Abstract
We have studied the CBK1 gene of Saccharomyces cerevisiae, which encodes a conserved protein kinase similar to the human myotonic dystrophy kinase. We have shown that the subcellular localization of the protein, Cbk1p, varies in a cell cycle-dependent manner. Three phenotypes are associated with the inactivation of the CBK1 gene: large aggregates of cells, round rather than ellipsoidal cells and a change from a bipolar to a random budding pattern. Two-hybrid and extragenic suppressor studies have linked Cbk1p with the transcription factor Ace2p, which is responsible for the transcription of chitinase. Cbk1p is necessary for the activation of Ace2p and we have shown that the aggregation phenotype is due to a lack of chitinase expression. The random budding pattern and the round cell phenotype of the CBK1 deletion strain show that in addition to its role in regulating chitinase expression via Ace2p, Cbk1p is essential for a wild-type morphological development of the cell.
Keywords: ACE2/budding pattern/CBK1 kinase/cell cycle/Saccharomyces cerevisiae
Introduction
Cell growth and morphogenesis is a complex process, which is tightly linked to the cell cycle. This has been studied extensively in the budding yeast Saccharomyces cerevisiae (see Madden and Snyder, 1998 for a detailed review) and involves a highly organized programme of gene transcription (Cho et al., 1998; Spellman et al., 1998), the specific transport of ‘raw materials’ to the site of growth, turnover and directed synthesis of the cell wall (Smits et al., 1999), selection of the new bud site (Chant and Pringle, 1995; reviewed in Chant, 1999) and an oscillation between apical and isotropic growth (Lew and Reed, 1993).
The organization of the actin cytoskeleton plays a critical role in several of these morphogenic processes (see Karpova et al., 1998 for a description of the actin cytoskeleton throughout the cell cycle). Actin patches are the site of plasma membrane invaginations and it is thought that these are the sites of growth, with the secretory vesicles containing cell wall and other components being delivered along actin cables (Mulholland et al., 1994). The actin cytoskeleton is also involved in bud site selection, at least for bipolar budding (Yang et al., 1997). Finally, the switch between apical and isotropic growth is determined by the timing of the depolarization of the actin cytoskeleton, which is regulated in a cell cycle-dependent manner by the PAK kinases Ste20p and Cla4p (Holly and Blumer, 1999).
Previously, we identified the CBK1 gene (YNL161w) and showed that it is a member of a large family of protein kinases that is conserved from yeast to man and includes the human myotonic dystrophy kinase. The functional data that are available concerning these kinases suggest that they are involved in the establishment, or maintenance, of a normal cellular morphology. In Schizosaccharomyces pombe mutations in orb6 result in the formation of small round cells (Verde et al., 1998), in Neurospora crassa mutations in Cot1 give a pseudo-colonial growth (Yarden et al., 1992) and in Ustilago maydis inactivation of the ukc1 gene leads to aggregates of round cells with hyphal extensions (Dürrenberger and Kronstad, 1999). In higher organisms the warts kinase of Drosophila is involved in the control of cell shape and proliferation (Justice et al., 1995) and mutants of let-502 in Caenorhabditis elegans block the elongation of the embryo (Wissmann et al., 1997). We present here a study of the S.cerevisiae CBK1 gene, we show that the subcellular localization of the protein is cell cycle-dependent and that it regulates CTS1 (chitinase) transcription via the activity of the transcription factor Ace2p. Also, we demonstrate that deletion of the CBK1 gene in strains that have a bipolar budding pattern gives rise to a random budding pattern and round rather than ellipsoidal cells, showing that Cbk1p is an important factor in the establishment of cellular morphology.
Results
Deletion of the CBK1 gene
We have shown previously that the S.cerevisiae gene CBK1 encodes a member of a large family of conserved protein kinases, including the human myotonic dystrophy kinase (Nasr et al., 1996). We decided to investigate the role of CBK1 in S.cerevisiae and to analyse some of its genetic and physical interactions. As a first step we deleted the gene as described in Materials and methods.
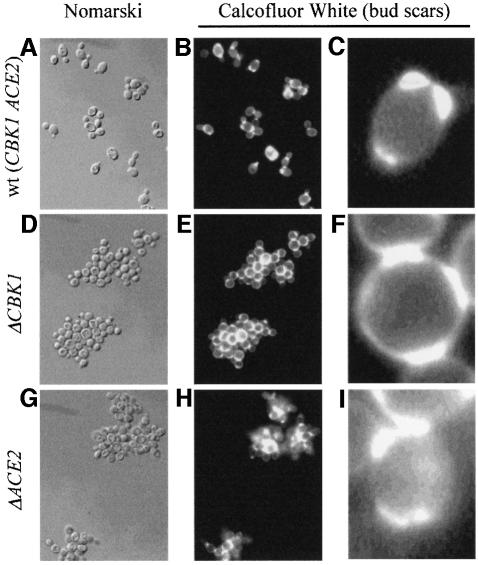
Deletion of the CBK1 gene showed that it is not essential. On complete solid medium the Δcbk1 strain gave rise to colonies that have a crinkled phenotype with many surface invaginations and a ‘dry’ appearance; in liquid culture it grew as large aggregates of cells (Figure 1A, B, D and E). These aggregates cannot easily be broken up with a micro-manipulator and repeated treatment with EDTA does not cause them to dissociate, suggesting that the cells are physically attached. A closer examination of the cells of the CBK1 deletion strain showed that they also presented a round cell phenotype compared with the ellipsoidal shape of the wild-type strain.
