Abstract
Relationships between genes and social behavior have historically been construed as a one-way street, with genes in control. Recent analyses have challenged this view by discovering broad alterations in the expression of human genes as a function of differing socio-environmental conditions. The emerging field of social genomics has begun to identity the types of genes subject to social regulation, the biological signaling pathways mediating those effects, and the genetic polymorphisms that moderate socio-environmental influences on human gene expression.
Keywords: Genetics, genomics, social factors, gene-environment interactions
GENETICS VS. GENOMICS
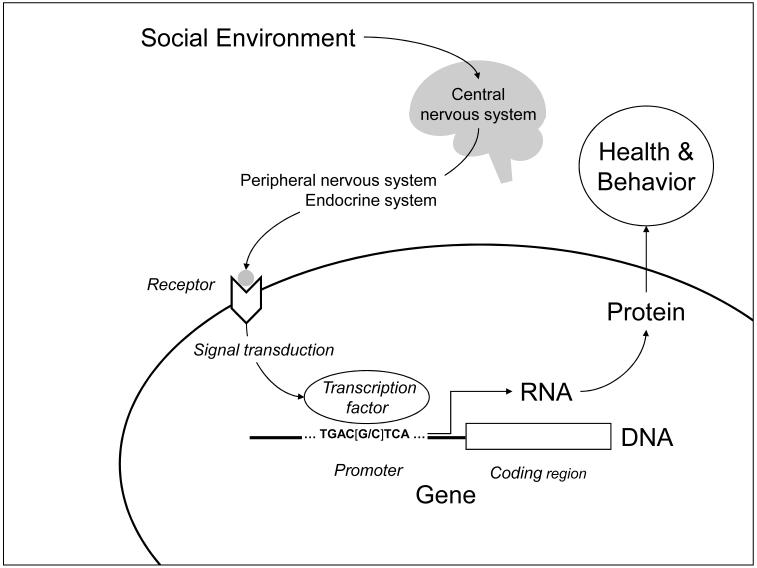
The conceptual relationship between genes and social behavior has shifted significantly during the past 20 years. As genes have come to be understood as concrete DNA sequences, rather than abstractions inferred from inheritance, it has become increasingly clear that social factors can play a significant role in regulating the activity of human genes. DNA encodes the potential for cellular behavior, but that potential is only realized if the gene is expressed - if its DNA is transcribed into RNA and translated into protein (Figure 1). Proteins shape the structure of a cell, and endow its characteristic behaviors such as movement, metabolism, and biochemical response to external stimuli (e.g., neurotransmission). Absent their transcription, DNA genes have no effect on health or behavioral phenotypes. With the advent of a sequenced human genome and the emergence of DNA microarray technologies, scientists can now survey the expression of all human genes simultaneously and map the specific subset of genes that are active in a given cell at a given point in time. One surprising finding from the field of “functional genomics” is that the expression of a specific gene is often more an exception than the rule. Cells are highly selective about which genes they express, and our DNA encodes a great deal more genetic potential than is realized in RNA and protein. Even more striking has been the discovery that the social world outside our bodies influences which genes are transcribed within the nuclei of our cells (the RNA “transcriptome”).
FIGURE 1.
Social signal transduction. Socio-environmental processes regulate human gene expression by activating central nervous system processes that subsequently influence hormone and neurotransmitter activity in the periphery of the body. Peripheral signaling molecules interact with cellular receptors to activate transcription factors, which bind to characteristic DNA motifs in gene promoters to initiate (or repress) gene expression. Only genes that are transcribed into RNA actually impact health and behavioral phenotypes. Individual differences in promoter DNA sequences (e.g., the [G/C] polymorphism shown here) can affect the binding of transcription factors, and thereby influence genomic sensitivity to socio-environmental conditions.
SOCIAL REGULATION OF GENE EXPRESSION
The possibility that social factors might regulate gene expression first emerged in the context of bio-behavioral health research. Social stress and isolation have long been known to affect the onset and progression of disease (Seeman, 1996). That effect is particularly strong for viral infections, where social factors have been linked to increased replication of cold-causing rhinoviruses (Cohen, Doyle, Skoner, Rabin, & Gwaltney, 1997), the AIDS virus, HIV-1 (Cole, 2008), and several cancer-related viruses (Antoni et al., 2006). Viruses are little more than small packages of 10-100 genes that hijack the protein production machinery of their host cells (us) to make more copies of themselves. As obligate parasites of our living cells, viruses have evolved within a micro-environment structured by our own genome. If social factors can regulate the expression of viral genes, that suggests that our own complement of ~22,000 genes is likely to be regulated in biologically significant ways as well.
