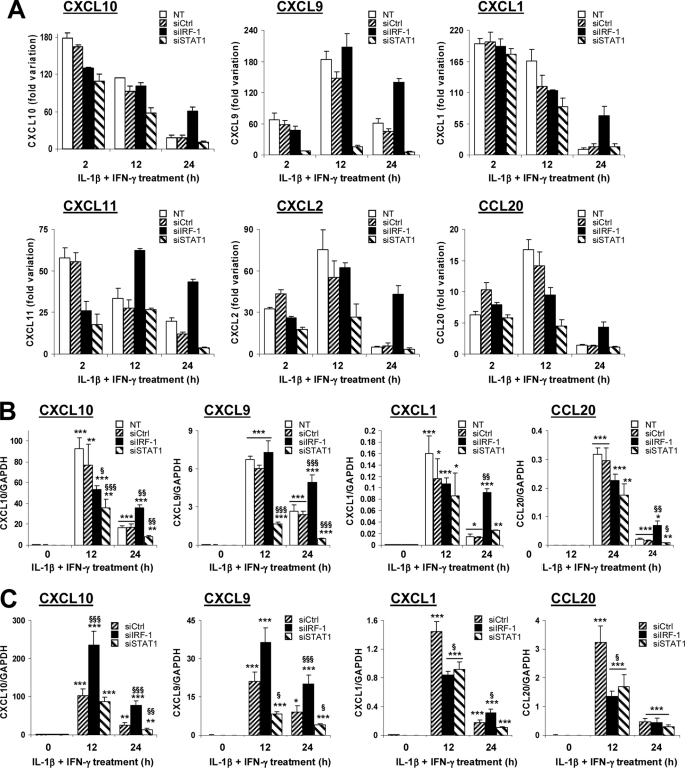
IRF-1 Provides a Negative Feedback on Cytokine-induced Chemokine Production
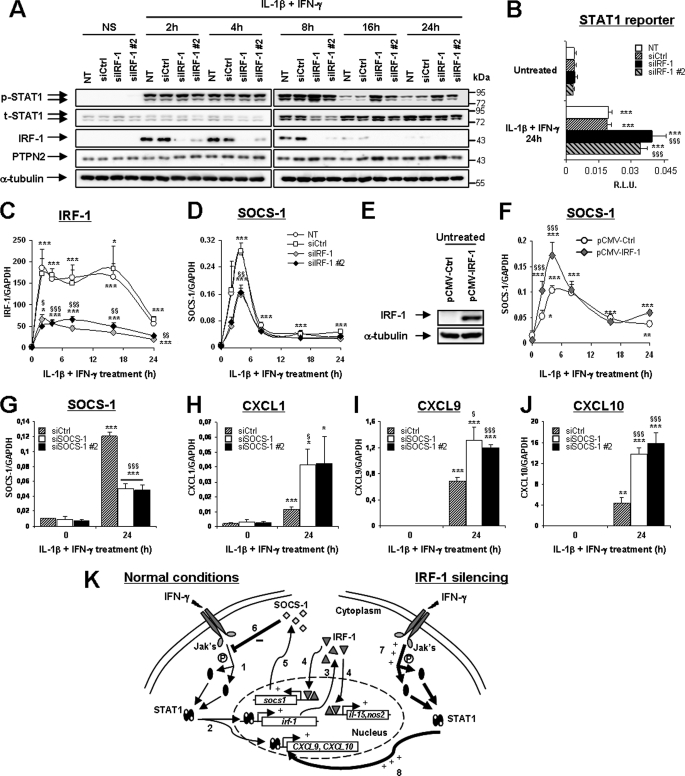
During islet inflammation, β-cells produce chemokines that further attract immune cells (7). Our microarray data demonstrated that IL-1β and IFN-γ exposure induced the early production of CXCL1, -2, -9, -10, -11, and CCL20 (Fig. 5A; for statistical analysis, see supplemental Table 1). STAT1 silencing down-regulated the production of all chemokines at 12 and/or 24 h of cytokine treatment (Fig. 5A), whereas IRF-1 silencing exacerbated the production of CXCL9 and -11 after 12 h and of all chemokines after 24 h (Fig. 5A). This was confirmed by real-time PCR experiments in cytokine-treated INS-1E cells and primary β-cells for the chemokines CXCL1, -9, -10, and CCL20 (Fig. 5, B and C). Again, IRF-1 silencing exacerbated cytokine-induced CXCL1, -9, and -10. This indicates that IRF-1 may provide a negative feedback on STAT1-driven chemokine production. To further investigate this phenomenon, we evaluated the influence of IRF-1 knockdown on STAT1 activation. IRF-1 expression was induced by cytokines until 8 h, and it was silenced by the two IRF-1-targeting siRNAs (Fig. 6A). STAT1 phosphorylation was equally induced in all conditions at 2 and 4 h but remained up-regulated until 16 and 24 h of cytokine exposure in IRF-1-silenced cells (Fig. 6A and supplemental Fig. 5A). This effect was independent of the total STAT1 content and on the activity of the nuclear phosphatase PTPN2 previously reported by our group to regulate IFN-γ-induced STAT1 activity in β-cells (Fig. 6A) (19). This prolonged STAT1 phosphorylation in IRF-1-silenced cells exacerbated STAT1 transcriptional activity because cytokines increased the activation of a STAT1 reporter in IRF-1-silenced cells as compared with controls (Fig. 6B), whereas an NF-κB reporter (used as an external control) was equally induced in all tested conditions (supplemental Fig. 5B). We next evaluated the effect of IRF-1 silencing