Fig. 1. Inactivation phenotypes of CBK1 and ACE2. Fresh overnight cultures of wild-type, Δcbk1 and Δace2 haploid strains in rich complete medium were stained with Calcofluor white (see Materials and methods). Inactivation of CBK1 and ACE2 leads to formation of large aggregates, which cannot be separated using a micro-manipulator. (A–C) The wild-type strain AW303-2b; (D–F) the Δcbk1 strain WR03-5d; (G–I) the Δace2 strain WR27-1a. (A), (D) and (G) Normaski, (B), (C), (E), (F), (H) and (I) Calcofluor white staining.
Two-hybrid partners of Cbk1p
In order to identify possible substrates of Cbk1p we decided to look for two-hybrid interactions (Fields and Sternglanz, 1994). To do this we cloned an N-terminal truncated version of cbk1p, which starts at T279. This construction corresponds to the part of the protein that is homologous to the protein kinases from other organisms and eliminates the first 278 amino acids, which contain several highly charged glutamine-rich regions that could give rise to non-specific two-hybrid interactions. This bait construction was confirmed by sequencing the fusion sites. Moreover, upon transformation of the yeast host strain, a CBK1 deletion phenotype was obtained, indicating that the Gal4DB–Cbk1p fusion protein is correctly expressed and suggesting that an important partner of Cbk1p is trapped and effectively removed from the cell. This phenotype was also observed when the strain was transformed with the Gal4DB domain fused to the complete Cbk1p.
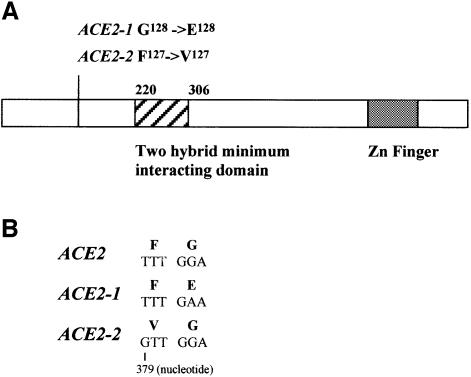
In total, six independent transformations yielded 107 primary transformants. After replica-plating these gave rise to 199 3-aminotriasol-positive clones, of which 152 were β-galactosidase positive. Prey plasmid DNA was isolated from all these clones, retransformed and these transformants were tested for 3-aminotriasol resistance and β-galactosidase activity. This resulted in the isolation of 119 true positives, which were identified by sequencing. A summary of the results is shown in Table I. Two proteins clearly appeared as preferred partners of Cbk1p in this two-hybrid analysis, the transcriptional activator Ace2p was isolated 23 times and Yol036wp 13. The next partner in the list was only identified six times followed by several others with five and four hits. A comparison of the different fragments of Ace2p selected in the two-hybrid screen showed that the region of the protein common to all fusions is only 87 amino acids that are situated in a domain that is distinct from the regions involved in DNA binding and regulated nuclear import (see Figure 2) (McBride et al., 1999).
Table I. Two-hybrid partners of Cbk1p.
| ORF name | Gene name | No. of isolates |
|---|---|---|
| YLR131c | ACE2 | 23 |
| YOL036w | 13 | |
| YCL051w | LRE1 | 6 |
| YNL272c | SEC2 | 5 |
| YER036c | 5 | |
| YDR293c | SSD1 | 5 |
| YIL038c | NOT3 | 4 |
| YFL035c | MOB2 | 4 |
A summary of the two-hybrid partners identified in our screen. In total 107 primary transformants were analysed, all clones have been confirmed by retransformation. Only proteins identified more than three times are shown.
Fig. 2. Schematic representation of Ace2p and the dominant suppressor mutations. (A) Schematic representation of Ace2p showing the position of the Zn finger DNA-binding domain, the minimal two-hybrid domain and the dominant suppressor mutations. (B) Sequence of the dominant suppressor mutations.
Extragenic suppressors of the CBK1 deletion
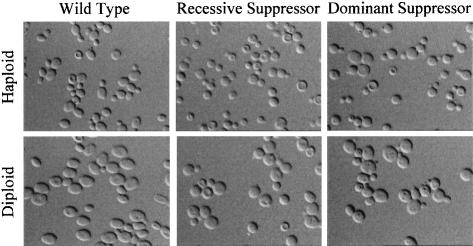
To further our knowledge of the genetic interactions of the CBK1 gene we have isolated extragenic suppressors of the aggregation and colony morphology phenotypes associated with its deletion. Twenty independent subclones of two Δcbk1 strains were UV mutagenized and suppressors were selected as described in Materials and methods. A genetic analysis showed that five of the suppressors were recessive and the other 15 dominant. Complementation studies showed that the recessive suppressors define a single gene and that the dominant suppressors are all closely linked and probably correspond to a single gene. Also, we have shown that the genes defined by the recessive and dominant suppressors are different. On solid complete media all the suppressors have a wild-type appearance. However, in liquid media it is clear that they are partial revertants, the large aggregates are no longer formed, but the cells are still round not ellipsoidal like the wild type (see Figure 3).
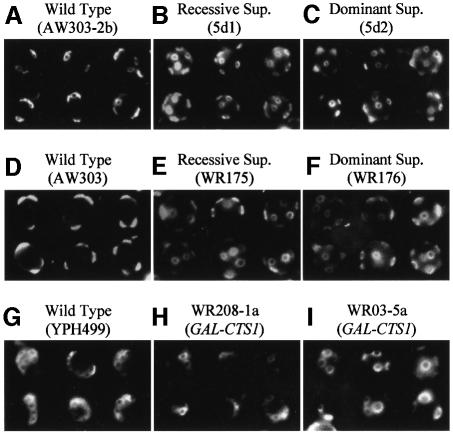
Fig. 3. Phenotype of the Δcbk1 suppressor strains. Fresh overnight cultures in rich complete medium of haploid and diploid wild-type (AW303-2b; AW303), Δcbk1 recessive suppressor (5d1; WR175) and Δcbk1 dominant suppressor (5d2; WR176) strains were observed by Nomarski interference microscopy. The suppressor strains no longer form large aggregates of cells, but are still round rather then ellipsoidal. This is most clearly seen in the diploid strains.