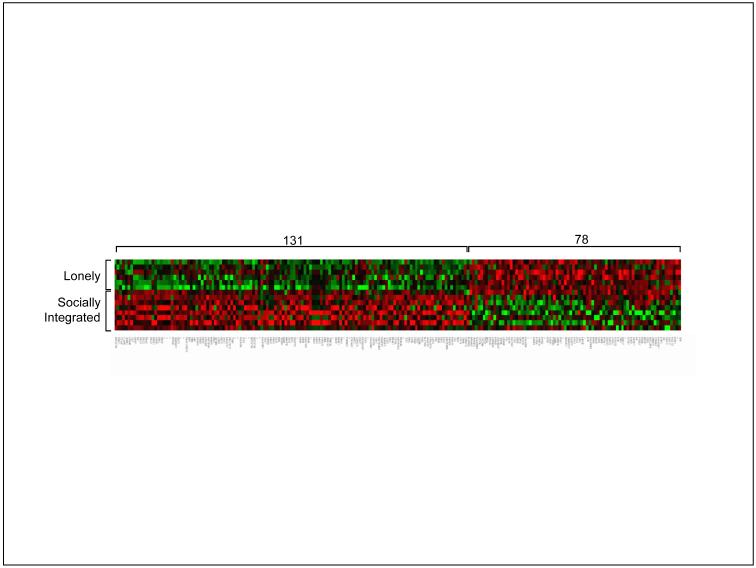
One of the first studies to analyze the relationship between social factors and human gene expression surveyed transcriptional profiles in white blood cells (leukocytes) from healthy older adults who differed in the extent to which they felt socially connected to others (Cole et al., 2007). Among the 22,283 genes assayed, 209 showed systematically different levels of expression in people who reported feeling lonely and distant from others consistently over the course of 4 years (Figure 2). These effects did not involve a random smattering of all human genes, but focally impacted three specific groups of genes. Genes supporting the early “accelerator” phase of the immune response – inflammation – were selectively up-regulated. However, two groups of genes involved in the subsequent “steering” of immune responses were down-regulated: genes involved in responses to viral infections (particularly Type I interferons), and genes involved in the production of antibodies by B lymphocytes. These results provided a molecular framework for understanding why socially isolated individuals show heightened vulnerability to inflammation-driven cardiovascular diseases (i.e., excessive non-specific immune activity) and impaired responses to viral infections and vaccines (i.e., insufficient immune responses to specific pathogens). A major clue about the psychological pathways mediating these effects came from the observation that differential gene expression profiles were most strongly linked to a person’s subjective sense of isolation, rather than their objective number of social contacts.
FIGURE 2.
Social regulation of gene expression in human immune cells. Expression of 22,283 human gene transcripts was assayed in 10 million blood leukocytes sampled from each of 14 older adults who showed consistent differences over 4 years in their level of subjective social isolation. 209 gene transcripts showed ≥ 30% difference in average expression level in leukocytes from 6 people experiencing chronic social isolation vs. 8 experiencing consistent social integration. In the heat-plot above, each row represents data from one of the 14 study participants, each column contains expression values for one of the 209 differentially active genes, and the coloring of each cell represents the relative level of that gene’s expression in a given participant’s leukocyte sample: Red = high expression, Black = intermediate expression, Green = low expression. (Adapted from Cole, et al., 2007).
Additional studies have identified transcriptional correlates of other socio-environmental conditions such as low socio-economic status (SES) (Chen et al., 2008) and the chronic threat of social loss (e.g., having a spouse with cancer) (Miller et al., 2008). These analyses also found up-regulated expression of leukocyte inflammatory genes, and identified specific psychological processes that appeared to contribute to those dynamics. For example, among children with asthma, those from a low-SES background tended to interpret ambiguous situations as threatening, and that perception of threat was more strongly linked to differential gene expression than was SES per se (Chen et al., 2008).