on the expression of SOCS-1, a negative regulator of STAT1 previously reported to be induced by IRF-1 in fibroblasts (29). The array data indicated that SOCS-1 expression was dependent on IRF-1 and STAT1 at 2 h and mostly STAT1-dependent at later time points (supplemental Tables 2, 3, and 4). Real-time PCR experiments showed that SOCS-1 induction reflected the cytokine-induced IRF-1 up-regulation in INS-1E cells, reaching a peak at 4 h and then slowly decreasing until 24 h (Fig. 6, C and D). IRF-1 knockdown decreased cytokine-induced SOCS-1 expression at all time points, suggesting that IRF-1 contributes to SOCS-1 expression in β-cells (Fig. 6D). To confirm these findings, we transfected INS-1E cells with an expression vector for rat IRF-1, resulting in increased basal- and cytokine-induced IRF-1 expression in these cells (Fig. 6E and supplemental Fig. 5C). Overexpression of IRF-1 exacerbated cytokine-driven SOCS-1 induction (Fig. 6F) while significantly inhibiting the up-regulation of STAT1 and chemokines CXCL1 and -9 after IL-1β and IFN-γ treatment (supplemental Fig. 5, D–F). To confirm the role of SOCS-1 for these inhibitory effects of IRF-1, we performed siRNA-mediated SOCS-1 silencing in INS-1E cells. As shown in Fig. 6G, the two SOCS-1-targeting siRNAs inhibited cytokine-induced SOCS-1 expression by 61–64% and, importantly, prolonged STAT1 phosphorylation as compared with si-control-transfected cells after 16 and 24 h of IL-1β and IFN-γ treatment (supplemental Fig. 5G). Moreover, SOCS-1 silencing by both siRNAs significantly augmented cytokine-induced CXCL1, -9, and -10 mRNA synthesis after 24 h of exposure (Fig. 6, H–J). Collectively, these data suggest that IRF-1 inhibition exacerbates cytokine-induced chemokine production in β-cells through a prolonged STAT1 activation resulting from a deficient SOCS-1 induction in IRF-1-silenced cells. These observations are summarized in Fig. 6K, which represents the proposed negative regulatory feedback loop by which IRF-1 modulates STAT1 activation.
FIGURE 5.