In order to determine if the dominant suppressors of the CBK1 deletion could also suppress the aggregation phenotype associated with the deletion of the ACE2 gene (see Figure 1G and H), we crossed them to a Δace2 strain. An analysis of the segregation pattern obtained from this diploid showed that the dominant suppressors were tightly linked to the ACE2 locus, suggesting that they might be alleles of ACE2. To test this hypothesis we amplified the ACE2 gene flanked by 150 bp from the wild-type strain and five dominant suppressors by high fidelity PCR. These fragments were cloned into a yeast shuttle vector and sequenced. The wild-type sequence was identical to that deposited in the Saccharomyces Genome Database at Stanford University. The sequence of the ACE2 alleles isolated from the dominant suppressors showed that in four out of five cases a single nucleotide was changed, all four mutations affected two adjacent residues in the protein sequence. The corresponding region was amplified from the other 10 dominant suppressors and the PCR fragments were sequenced directly. In total, we have obtained unambiguous sequence data from 10 independent dominant suppressors, one leads to the substitution F127→V127 and the other nine to the substitution G128→E128 (see Figure 2).
Cbk1p controls the expression of chitinase
At the present time two independent screens have linked ACE2 and CBK1. ACE2 encodes a transcription factor that is homologous to Swi5p (Butler and Thiele, 1991). One of the best-characterized targets of Ace2p is CTS1, which encodes chitinase (Dohrmann et al., 1992). The inactivation of ACE2 leads to the formation of aggregates similar to those seen in Δcbk1 strains. In the case of the Δace2 strains this is because chitinase is no longer transcribed and the chitin plate that separates the mother and daughter cell after division cannot be digested (see Figure 1E, F, H and I, where the chitin plate is revealed by staining with Calcofluor white). Given the similar phenotypes and the links we have shown between CBK1 and ACE2, we decided to look at the expression of CTS1 in our strains. Total RNA was prepared from wild type, Δcbk1, Δace2, dominant (ACE2-1) and recessive suppressors and probed with CTS1 in a northern analysis. The results in Figure 4A show that CTS1 transcription is almost undetectable in the Δcbk1 and Δace2 strains but restored to wild-type levels in the dominant and recessive suppressors. This suggests that Cbk1p regulates the transcription of CTS1 via Ace2p.
Fig. 4. Expression of the CTS gene. (A) Total RNA was extracted from wild-type (AW303-2b), Δcbk1 (WR03-5d), Δace2 (WR27-1a) Δcbk1 dominant suppressor (5d2) and Δcbk1 recessive suppressor (5d1) strains. Aliquots of 10 µg were used in a northern analysis, probed with CTS1, the ACT1 gene was used as an internal standard. CTS1 transcription is almost undetectable in the Δcbk1 and Δace2 strains but is re-established in the suppressor strains. (B) Haploid strains with a deleted CBK1 gene derived from W303 (WR05-5a) and S288c (WR208-1a) were transformed by a plasmid where the expression of the CTS1 gene is under the control of the GAL promotor (YEpWJR064). Transient expression of chitinase on galactose medium causes the large aggregates associated with the deletion of the CBK1 gene to separate.
To confirm that the aggregation phenotype was due to a lack of chitinase expression, the CTS1 gene was cloned under the control of the GAL promotor and transformed into a strain deleted for CBK1. The transformants were grown on glucose medium, transferred to raffinose medium for 8 h and then to galactose medium in order to induce expression of the CTS1 gene. After 2 h in galactose medium the large aggregates separated to small aggregates and individual cells (see Figure 4B). Further incubation in galactose medium led to the formation of large morphologically aberrant cells, probably caused by the overexpression of chitinase.
ACE2 and CTS1 transcription is still cell cycle- dependent in ACE2-1, the dominant suppressor of Δcbk1
Ace2p is cell cycle-regulated in two ways: the gene is transcribed specifically in G2 and the protein is excluded from the nucleus, probably by Cdc28p-dependent phosphorylation, except in late M/early G1 (Dohrmann et al., 1992; O’Conallanin et al., 1999). As a result of this CTS1 transcription is restricted to G1. As the dominant suppressors of Δcbk1 affect Ace2p, we decided to determine if the programme of ACE2 and CTS1 transcription was altered in a strain carrying one of these mutations.
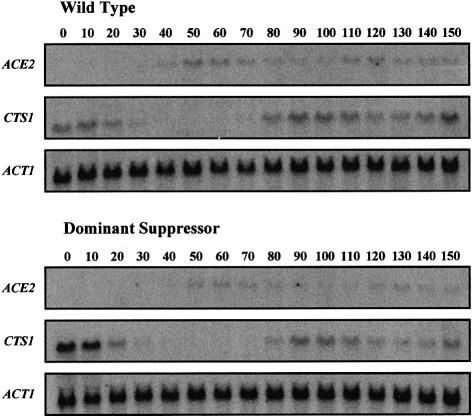
Young cultures (0.2 OD600) of MATa wild-type and Δcbk1-ACE2-1 strains were synchronized with α factor (Breeden, 1997), samples were taken every 10 min and stopped by adding 2 vols of ethanol at –60°C. Total RNA was extracted and the samples were analysed by northern blot using ACE2, CTS1 and ACT1 probes. The results are presented in Figure 5 and show that there is essentially no difference in the pattern of ACE2 and CTS1 transcription in the wild-type and Δcbk1-ACE2-1 strains. Both genes are cell cycle-regulated with the ACE2 transcripts being detected just before the CTS1 transcripts.