Several studies have shown that social influences can penetrate remarkably deeply into our bodies. The nervous system plays a key role in perceiving and responding to social stimuli, and social conditions have been found to regulate the expression of neural genes such as the Nerve Growth Factor NGF gene (Sloan et al., 2007) and the glucocorticoid receptor gene (Zhang et al., 2006). More surprising is the discovery that key immune system genes are also sensitive to social conditions (Sloan et al., 2007). Immune cells exert selective pressure on the evolution of viral genomes, and many viruses also appear to have developed a genomic sensitivity to our social conditions (as reviewed above). However, even pathogens that escape our immune system may still modulate gene transcription in response to host stress and social conditions. Most human cancers are invisible to the immune system, but some still change gene expression patterns in response to social stress (Antoni et al., 2006). One recent study of women with ovarian cancer found more than 220 genes to be selectively up-regulated in tumors from women with low levels of social support and high depressive symptoms (Lutgendorf et al., 2008). If our socially sensitive immune system is not conveying those effects, how do social influences reach into the damaged genome of a cancer cell? New insights have come from bioinformatic analyses of “social signal transduction.”
SOCIAL SIGNAL TRANSDUCTION
Molecular biologists construe signal transduction as a local process by which signaling molecules outside the cell interact with cellular receptors to initiate a cascade of biochemical reactions inside the cell, ultimately stimulating a protein “transcription factor” to activate gene expression (Figure 1). Transcription factors flag a particular stretch of DNA (the “coding region” of a gene) for transcription into RNA. Which genes can be activated by a given transcription factor is determined by the nucleotide sequence of the gene’s promoter – the stretch of DNA lying upstream of the coding region. For example, the transcription factor NF-κB binds to the nucleotide motif GGGACTTTCC, whereas CREB/ATF transcription factors target the motif TGACGTCA. These two transcription factors are activated by different receptor-mediated signal transduction pathways, providing distinct molecular channels by which extra-cellular events can regulate intracellular genomic response. The distribution of transcription factor-binding motifs across our ~22,000 gene promoters constitutes a “wiring diagram” that maps micro-environmental processes onto genome-wide transcriptional responses.
Transduction of socio-environmental influences into functional genomic responses is mediated by the brain’s perception of social conditions, and its subsequent regulation of hormones, neurotransmitters, and other signaling molecules that disseminate throughout the body to activate cellular receptors and transcription factors. For example, the sympathetic nervous system (SNS) and the hypothalamic-pituitary-adrenal (HPA) axis represent two major pathways by which central nervous system (CNS) perceptions of negative social conditions can regulate gene transcription in a wide array of somatic cells (Sapolsky, 1994). Positive psychological states may also regulate human gene expression (Dusek et al., 2008), although their molecular mediators are less well understood.
Links between social experiences and neural / endocrine responses have long been recognized, but the breadth of their impact on gene expression has only recently become apparent following the sequencing of the human genome. Early computational analyses of the human genome sequence suggested that promoter DNA sequences might provide for psychologically specific transcriptional responses. For example, any gene bearing the motif GGTACAATCTGTTCT in its promoter might potentially be stimulated by severe, overwhelming stress experiences that release cortisol, because the cortisol-stimulated glucocorticoid receptor (GR) binds specifically to that DNA motif. In contrast, genes bearing the CREB/ATF promoter motif TGACGTCA would be predicted to activate in response to active-coping, fight-or-flight stress responses associated with catecholamine release and beta-adrenergic receptor signaling. Based on the distribution of these promoter motifs across the human genome, it appears that these two distinct psychological stress experiences may trigger very different transcriptional responses. Genes predicted to be cortisol-responsive disproportionately encode receptors and other molecules involved in a cell’s “perception” of its local environment. In contrast, putative catecholamine-responsive genes include few receptors, but a high concentration of signal transduction molecules and transcription factors involved in cellular “decision-making” (converting receptor-mediated perception into changes in gene expression and cellular behavior). Thus, severe, overwhelming stress may trigger a cellular form of “denial” (altering perception), whereas active-coping challenges induce something more akin to “sublimation” (altering responses to perceptions).