IRF-1 provides a negative feedback on cytokine-induced chemokine production. INS-1E cells (A and B) or primary FACS-sorted rat β-cells (C) were left untransfected (NT), or transfected with 30 nm si-control, siIRF-1, or siSTAT1. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 2, 12, or 24 h as indicated. A, expression of the chemokines CXCL10, CXCL9, CXCL1, CXCL11, CXCL2, and CCL20 were analyzed by microarray. Results represent the mean fold variations ± S.E. of the genes as compared with untreated controls at the same time points (n = 3). Statistical analyses for the represented genes are described in supplemental Table 1. B and C, confirmation experiments in INS-1E cells (B) and primary rat β-cells (C). CXCL10, CXCL9, CXCL1, and CCL20 mRNA expressions were assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of four to five independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus untreated (i.e. not treated with cytokines) or untreated transfected with the same siRNA. §, p < 0.05; §§, p < 0.01; and §§§, p < 0.001 versus untransfected and si-control-treated with cytokines at the same time point, ANOVA followed by Student's t test with Bonferroni correction.
FIGURE 6.
IRF-1 hampers STAT1 activation through the induction of SOCS-1. A, C, and D, INS-1E cells were left untransfected (NT) or transfected with 30 nm si-control, siIRF-1, or siIRF-1 primer 2. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 2, 4, 8, 16, or 24 h as indicated. A, phospho-STAT1, total STAT1, IRF-1, PTPN2, and α-tubulin protein expressions were evaluated by Western blot. Pictures are representative of five independent experiments. B, INS-1E cells were co-transfected with a STAT1 reporter and pRL-CMV alone (NT) or in combination with si-control, siIRF-1, or siIRF-1 primer 2. After 1 day of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 24 h as indicated. Results are mean relative luciferase unit (R.L.U.) ± S.E. of five independent experiments. C and D, IRF-1 and SOCS-1 mRNA expressions were assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of four independent experiments. E and F, INS-1E cells were transfected with pCMV-Ctrl or pCMV-IRF-1. After overnight incubation, the cells were left untreated (time 0) or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 2, 4, 8, 16, or 24 h as indicated. E, IRF-1 and α-tubulin protein expressions were evaluated by Western blot in untreated cells 24 h after transfection. Pictures are representative of four independent experiments. F, SOCS-1 mRNA expression was assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of four independent experiments. G–J, INS-1E cells were transfected with 30 nm si-control, siSOCS-1, or siSOCS-1 primer 2. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 24 h as indicated. SOCS-1, CXCL1, CXCL9, and CXCL10 mRNA expression was assayed by real-time RT-PCR and normalized for the housekeeping gene GAPDH. Results are mean ± S.E. of four independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus respective untreated control. §, p < 0.05; §§, p < 0.01; and §§§, p < 0.001 versus respective control treated with cytokines at the same time point, ANOVA followed by Student's t test with Bonferroni correction. K, schematic representation of the suggested regulatory loop controlled by IRF-1 in β-cells. IFN-γ binding to its receptor induces Jak-mediated STAT1 phosphorylation and dimerization (1) and its subsequent migration to the nucleus (2), where it stimulates the transcription of many genes, including chemokines and IRF-1. Once synthesized in the cytoplasm (3), IRF-1 also migrates to the nucleus (4) to stimulate the transcription of several genes including SOCS-1 (5). SOCS-1 may then interfere with Jak-mediated STAT1 phosphorylation (6), hence hampering STAT1 activation over time. In the absence of IRF-1 signaling, the defective SOCS-1 induction allows prolonged Jak-mediated STAT1 phosphorylation (7) and sustained STAT1 activity (8).