Fig. 5. Cell cycle-dependent expression of ACE2 and CTS1. MATa haploid cultures of wild-type (AW303-2a) and Δcbk1 dominant suppressor (5a3) were synchronized with α factor, samples were taken at 10 min intervals and stopped by adding 2 vols of ethanol at –60°C. Total RNA was extracted and used in a northern analysis probed with ACE2 and CTS1, the ACT1 gene was used as an internal standard. No difference in the cell cycle-coordinated expression of ACE2 and CTS1 could be detected between the wild type and Δcbk1 dominant suppressor.
Deletion of the CBK1 gene leads to a loss of polarity in bipolar budding cells
A closer examination of the results presented in Figure 1 shows that although the deletion of CBK1 and ACE2 led to a similar aggregation phenotype, the aggregates are not identical. The difference is most clearly seen in the close-ups of the Calcofluor white staining (Figure 1C, F and I). The Δcbk1 strain appears to have a random budding pattern whereas the Δace2 strain maintains a bipolar budding pattern the same as the wild type; this gives a more open appearance to the Δcbk1 aggregates.
As the budding pattern is difficult to follow in the large aggregates of the Δcbk1 strain, we decided to examine the budding pattern in wild-type and the Δcbk1 suppressor strains using Calcofluor white to stain the bud scars. An example of the results of this analysis can be seen in Figure 6A–F. The first thing to note is that haploids derived from W303 show a bipolar budding pattern more normally associated with diploid cells. The details of the budding pattern for both haploid and homozygous diploid strains are presented in Table II. In both dominant and recessive suppressors ∼90% of the cells show a random budding pattern, independent of ploidy, compared with ∼2–4% in the wild-type strain.
Fig. 6. Budding pattern of wild-type and Δcbk1 recessive and dominant suppressors. Old cultures of haploid and diploid: wild-type [AW303-2b (A); AW303 (D)], Δcbk1 recessive suppressor [5d1 (B); WR175 (E)] and Δcbk1 dominant suppressor [5d2 (C); WR17 (F)] all derived from W303 and the wild-type haploid YPH499 derived from S288c (G) were stained with Calcofluor white to reveal the bud scars and observed by fluorescence microscopy. The W303-derived haploid and diploid wild-type cells show a bipolar budding pattern, whereas the S288c-derived wild-type haploid shows an axial budding pattern. All suppressor strains show a random budding pattern. The remaining panels show cells liberated from aggregates of haploid strains with a deleted CBK1 gene derived from S288c [WR208-1a (H)] and W303 [WR05-5a (I)] during transient expression of chitinase. In the S288c-derived cells the axial budding pattern is maintained, while in the W303-derived cells the budding pattern is random.
Table II. Budding pattern of wild-type, Δcbk1 suppressor strains and Cbk1p–GFP fusion strains.
| Strains | Total cells | Identifiable budding | Bipolar budding | Random budding | % random budding |
|---|---|---|---|---|---|
| Haploid | |||||
| wild type | 384 | 126 | 121 | 5 | 4 |
| recessive suppressor | 324 | 95 | 9 | 86 | 90 |
| dominant suppressor | 386 | 91 | 8 | 83 | 91 |
| Cbk1p–GFP fusion | 277 | 72 | 67 | 5 | 7 |
| Diploid | |||||
| wild type | 240 | 153 | 150 | 3 | 2 |
| recessive suppressor | 276 | 97 | 10 | 87 | 89 |
| dominant suppressor | 336 | 112 | 11 | 101 | 90 |
| Cbk1p–GFP fusion | 151 | 99 | 97 | 2 | 2 |
Old cultures in rich complete medium were stained with Calcofluor white to reveal the bud scars and observed by fluorescence microscopy. All cells in a given field were counted and the analysis was performed on those cells whose budding pattern could be unambiguously determined.
To clarify the effect of the deletion of CBK1 on budding pattern, we have inactivated the gene in YPH449 that is a haploid derivative of S288c (Sikorski and Hieter, 1989) and has an axial budding pattern (see Figure 6G). The results in Figure 4B show that in this strain, inactivation of CBK1 also leads to the formation of aggregates that separate upon the expression of chitinase. We have examined the budding pattern in haploid Δcbk1 strains from both backgrounds during the transient expression of chitinase from the GAL promotor. It is clear that whilst the strain derived from W303 presents a random budding pattern, the strain derived from S288c maintains an axial budding pattern (see Figure 6H and I).
In summary, these data show that the deletion of the CBK1 gene does not affect strains with an axial budding pattern but leads to a loss of polarity and random budding in strains with a bipolar budding pattern.