A sequenced human genome also provided new analytic infrastructure for mapping the molecular signaling pathways that convert socio-environmental conditions into differential gene expression. One approach reverses the normal flow of biological information from the environment, through transcription factor activity, and into gene expression (Figure 1). This analysis scans the promoters of differentially expressed genes to identify transcription factor-binding motifs that are over-represented in activated promoters, and thus reflect which specific transcription factors drove the observed differences in gene expression (Cole, Yan, Galic, Arevalo, & Zack, 2005). This approach has uncovered some surprising differences between the transcriptional signals “sent” by the brain, and the transcriptional signals “heard” by the human genome. In studies of chronic loneliness and threat of social loss (Cole et al., 2007; Miller et al., 2008), analyses indicated that the inflammation-driving NF-κB transcription factor played a key role in orchestrating both patterns of differential gene expression. Results also suggested that the GR was failing to inhibit NF-κB’s pro-inflammatory activity as it should. Neither study found decreases in circulating cortisol levels that might explain the reduced GR activity. If the HPA axis were sending the proper anti-inflammatory cortisol signal, why would stressed people’s leukocytes not down-regulate NF-κB transcription of inflammatory genes? The answer appears to involve a reduction in the GR’s sensitivity to cortisol – rendering the leukocyte transcriptome deaf to the brain’s request to down-regulate pro-inflammatory genes (Cole et al., 2007; Miller et al., 2008). Both chronic loneliness and threat of social loss appear to disconnect this key physiologic feedback system, and may thereby increase the risk of inflammation-related disease (Seeman, 1996). Similar analyses have identified other alterations in transcription factor activity that may connect low SES to inflammatory gene expression in asthma (Chen et al., 2008), and connect low social support and depression to altered gene expression in ovarian cancer (Lutgendorf et al., 2008).
REMODELING THE BODY
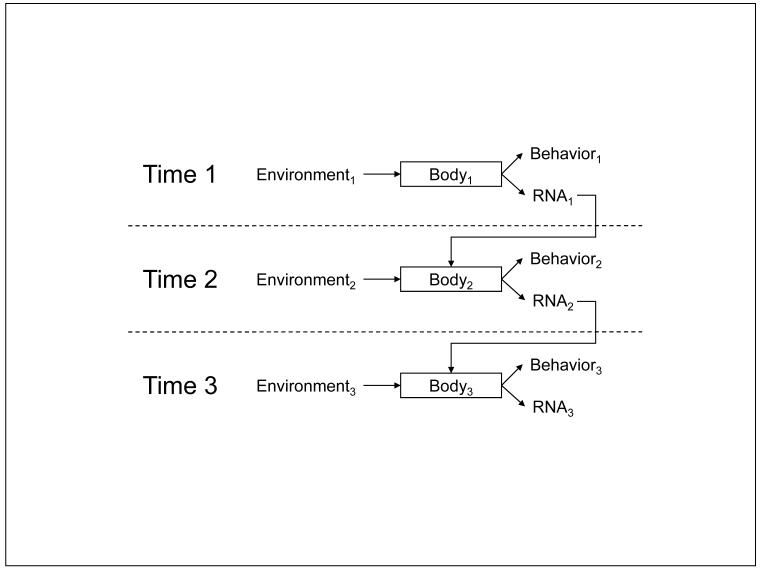
Because RNA transcription shapes the protein complement of our cells, and those proteins mediate cellular function (Figure 1), psychological regulation of gene expression implies that the social world can remodel the functional characteristics of the human body. Consider the ability of chronic social stress to increase NGF gene expression, and thereby enhance the growth of SNS neural fibers in the lymph node tissues that structure immune responses (Sloan et al., 2007). This socio-environmental remodeling of lymph node innervation at Time1 can persist, providing a denser neural network through which subsequent stressful exposure at Time2 distributes SNS neurotransmitters into the lymph node. As a consequence, the immune system mounts a poorer response to viral infection at Time2 solely because lymph node innervation was remodeled by differing social conditions at Time1. In the model of Figure 3, social stress at Time1 (Environment1) is transmitted through the nervous system (Body1) into behavioral stress responses (Behavior1) and increased NGF gene expression (RNA1). Up-regulated NGF increases SNS innervation of the lymph node, and thereby alters the functional relationship between the nervous and immune systems (Body2). When that functionally remodeled Body2 encounters a new viral infection in Environment2, increased SNS neurotransmitter release can inhibit transcription of Type I interferon genes (RNA2). As a consequence of that impaired anti-viral response, intensified disease alters physical tissue characteristics and behavioral capacities in the future (RNA3 and Behavior3). In this way, the experience of Environment1 not only “gets inside the body” but “stays there” in a concrete molecular sense that propagates through multiple gene transcriptional responses, physiologic systems, and time epochs.
FIGURE 3.