DISCUSSION
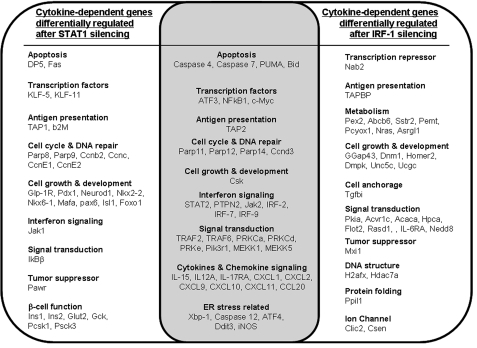
Data-driven models based on array analysis identify large numbers of potential correlations, making it difficult to perturb each individual pathway experimentally. One alternative, followed in the present study, is to target key transcription factors regulating relevant gene networks. Cytokine-induced STAT1 activation in β-cells is associated with the induction of apoptosis and diabetes progression in murine models of T1D (14, 15, 30), whereas the STAT1 downstream transcription factor IRF-1 may decrease islet inflammation without directly regulating β-cell death (16, 31). To explain these apparently divergent effects, we have presently combined siRNA-mediated STAT1- or IRF-1 silencing with global gene expression profiling. The siRNAs utilized (two independent ones for each target gene) were validated by showing their inhibitory effect on the target genes, by the protection they induced against cytokine-triggered β-cell death and by the good agreement between their effects and the observed effects in islets from STAT1 and IRF-1 KO mice (15, 16). Importantly, the nonspecific siRNA used as control did not affect β-cell viability (present study and 19) or function (32) and had only minimal effect (<0.5%) on gene expression, as evaluated by array analysis (present study). Using this well controlled approach, we observed that cytokine-induced STAT1 and IRF-1 expression regulate gene networks associated with cell cycle, signal transduction, apoptosis, endoplasmic reticulum stress, and inflammation in β-cells. A list of some of the key regulated genes is shown in Fig. 7. IRF-1 silencing affected the expression of nearly 800 cytokine-induced genes independently of STAT1 expression (supplemental Table 3 and Fig. 7); this is somewhat surprising as IRF-1 is usually considered a downstream transcription factor of STAT1 (17). We presently show that early transcriptional control of IRF-1 expression in β-cells is also dependent on NF-κB (that is rapidly activated by IL-1β or TNF-α in β-cells (10)), as NF-κB blockade inhibits cytokine-induced IRF-1 up-regulation (supplemental Fig. 1 and Tables 2, 3, and 4). Moreover, IL-1β and IFN-γ-induced IRF-1 expression was not decreased by STAT1 silencing in primary rat β-cells (supplemental Fig. 1B), confirming that IRF-1 may act independently of STAT1.
FIGURE 7.
Schematic representation of selected cytokine-dependent genes differentially regulated by the transcription factors STAT1 (left), IRF-1 (right), or both STAT1 and IRF-1 (center). Of note, some genes (e.g. chemokines) are regulated, at least in part, in opposite directions by STAT1 and IRF-1.
Detailed examination of the array data, either manually or by unbiased analysis using the Ingenuity Pathways Analysis software, indicated three key pathways for β-cell survival and function, and local inflammation, potentially modulated by STAT1 and, for one of them, also by IRF-1. The first pathway is related to β-cell function and differentiation. Loss of a differentiated β-cell phenotype occurs during insulitis (5), and our group has reported recently that exposure of primary β-cells to IL-1β and IFN-γ or TNF-α and IFN-γ down-regulates several genes involved in β-cell-differentiated functions (e.g. insulin, glucokinase, Glut2, prohormone convertases, etc.) as well as many transcription factors involved in the differentiation and maintenance of β-cell phenotype (e.g. Pdx1, MafA, Nkx2.2, etc.) (24). Recent evidence points to the central role of IFN-γ in vivo in this inhibitory effect of inflammation in BB rat β-cells (33). The present array confirmed these findings and indicated that STAT1 but not IRF-1 silencing partially protects β-cells against the “dedifferentiating” effects of proinflammatory cytokines. This is in line with observations in other tissues, suggesting that STAT1 inhibits the differentiation of osteoblasts, myoblasts, and human adipocytes (34–36).
It has been reported that re-expressing a combination of three key developmental transcription factors (Ngn3, Pdx1, and mafA) in adult mouse pancreas reprograms pancreatic exocrine cells into cells that closely resemble β-cells (25). People expected to benefit most from reprogramming of pancreatic exocrine cells to β-cells are patients with T1D. Insulin epitopes are targets of the immune assault in T1D (37), and new insulin-producing cells will be recognized and attacked by the immune system (38). Our present and previous data (39) suggest that immune mediators of insulitis such as cytokines can “push back” newly developed β-cells into a dedifferentiated state. Unless novel strategies are found to prevent these cytokine effects, β-cell reprogramming will remain an interesting research finding with limited translational potential. The present data indicate that STAT1 is a promising target for this approach, and it will be of high interest to test whether combinations of key developmental regulators (25) with blockers of STAT1 (present study) or NF-κB (3, 9) restore and maintain β-cell mass in animal models of autoimmune diabetes.