Subcellular localization of Cbk1p
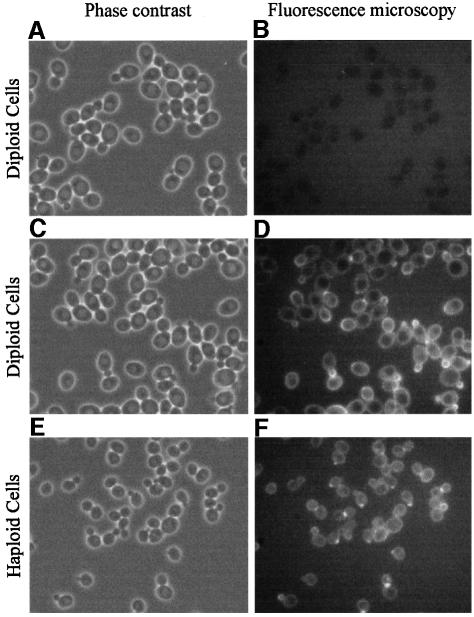
We have shown that Cbk1p regulates the activity of Ace2p, a protein whose localization is cell cycle-regulated (Dohrmann et al., 1992; O’Conallanin et al., 1999). In order to try and clarify the level at which Cbk1p regulates Ace2p we decided to determine the subcellular localization of Cbk1p. To do this we constructed CBK1–GFP fusions integrated at the CBK1 locus and under the control of the CBK1 regulatory sequences (see Materials and methods). These give rise to a derivative of Cbk1p with GFP fused to the C-terminus. As far as we can judge, this fusion protein is fully active and the cells have a wild-type morphology and bipolar budding pattern (see Table II). The strains carrying the fusion construction (either haploid or homozygous diploids) were examined by fluorescence microscopy and an example of the results is presented in Figure 7. Three observations can be made from these results. First, there is a difference between the haploid and diploid strain, but in our opinion this is a quantitative difference in the amount of the fusion protein detected, not a qualitative difference in localization. Secondly, there appears to be some general cytoplasmic staining and third, there is a specific cell cycle-dependent localization of the fusion protein.
Fig. 7. Subcellular localization of CBK1p. Fresh overnight cultures of control diploid cells (AW303) and diploid and haploid cells (WR154; WR152-1a) expressing the Cbk1p–GFP fusion protein were grown in complete medium supplemented with a 4-fold excess of adenine and observed by phase contrast and fluorescence microscopy. No GFP fluorescence could be detected in the control cells. In both diploid and haploid cells some cytoplasmic staining can be seen and specific staining of different parts of the cell depending on their position in the cell cycle. The staining is brighter in the diploid strain.
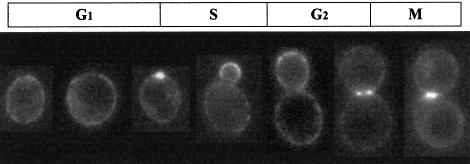
In Figure 8, representative cells from different stages of the cell cycle have been assembled to show the localization of the Cbk1p–GFP fusion protein throughout the cycle in a haploid strain. A punctate pattern at the cell periphery can be seen at all stages; at the first signs of bud formation, Cbk1p appears to concentrate at the bud site. Later, there is a high level at the tip of the young bud; as the bud grows this diminishes to the level seen at the periphery of the mother cell. At this point a ring of Cbk1p appears at the bud neck. Just before cytokinesis Cbk1p seems to form two symmetrical plates or rings, one in the mother cell and the other in the daughter. These results show that although the transcription of CBK1 is not regulated in a cell cycle-dependent manner (Cho et al., 1998; Spellman et al., 1998) the protein is probably involved in cell cycle-dependent regulation as its specific localization changes throughout the cell cycle.
Fig. 8. Cell cycle-dependent localization of Cbk1p. Representative individual haploid cells expressing the Cbk1p–GFP fusion, taken from the experiment presented in Figure 7 have been selected to show the localization of Cbk1p at different stages of the cell cycle. During G1 the protein is seen in a punctate pattern around plasma membrane as well as in the cytoplasm. The protein concentrates at the emerging bud site, then at the tip of the bud, dispersing around the bud as it grows. In cells with a large bud the majority of the protein forms a ring around the bud neck and just before cell separation, a plate or ring is present in both the mother and daughter cell, either side of the cell junction.
Discussion
We have studied the role of CBK1 in S.cerevisiae and shown that at least three phenotypes are associated with the deletion of the gene: large aggregates of cells, the conversion of a bipolar budding pattern into a random budding pattern and round rather than ellipsoidal cells. Most of our strains are derived from W303 and it should be noted that haploid derivatives of W303 present a bipolar budding pattern typical of diploids rather than the normal haploid axial budding pattern. Mutations in AXL1, BUD3, BUD4 and BUD10 (AXL2) give rise to a bipolar budding pattern in haploids (Madden and Snyder, 1998); thus, it is possible that W303 carries a mutation in one of these genes or in a related gene. Deletion of CBK1 in YPH499, a haploid strain with an axial budding pattern (derived from S288c; Sikorski and Hieter, 1989) leads to an aggregation phenotype but does not alter the budding pattern.
As we described previously (Nasr et al., 1996), Cbk1p is part of a family of kinases conserved from yeast to man. The limited data available suggests an underlying conservation of function amongst these protein kinases. To test these functional relationships we cloned the cDNAs corresponding to the human myotonic dystrophy kinase, the Arabidopsis thaliana kinase and Orb6p. The human NDR kinase cDNA was kindly provided by Professor Hemmings (Millward et al., 1995). These four cDNAs have been tested to see if they are able to replace CBK1. None of them were able to complement any of the phenotypes associated with the deletion of CBK1, or to interact with Ace2p in two-hybrid experiments (data not shown).
Cbk1p and the transcriptional activator Ace2p have been linked by two independent approaches, they interact strongly in two-hybrid experiments and mutations in ACE2 can act as dominant suppressors of the aggregation phenotype of the CBK1 deletion strain. Ace2p is responsible for the transcription of CTS1 and is very similar to another transcriptional activator, Swi5p (Butler and Thiele, 1991), which is responsible for the transcription of the HO gene (Stillman et al., 1988). In vitro both proteins bind to the same promotor sequence (Dohrmann et al., 1996), thus in vivo, promotor specificity probably results from interactions with other proteins. The region of Ace2p identified in our two-hybrid experiments immediately precedes the domain shown by McBride et al. (1999) to be specifically required for CTS1 transcription. Given the high level of similarity between Ace2p and Swi5p it is important to note that Swi5p was never identified as a partner of Cbk1p in our two-hybrid study.