RNA as a molecular medium of recursive development. Social conditions at one point in time (Environment1) are transduced into changes in behavior (Behavior1) and gene expression (RNA1) via CNS perceptual processes that trigger systemic neural and endocrine responses (mediated by Body1). Those RNA transcriptional dynamics may alter molecular characteristics of cells involved in environmental perception or response, resulting in a functionally altered Body2. Body2 may respond differently to a given environmental challenge that would the previous Body1, resulting in different behavioral (Behavior2) and RNA transcriptional responses (RNA2). The persisting effect of RNA transcriptional dynamics on cellular protein and functional characteristics provides a molecular framework for understanding how socio-environmental conditions in the past may continue to affect current behavior and health, and how those historical conditions interact with current environments to shape our future trajectories (e.g., Body3, Behavior3, RNA3). Because gene transcription serves as both a cause of social behavior (by shaping Body) and a consequence of social behavior (a product of Environment x Body), RNA can serve as the physical medium for a recursive developmental trajectory that integrates genetic characteristics and historical-environmental regulators to understand individual biological and behavioral responses to current environmental conditions.
Socio-environmental conditions can also regulate the molecular composition of CNS cells, and thereby alter psychological and behavioral responses to future environments (Zhang et al., 2006). Because the molecular composition of our cells constitutes the physical machinery by which we perceive and respond to the world around us (“Body” in Figure 3), and that molecular composition is itself subject to remodeling by socoio-environmental influences, gene expression constitutes both a cause and a consequence of behavior. RNA can be construed as the physical medium of a recursive developmental system in which social, behavioral, and health outcomes at one point in time also constitute inputs that shape our future responses to the environment (e.g., as in Heckman’s model of human capability development, which analyzes how capacities developed at Time1 impact our ability to capitalize on environmental opportunities at Time2 (Heckman, 2007)). Future research will push these models out of accessible immune cells and into the more sensitive CNS structures that shape social, cognitive, and affective processes. It will also be critical to define the particular features of social environments that trigger transcriptional remodeling of specific cells. Given the key role of neuroendocrine responses in mediating these effects, the most decisive influences may involve our psychological reactions to social conditions rather than the properties of the external condition per se. After all, it is the subjective perception of conditions as threatening or uncertain that directly triggers SNS and HPA responses (Sapolsky, 1994). Our genome’s social sensitivity ultimately stems from the capacity of social conditions to affect CNS perceptions of safety vs. threat (Dickerson & Kemeny, 2004), and thereby trigger biological stress responses that alter gene transcription.
THE NEW GENETICS
With genes and environments now operating in parallel to shape our RNA-driven bodies, the integration of those two streams of influence has become a central challenge in biological analyses of human health and behavior. The regulatory paradigm outlined in Figure 1 provides a framework for analyzing their interplay in the context of Gene x Environment interactions. For example, variations in the DNA sequence of the promoter regulating the serotonin transporter gene (5HTT) can affect the binding of environmentally sensitive transcription factors, and thereby buffer the effects of adverse social environments on the risk of depression and other affective behaviors (Caspi et al., 2003; Champoux et al., 2002). These effects extend into the realm of immune response and survival (Capitanio et al., 2008), and thus may also shape the evolutionary trajectory of our DNA genome at a population level. In integrating the molecular biology of gene structure, the environmental control of gene expression, and the social biology of individual behavior and survival, the 5HTT promoter polymorphism exemplifies the new “environmentally conscious” conception of genetics in which cellular and organismic behavior constitute the fundamental units of evolutionary selection, and genes and environments depend mutually on one another to shape that behavior by structuring our brains and bodies.
Research in social genomics has now clearly established that our interpersonal world exerts biologically significant effects on the molecular composition of the human body. These effects typically target a non-random ~1% of the human genome (though often a different 1% depending upon the social circumstances and cell type studied). Major topic for future exploration will involve determining which particular genes are subject to social regulation, what types of social conditions elicit such dynamics, which psychological and biological pathways mediate those effects, and which DNA polymorphisms moderate or intensify their impact.
ACKNOWLEDGEMENTS
Supported by R01 CA116778 from the National Cancer Institue.