The second pathway studied is related to β-cell apoptosis and focused on DP5. DP5 (Hrk) is a proapoptotic BH3-only member of the Bcl-2 family, playing a crucial role for apoptosis in neurons (40–42). Recent work from our laboratory established that DP5 is central for cytokine- and endoplasmic reticulum-stress-induced β-cell death (27). The present data demonstrate that silencing of STAT1 but not IRF-1 prevents, to a large extent, cytokine-induced DP5 mRNA up-regulation in β-cells, whereas STAT1 overexpression results in exacerbated DP5 induction upon IL-1β and IFN-γ exposure. This is probably a transcriptional effect, as siSTAT1 (but not IRF-1) also prevents cytokine-induced activation of a DP5 reporter promoter. However, we found no putative binding sites for STAT1 (GAS sequences) in the promoter region of the rat DP5 gene (data not shown), suggesting that STAT1 regulates DP5 expression in an indirect manner. Collectively, these observations are in line with the present and previous observations that inhibition of STAT1 activity but not of IRF-1 protects β-cells against cytokine-induced apoptosis (Fig. 1) (15, 16), suggesting that up-regulation of DP5 is an important mechanism by which STAT1 leads to β-cell apoptosis. Of note, several other apoptosis-related genes such as Puma, CHOP (Ddit3), Bax, Bid, and caspase-4, -7, and -12 were also differentially regulated by cytokines following STAT1 knockdown (supplemental Table 1) and may contribute for cytokine-induced β-cell apoptosis.
The third pathway identified as STAT1/IRF-1-modulated is related to islet inflammation. During insulitis, locally produced cytokines both contribute to β-cell apoptosis (5, 7) and stimulate the production of several chemokines by β-cells, further recruiting activated immune cells to the site of inflammation (7, 43). This local production of chemokines may be crucial in early T1D, as transgenic expression of CCL2 in β-cells induces spontaneous diabetes (44), whereas KO of CXCR3 delays diabetes by preventing attraction of CXCR3-expressing T cells (45). Furthermore, recent findings in new onset T1D patients show islet expression of CXCL10, whereas infiltrating lymphocytes expressed its receptor, CXCR3 (46). Our present findings indicate that STAT1 partially regulates cytokine-induced secretion of several chemokines by β-cells, including CXCL9, CXCL10, CXCL11, and CCL20. These results are in agreement with our previous observations that islets from STAT1 KO mice have decreased production of CXCL10 upon cytokine exposure in vitro and in vivo (15). In contrast to the inhibitory effect of STAT1 silencing, IRF-1 inhibition exacerbated cytokine-induced chemokine production in β-cells, especially at later time points (12 h and particularly 24 h), whereas IRF-1 overexpression hampered STAT1 induction and chemokine production after cytokine exposure (supplemental Fig. 5). These data provide a molecular explanation for our previous in vivo observations in mice, which showed increased primary non-function and rejection of grafted IRF-1−/− islets, which was accompanied by augmented infiltration by macrophages and T cells (16). We thus suggest that IRF-1 provides a negative feedback on STAT1-induced chemokine production, which is probably mediated via SOCS-1 up-regulation and STAT1 dephosphorylation. Indeed, SOCS-1 silencing also prolongs STAT1 phosphorylation and exacerbates CXCL1, -9, and -10 production in INS-1E cells (Fig. 6). Transgenic expression of SOCS-1 in β-cells reduces diabetes development in non-obese diabetic mice (47) and protects β-cells against the deleterious effects of infiltrating CD8+ T cells in the same model (48), reinforcing the role of SOCS-1 downstream of IRF-1. Interestingly, a similar role has been described for STAT3 in myeloid cells, where IFN-α-induced STAT3 expression represses STAT1-driven CXCL9 and CXCL10 induction through heterodimerization and suppression of formation of STAT1 homodimers (49). This mechanism and the presently described IRF-1-mediated negative feedback on STAT1 are probably part of elaborate “defense” mechanisms utilized by long-lived cells, such as pancreatic β-cells, to down-regulate local inflammation and thus limit tissue damage (7). Analysis of the array data indicates that the negative feedback by IRF-1 on STAT1 activity is mostly restricted to up-regulation of chemokines; additional studies are required to clarify the mechanisms for this specificity.