We have shown that in the CBK1 deletion strain, as in the ACE2 deletion strain, CTS1 transcription is almost undetectable and is restored in both the dominant and recessive suppressors (see Figure 4). Also transient expression of chitinase causes the aggregates to separate. This suggests that transcription of CTS1 by Ace2p is dependent upon Cbk1p, possibly in a phosphorylation-dependent manner. Thus, the aggregation phenotype is caused by the inability to digest the chitin plate that separates the mother and daughter cells after division. According to this model, the dominant ACE2 suppressor alleles would be constitutively active and independent of Cbk1p. This is consistent with the very specific nature of the ACE2 suppressor mutations (see Figure 2). So far we have been unable to clone the gene corresponding to the recessive suppressors. However, we know that ACE2 is required for the activity of the recessive suppressors (data not shown), so it is tempting to speculate that these mutations might affect another factor involved in CTS1 transcription and facilitate the interaction of non-phosphorylated Ace2p with the transcription complex. The observation that ACE2 is a multi-copy suppressor of the CBK1 deletion aggregation phenotype (data not shown), may be taken to support this hypothesis.
Ace2p activity is linked to the cell cycle in two ways: the protein is excluded from the nucleus except in late M/early G1 and the gene is specifically transcribed in G2 (Dohrmann et al., 1992; O’Conallanin et al., 1999). In this way CTS1 transcription is restricted to G1. We have shown that the cell cycle-regulated transcription of ACE2 and CTS1 is not affected in the dominant ACE2 suppressor alleles. CBK1 transcription is not cell cycle-regulated (Cho et al., 1998; Spellman et al., 1998) but Cbk1p is essential for the activity of a cell cycle-regulated transcription factor. Using a fully active Cbk1p–GFP fusion protein we have shown that the localization of a significant proportion of the protein is controlled in a cell cycle-dependent manner (see Figure 8). Some Cbk1p–GFP fusion protein is always seen in the cytoplasm, and we assume that this is where the Cbk1p–Ace2p interaction occurs. In general terms, the specific localization is characterized by the presence of elevated levels of Cbk1p at points of active growth; cell periphery during G1, followed by the bud site, then the bud tip, dispersion at the apical isotropic switch and concentration at the mother–daughter junction just before cytokinesis.
The cell cycle-dependent localization of Cbk1p may also be the key to understanding the round cell and random budding phenotypes associated with the deletion of CBK1 in strains that normally have a bipolar budding pattern. It is clear that these phenotypes are independent of ACE2, as they are not seen in the ACE2 deletion strain and are still observed both dominant and recessive suppressors (see Figures 1, 3 and 6). The shape of a cell is determined in part by the timing of the switch from apical to isotropic growth (Lew and Reed, 1993) and this in turn is controlled by the timing of the depolarization of the actin cytoskeleton, a structure that is also involved in bud site selection (Yang et al., 1997). In S.cerevisiae, actin polarization is regulated throughout the cell cycle by the PAK kinases Ste20p and Cla4p (Holly and Blumer, 1999) and Ste20p is known to localize to sites of cell surface growth (Peter et al., 1996). This is significant as mutations in the S.pombe equivalents of CBK1 and STE20 (Orb6 and Pak1, respectively) are co-lethal (Verde et al., 1998). Orb6p, like Cbk1p, localizes to sites of cell growth, but there are significant differences between the S.pombe and S.cerevisiae systems as Orb6 and Pak1 are both essential genes, whereas CBK1 and STE20 are not. Also the double deletion of CBK1 and STE20 does not show co-lethality (data not shown).
It has been suggested that Cbk1p can interact directly with several components of the pheromone cascade including Ste20p (Geyer et al., 1999). None of these genes, or any other known component of the pheromone cascade were identified in our two-hybrid screen. Subsequent PCR amplification on the two-hybrid bank has shown that STE5, STE11, STE20 and STE50 are present. These genes have been examined in direct tests. A weak interaction between Cbk1p (prey) and the scaffold protein Ste5p (bait) was detected, but the significance of this is not clear as the Ste5p bait plasmid also shows weak auto-activation. Geyer et al. (1999) also reported that the deletion of CBK1 leads to an increased resistance to α factor. We have confirmed this observation, which might be assumed to imply a link between Cbk1p and the pheromone cascade. However, this interpretation is open to question, the deletion of CBK1 leads to a lack of CTS1 transcription and changes in the composition of the cell wall, so the increased resistance to α factor may be due simply to a permeability problem. This hypothesis is supported by the observation that the dominant ACE2-1 suppressor shows no increased α factor resistance (data not shown). Homozygous Δcbk1/Δcbk1 diploids sporulate poorly, but Δcbk1 haploids and both the dominant and recessive suppressor strains show no obvious mating defect, suggesting that the pheromone cascade is not seriously affected.
It is clear from the round cell and random budding phenotypes associated with the deletion of the CBK1 gene in bipolar budding strains, that in addition to regulating chitinase expression by the transcriptional activator Ace2p, Cbk1p also plays a role in the control, or the establishment, of cell morphology. As the actin cytoskeleton is essential for both polarized growth and bud site selection (see Madden and Snyder, 1998 for a review) it is tempting to speculate that Cbk1p, either directly or via one of its two-hybrid partners, might be involved in the organization of the actin cytoskeleton during polarized growth.
Materials and methods
Strains and media
Escherichia coli strains. For standard DNA propagation the strain XL2-Blue (Stratagene) was used. For the isolation of prey plasmids in the 2-hybrid screen MC1066 [Δ(lac)X74 hsdR hsdM rspsL galU galK trpC9830 leuB600 pyrF::Tn5] was used (Casadaban et al., 1983).