REFERENCES
- Antoni MH, Lutgendorf SK, Cole SW, Dhabhar FS, Sephton SE, McDonald PG, et al. The influence of bio-behavioural factors on tumour biology: pathways and mechanisms. Nat Rev Cancer. 2006;6(3):240–248. doi: 10.1038/nrc1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio JP, Abel K, Mendoza SP, Blozis SA, McChesney MB, Cole SW, et al. Personality and serotonin transporter genotype interact with social context to affect immunity and viral set-point in simian immunodeficiency virus disease. Brain Behav Immun. 2008;22(5):676–689. doi: 10.1016/j.bbi.2007.05.006. Epub 2007 Aug 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. A landmark paper identifying a transcription-regulating DNA polymorphism as a molecular vulnerability factor for the development of depression in response to psychological stress.
- Champoux M, Bennett A, Shannon C, Higley JD, Lesch KP, Suomi SJ. Serotonin transporter gene polymorphism, differential early rearing, and behavior in rhesus monkey neonates. Mol Psychiatry. 2002;7(10):1058–1063. doi: 10.1038/sj.mp.4001157. [DOI] [PubMed] [Google Scholar]
- Chen E, Miller GE, Walker HA, Arevalo JM, Sung CY, Cole SW. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2008 doi: 10.1136/thx.2007.095091. doi:10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Doyle WJ, Skoner DP, Rabin BS, Gwaltney JM. Social ties and susceptibility to the common cold. Journal of the American Medical Association. 1997;227:1940–1944. [PubMed] [Google Scholar]
- Cole SW. Psychosocial influences on HIV-1 disease progression: neural, endocrine, and virologic mechanisms. Psychosom Med. 2008;70(5):562–568. doi: 10.1097/PSY.0b013e3181773bbd. [DOI] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. This study provided the first indication that social factors might systematically regulate human genome activity in the immune system, and it pioneered novel strategies for identifying the social signal transduction pathways involved.
- Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: The TELiS database. Bioinformatics. 2005;21(6):803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- Dickerson SS, Kemeny ME. Acute stressors and cortisol responses: a theoretical integration and synthesis of laboratory research. Psychol Bull. 2004;130(3):355–391. doi: 10.1037/0033-2909.130.3.355. [DOI] [PubMed] [Google Scholar]
- Dusek JA, Otu HH, Wohlhueter AL, Bhasin M, Zerbini LF, Joseph MG, et al. Genomic counter-stress changes induced by the relaxation response. PLoS ONE. 2008;3(7):e2576. doi: 10.1371/journal.pone.0002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman JJ. The economics, technology, and neuroscience of human capability formation. Proc Natl Acad Sci U S A. 2007;104(33):13250–13255. doi: 10.1073/pnas.0701362104. Epub 12007 Aug 13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgendorf SK, Degeest K, Sung CY, Arevalo JM, Penedo F, Lucci J, 3rd, et al. Depression, social support, and beta-adrenergic transcription control in human ovarian cancer. Brain Behav Immun. 2008;10:10. doi: 10.1016/j.bbi.2008.04.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R, et al. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. Epub 2008 Apr 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky RM. Why zebras don’t get ulcers: A guide to stress, stress-related diseases, and coping. Freeman; New York: 1994. [Google Scholar]
- Seeman TE. Social ties and health: the benefits of social integration. Ann Epidemiol. 1996;6(5):442–451. doi: 10.1016/s1047-2797(96)00095-6. [DOI] [PubMed] [Google Scholar]
- Sloan EK, Capitanio JP, Tarara RP, Mendoza SP, Mason WA, Cole SW. Social stress enhances sympathetic innervation of primate lymph nodes: mechanisms and implications for viral pathogenesis. J Neurosci. 2007;27(33):8857–8865. doi: 10.1523/JNEUROSCI.1247-07.2007. A key experimental study showing that social stress can activate neural growth genes in non-human primates, and thereby remodel the neural structures that transmit social influences into gene expression in the immune system and viral pathogens.
- Zhang TY, Bagot R, Parent C, Nesbitt C, Bredy TW, Caldji C, et al. Maternal programming of defensive responses through sustained effects on gene expression. Biol Psychol. 2006;73(1):72–89. doi: 10.1016/j.biopsycho.2006.01.009. Epub 2006 Feb 2028. [DOI] [PubMed] [Google Scholar]
- Miller G, Chen E, Cole SW. Health psychology: Developing biologically plausible models linking the social world and physical health. Ann. Rev. Psychol. 2009;60:501–524. doi: 10.1146/annurev.psych.60.110707.163551. This article provides a deeper review of recent research on psychological regulation of gene expression in human health.