In conclusion, we have presently combined RNA interference and array analysis to clarify the gene networks regulated by the IFN-γ-STAT1-IRF-1 pathway in β-cells. This enabled the identification of three key pathways that may play a role for loss of functional β-cell mass in T1D: 1) β-cell dedifferentiation, an effect mediated by STAT1; 2) β-cell apoptosis, an effect mediated by STAT1 via DP5 up-regulation; and 3) induction and modulation of chemokine production, an effect mediated by STAT1 but with IRF-1 providing a negative feedback through SOCS-1 induction. This and the discovery of a large number of additional STAT1- and IRF-1-regulated genes in β-cells, broadens our understanding of β-cell dysfunction and death and opens new possibilities for prevention of loss of functional β-cell mass in early T1D.
Supplementary Material
Acknowledgments
We thank G. Vandenbroeck, R. Makhnas, A. M. Musuaya, S. Mertens, M. A. Neef, M. Urbain, and M. Pangerl (Laboratory of Experimental Medicine, Université Libre de Bruxelles) for excellent technical support.
This work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS-FRSM) Belgium, the Communauté Française de Belgique (Actions de Recherche Concertées), the European Union (NAIMIT, Health F22009-241447; in the Framework Programme 7 of the European Community) and the Belgium Program on Interuniversity Poles of Attraction initiated by the Belgian government (IUAP P6/40).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–5 and Figs. 1–5.
- T1D
- type 1 diabetes
- ANOVA
- analysis of variance
- Ad
- adenovirus
- si
- small interfering
- F
- forward
- R
- reverse
- RT
- real-time (PCR)
- Std
- standard.
REFERENCES
- 1. Cai L., Dalal C. K., Elowitz M. B. (2008) Nature 455, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Estruch F. (2000) FEMS Microbiol. Rev. 24, 469–486 [DOI] [PubMed] [Google Scholar]
- 3. Eizirik D. L., Moore F., Flamez D., Ortis F. (2008) Biochem. Soc. Trans. 36, 321–327 [DOI] [PubMed] [Google Scholar]
- 4. Gepts W. (1965) Diabetes 14, 619–633 [DOI] [PubMed] [Google Scholar]
- 5. Cnop M., Welsh N., Jonas J. C., Jörns A., Lenzen S., Eizirik D. L. (2005) Diabetes 54, S97–107 [DOI] [PubMed] [Google Scholar]
- 6. Eizirik D. L., Mandrup-Poulsen T. (2001) Diabetologia 44, 2115–2133 [DOI] [PubMed] [Google Scholar]
- 7. Eizirik D. L., Colli M. L., Ortis F. (2009) Nat. Rev. Endocrinol. 5, 219–226 [DOI] [PubMed] [Google Scholar]
- 8. Thomas H. E., McKenzie M. D., Angstetra E., Campbell P. D., Kay T. W. (2009) Apoptosis. 14, 1389–1404 [DOI] [PubMed] [Google Scholar]
- 9. Eldor R., Yeffet A., Baum K., Doviner V., Amar D., Ben-Neriah Y., Christofori G., Peled A., Carel J. C., Boitard C., Klein T., Serup P., Eizirik D. L., Melloul D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5072–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ortis F., Pirot P., Naamane N., Kreins A. Y., Rasschaert J., Moore F., Théâtre E., Verhaeghe C., Magnusson N. E., Chariot A., Orntoft T. F., Eizirik D. L. (2008) Diabetologia 51, 1213–1225 [DOI] [PubMed] [Google Scholar]