Yeast strains. All the S.cerevisiae strains are listed in Table III. AW303-2b carries a partial deletion of the ADE3 gene and was derived from W303 (Thomas and Rothstein, 1989) in our laboratory.
Table III. Yeast strains used in this study.
| Strain | Genotype |
|---|---|
| AW303-2a | MATa ade3 |
| AW303-2b | MATα ade3 |
| AW303 | MATa/α ade3/ade3 |
| FN40-4a | MATa Δcbk1::HIS3 |
| WR03-5d | MATα ade3 Δcbk1::HIS3 |
| WR27-1a | MA α Δace2::kanMX4 |
| WR28-3b | MATα Δcbk1::HIS3 Δace2::kanMX4 |
| 5d2 | MATα ade3 Δcbk1::HIS3 SUPdom |
| 5d1 | MATα ade3 Δcbk1::HIS3 suprec |
| 5a3 | MATa ade3 Δcbk1::HIS3 SUPdom |
| WR152-1a | MATa ade3 CBK1-GFP |
| WR154 | MATa/α ade3/ade3 CBK1-GFP/CBK1-GFP |
| WR175 | MATa/α ade3/ade3 Δcbk1::HIS3/Δcbk1::HIS3 suprec/suprec |
| WR176 | MATa/α ade3/ade3 Δcbk1::HIS3/Δcbk1::HIS3 SUPdom/SUPdom |
| Y190 | MATa trp1-901 ade2-201 leu2-3,112 his3-11,15 URA3::UAS GAL1-lacZ gal4Δ gal80Δ LYS2::UAS GAL1-HIS3 cych2r |
| YPH449 | MATa ade2-201 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 |
| WR208-1a | MATa ade2-201 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 Δcbk1::kanMX4 |
The strain Y190 was provided by Dr P.Legrain and YPH449 was provided by Dr B.Giuard. All other strains used in this study were derived from the homozygous diploid W303 (MATαΔMATa, ade2-1, his3-11, 15, leu2-3, 12, trp1-1, ura3-1, can1-100; Thomas and Rothstein, 1989). Only the genetic markers that differ from W303 are indicated.
Media
All media for bacteria were prepared as described in Sambrook et al. (1989). Yeast media were prepared according to Dujardin et al. (1980). Complete synthetic medium and the minimal media used for the co-lethal screen were supplemented with 5-fluoro-orotic acid to 0.1%. Minimal medium used for two-hybrid positive clone isolation was supplemented with 3-aminotriasol (Sigma) to a concentration of 60 mM.
Plasmids
Plasmids were constructed by standard methods (Sambrook et al., 1989), or by ‘gap repair’ (Orr-Weaver et al., 1983), and are listed in Table IV.
Table IV. Yeast plasmids.
| Plasmid | Insert | Reference |
|---|---|---|
| pGBT9 | Two-hybrid bait plasmid with TRP1 marker | Bartel et al. (1993) |
| pACT2 | Two-hybrid prey plasmid with LEU2 marker | Li and Lagarias (1994) |
| pYeDP1/10 | GAL expression plasmid | Pompon et al. (1996) |
| YEpWJR064 | CTS1 cloned in pTeDP1/10 under the control of the GAL promotor | |
| pYM12 | yEGFP | Knop et al. (1999) |
| YCpFNG061 | pFL36 with CBK1 gene under native regulatory sequences | |
| YEpWJR003 | pGBT9 with C-terminal part of CBK1 starting at T279 |
All plasmids were constructed by standard techniques or by gap repair.
Primers
Inactivation primers. All inactivation primers were composed of ∼50 nucleotides of 5′ and 3′ non-coding sequences, followed by ∼20 nucleotides of HIS3, KanMX4 or HIS3MX4 cassettes (Wach et al., 1994).
Cloning primers. All PCRs were performed on genomic DNA of W303. ACE2 with its native regulatory sequences was amplified using the primers: 5′-AATTTCACACAGGAAACAGCTATGACCATGATTAC GCCAAGCTTGCATGCTAGTGTTATAGGAATAGCCG-3′ and 5′-GTA AAACGACGGCCAGTGAATTCGAGCTCGGTACCCGGGGATCCT CTAGAACTTTCGAATATGCAGTGTC-3′, and gap-repaired into YCplac33 to give the plasmid YCpWJR007.
GFP fusion primers. The CBK1–GFP fusion was constructed as described by Wach et al. (1997) using the plasmid pFA6a-5GA GFP super bright (Knop et al., 1999) and the primers: 5′-TTGGCTACA CTTACTCCAGATTTGACTATTTGACAAGAAAAAATGCGTTGG GATCCGGAGCAGGTGCTGGTGC-3′ and 5′-GGTAAAAAAACCAGCCATCACCGAGATTGTGCTATTGTGCGTGGATGTATATCGAT GAATTCGAGCTCG-3′.
CTS1 expression primers. The CTS1 coding sequence and native 3′ region was amplified by PCR using the primers: 5′-ATAGAC ACGCAAACACAAATACACACACTAAATTAATAATGACCGGAT CCATGTCACTCCTTTACATCATTCTTC-3′ and 5′-AAAAGAAAA AAATTGATCTATCGATTTCAATTCAATTCAATGGGAGATCTTG GCATTCTCAACAATCATTTTTAT-3′ and cloned in the galactose-induced expression vector pYeDP1/10 to give the plasmid YEpWRJ064.