- 11. Cardozo A. K., Heimberg H., Heremans Y., Leeman R., Kutlu B., Kruhøffer M., Ørntoft T., Eizirik D. L. (2001) J. Biol. Chem. 276, 48879–48886 [DOI] [PubMed] [Google Scholar]
- 12. Cardozo A. K., Kruhøffer M., Leeman R., Orntoft T., Eizirik D. L. (2001) Diabetes 50, 909–920 [DOI] [PubMed] [Google Scholar]
- 13. Cardozo A. K., Ortis F., Storling J., Feng Y. M., Rasschaert J., Tonnesen M., Van Eylen F., Mandrup-Poulsen T., Herchuelz A., Eizirik D. L. (2005) Diabetes 54, 452–461 [DOI] [PubMed] [Google Scholar]
- 14. Callewaert H. I., Gysemans C. A., Ladrière L., D'Hertog W., Hagenbrock J., Overbergh L., Eizirik D. L., Mathieu C. (2007) Diabetes 56, 2169–2173 [DOI] [PubMed] [Google Scholar]
- 15. Gysemans C. A., Ladrière L., Callewaert H., Rasschaert J., Flamez D., Levy D. E., Matthys P., Eizirik D. L., Mathieu C. (2005) Diabetes 54, 2396–2403 [DOI] [PubMed] [Google Scholar]
- 16. Gysemans C., Callewaert H., Moore F., Nelson-Holte M., Overbergh L., Eizirik D. L., Mathieu C. (2009) Diabetologia. 52, 2374–2384 [DOI] [PubMed] [Google Scholar]
- 17. Kröger A., Köster M., Schroeder K., Hauser H., Mueller P. P. (2002) J. Interferon Cytokine Res. 22, 5–14 [DOI] [PubMed] [Google Scholar]
- 18. Liu L., Paul A., MacKenzie C. J., Bryant C., Graham A., Plevin R. (2001) Br. J. Pharmacol. 134, 1629–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moore F., Colli M. L., Cnop M., Esteve M. I., Cardozo A. K., Cunha D. A., Bugliani M., Marchetti P., Eizirik D. L. (2009) Diabetes 58, 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ortis F., Cardozo A. K., Crispim D., Störling J., Mandrup-Poulsen T., Eizirik D. L. (2006) Mol. Endocrinol. 20, 1867–1879 [DOI] [PubMed] [Google Scholar]
- 21. Magnusson N. E., Cardozo A. K., Kruhøffer M., Eizirik D. L., Ørntoft T. F., Jensen J. L. (2005) BMC Bioinformatics 6, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heimberg H., Heremans Y., Jobin C., Leemans R., Cardozo A. K., Darville M., Eizirik D. L. (2001) Diabetes 50, 2219–2224 [DOI] [PubMed] [Google Scholar]
- 24. Ortis F., Naamane N., Flamez D., Ladrière L., Moore F., Cunha D. A., Colli M. L., Thykjaer T., Thorsen K., Orntoft T. F., Eizirik D. L. (2010) Diabetes 59, 358–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008) Nature 455, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 27. Gurzov E. N., Ortis F., Cunha D. A., Gosset G., Li M., Cardozo A. K., Eizirik D. L. (2009) Cell Death. Differ. 16, 1539–1550 [DOI] [PubMed] [Google Scholar]
- 28. Ma C., Ying C., Yuan Z., Song B., Li D., Liu Y., Lai B., Li W., Chen R., Ching Y. P., Li M. (2007) J. Biol. Chem. 282, 30901–30909 [DOI] [PubMed] [Google Scholar]
- 29. Saito H., Morita Y., Fujimoto M., Narazaki M., Naka T., Kishimoto T. (2000) J. Immunol. 164, 5833–5843 [DOI] [PubMed] [Google Scholar]
- 30. Kim S., Kim H. S., Chung K. W., Oh S. H., Yun J. W., Im S. H., Lee M. K., Kim K. W., Lee M. S. (2007) Diabetes 56, 2561–2568 [DOI] [PubMed] [Google Scholar]
- 31. Gysemans C., Callewaert H., Overbergh L., Mathieu C. (2008) Biochem. Soc. Trans. 36, 328–333 [DOI] [PubMed] [Google Scholar]
- 32. Allagnat F., Christulia F., Ortis F., Pirot P., Lortz S., Lenzen S., Eizirik D. L., Cardozo A. K. (2010) Diabetologia 53, 1120–1130 [DOI] [PubMed] [Google Scholar]
- 33. Pérez-Arana G., Blandino-Rosano M., Prada-Oliveira A., Aguilar-Diosdado M., Segundo C. (2010) Endocrinology 151, 2538–2546 [DOI] [PubMed] [Google Scholar]
- 34. McGillicuddy F. C., Chiquoine E. H., Hinkle C. C., Kim R. J., Shah R., Roche H. M., Smyth E. M., Reilly M. P. (2009) J. Biol. Chem. 284, 31936–31944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun L., Ma K., Wang H., Xiao F., Gao Y., Zhang W., Wang K., Gao X., Ip N., Wu Z. (2007) J. Cell Biol. 179, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tajima K., Takaishi H., Takito J., Tohmonda T., Yoda M., Ota N., Kosaki N., Matsumoto M., Ikegami H., Nakamura T., Kimura T., Okada Y., Horiuchi K., Chiba K., Toyama Y. (2010) J. Orthop. Res. 28, 937–941 [DOI] [PubMed] [Google Scholar]
- 37. Kent S. C., Chen Y., Bregoli L., Clemmings S. M., Kenyon N. S., Ricordi C., Hering B. J., Hafler D. A. (2005) Nature 435, 224–228 [DOI] [PubMed] [Google Scholar]
- 38. Sibley R. K., Sutherland D. E., Goetz F., Michael A. F. (1985) Lab. Invest. 53, 132–144 [PubMed] [Google Scholar]
- 39. Darville M. I., Eizirik D. L. (2006) Biochem. Biophys. Res. Commun. 339, 1063–1068 [DOI] [PubMed] [Google Scholar]
- 40. Imaizumi K., Tsuda M., Imai Y., Wanaka A., Takagi T., Tohyama M. (1997) J. Biol. Chem. 272, 18842–18848 [DOI] [PubMed] [Google Scholar]
- 41. Imaizumi K., Benito A., Kiryu-Seo S., Gonzalez V., Inohara N., Lieberman A. P., Kiyama H., Nuñez G. (2004) J. Neurosci. 24, 3721–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kanazawa K., Imaizumi K., Mori T., Honma Y., Tojo M., Tanno Y., Yokoya S., Niwa S., Tohyama M., Takagi T., Wanaka A. (1998) Brain Res. Mol. Brain Res. 54, 316–320 [DOI] [PubMed] [Google Scholar]
- 43. Cardozo A. K., Proost P., Gysemans C., Chen M. C., Mathieu C., Eizirik D. L. (2003) Diabetologia. 46, 255–266 [DOI] [PubMed] [Google Scholar]
- 44. Martin A. P., Rankin S., Pitchford S., Charo I. F., Furtado G. C., Lira S. A. (2008) Diabetes 57, 3025–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frigerio S., Junt T., Lu B., Gerard C., Zumsteg U., Holländer G. A., Piali L. (2002) Nat. Med. 8, 1414–1420 [DOI] [PubMed] [Google Scholar]
- 46. Roep B. O., Kleijwegt F. S., van Halteren A. G., Bonato V., Boggi U., Vendrame F., Marchetti P., Dotta F. (2010) Clin. Exp. Immunol. 159, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flodström-Tullberg M., Yadav D., Hägerkvist R., Tsai D., Secrest P., Stotland A., Sarvetnick N. (2003) Diabetes 52, 2696–2700 [DOI] [PubMed] [Google Scholar]
- 48. Chong M. M., Chen Y., Darwiche R., Dudek N. L., Irawaty W., Santamaria P., Allison J., Kay T. W., Thomas H. E. (2004) J. Immunol. 172, 5714–5721 [DOI] [PubMed] [Google Scholar]
- 49. Ho H. H., Ivashkiv L. B. (2006) J. Biol. Chem. 281, 14111–14118 [DOI] [PubMed] [Google Scholar]
- 50. Rasschaert J., Ladrière L., Urbain M., Dogusan Z., Katabua B., Sato S., Akira S., Gysemans C., Mathieu C., Eizirik D. L. (2005) J. Biol. Chem. 280, 33984–33991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.