Nucleic acid manipulation
Transformation of E.coli, plasmid isolation, small scale genomic DNA isolation, gel electrophoresis, Southern and northern blot analysis were performed as described by Sambrook et al. (1989). Yeast was transformed as described by Gietz et al. (1992). Radioactive probes were prepared by random priming (Gibco-BRL) or by nick translation (Boehringer Mannheim) according to manufacturers’ instructions. DNA sequencing was performed using either a Pharmacia ALF or an ABI 310 automatic sequencer.
RNA isolation. Total RNA was prepared by the ‘hot phenol’ method. Cells (5 OD600) were collected and resuspended in 400 µl of AE buffer (50 mM sodium acetate pH 5.3, 10 mM EDTA pH 8.0); to this was added 40 µl 10% SDS and 440 µl phenol saturated with AE buffer. The suspension was successively vortexed for 5 min, incubated at 65°C for 5 min, frozen in ethanol/dry ice, vortexed for 5 min and centrifuged for 2 min. The supernatant was collected, phenol extracted and the RNA was precipitated with ethanol, washed with 80% ethanol, dried and resuspended in 50 µl of sterile water.
Microscopy
Staining for budscars (Calcofluor white) was performed according to Adams et al. (1997). Fluorescence microscopy was performed with a Leica DMRD microscope linked to a Princeton Instruments inc. Micro MAX camera.
Genetic techniques
Two-hybrid screening. Experiments were performed as described by Fields and Sternglanz (1994) using the bank prepared by Fromont-Racine et al. (1997). 3-aminotriasol-positive clones were collected between 4 and 7 days and streaked to single colonies on 3-aminotriasol medium. These were grown in minimal medium and tested for the expression of β-galactosidase. Prey plasmid DNA from clones positive in both tests was isolated after a passage through E.coli MC1066. This DNA was used to retransform yeast. DNA that gave 3-aminotriasol-resistant and β-galactosidase-positive clones upon retransformation was sequenced.
Isolation of extragenic suppressors of the deletion of CBK1. Twenty independent subclones (10 for each mating type) of Δcbk1 strains were grown on YPGA plates for 2 days, then left in the dark at 4°C for a week. A patch of cells was plated on to fresh YPGA plates to obtain a lawn of cells and UV mutagenized for 40 s (∼10% survival). Plates were grown in the dark for 2 days, yeast was washed off the plates and resuspended in 5 ml of YPGA medium. One-tenth of this suspension was inoculated into 20 ml of YPGA medium, grown overnight, then transferred to 20 ml tubes and left to sediment. After 4 h the topmost 0.5 ml clear fraction of the cultures was collected and re-inoculated into 20 ml of YPGA medium. This enrichment step was carried out 10 times. Finally, the culture was diluted and plated on YPGA plates to observe the colony morphology phenotype.
Acknowledgments
Acknowledgements
We would like to thank Mme D.Menay for the synthesis of the oligonucleotides, Dr G.Dujardin and Professor F.Lacroute for critically reading the manuscript, Professor B.Hemmings for providing a cDNA clone of the human NDR kinase and Dr P.Legrain for providing the strain Y190 and the two-hybrid bank. We would also like to thank Dr C.Mann for invaluable assistance with the fluorescence microscopy. This work was supported in part by a grant from the Association Française contre des Myopathies.
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bartel P., Chien,C.T., Sternglanz,R. and Fields,S. (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques, 14, 920–924. [PubMed] [Google Scholar]
- Breeden L.L. (1997) α-factor synchronization of budding yeast. Methods Enzymol., 283, 332–341. [DOI] [PubMed] [Google Scholar]
- Butler G. and Thiele,D.J. (1991) ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol. Cell. Biol., 11, 476–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M.J., Martinez-Arias,A., Shapira,S.K. and Chou,J. (1983) β-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol., 100, 293–308. [DOI] [PubMed] [Google Scholar]
- Chant J. (1999) Cell polarity in yeast. Annu. Rev. Cell Dev. Biol., 15, 365–391. [DOI] [PubMed] [Google Scholar]
- Chant J. and Pringle,J.R. (1995) Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol., 129, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.J. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- Dohrmann P.R., Butler,G., Tamai,K., Dorland,S., Greene,J.R., Thiele,D.J. and Stillman,D.J. (1992) Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev., 6, 93–104. [DOI] [PubMed] [Google Scholar]
- Dohrmann P.R., Voth,W.P. and Stillman,D.J. (1996) Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol. Cell. Biol., 16, 1746–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G., Pajot,P., Groudinsky,O. and Slonimski,P.P. (1980) Long range control circuits within mitochondria and between the nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol. Gen. Genet., 179, 469–482. [DOI] [PubMed] [Google Scholar]
- Durrenberger F. and Kronstad,J. (1999) The ukc1 gene encodes a protein kinase involved in morphogenesis, pathogenicity and pigment formation in Ustilago maydis. Mol. Gen. Genet., 261, 281–289. [DOI] [PubMed] [Google Scholar]
- Fields S. and Sternglanz,R. (1994) The two-hybrid system: an assay for protein–protein interactions. Trends Genet., 10, 286–292. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain,J.C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Geyer C.R., Colman-Lerner,A. and Brent,R. (1999) ‘Mutagenesis’ by peptide aptamers identifies genetic network members and pathway connections. Proc. Natl Acad. Sci. USA, 96, 8567–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly S.P. and Blumer,K.J. (1999) PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol., 147, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice R.W., Zilian,O., Woods,D.F., Noll,M. and Bryant,P.J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev., 9, 534–546. [DOI] [PubMed] [Google Scholar]
- Karpova T.S., McNally,J.G., Moltz,S.L. and Cooper,J.A. (1998) Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol., 142, 1501–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Lew D.J. and Reed,S.I. (1993) Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol., 120, 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. and Lagarias,J.C. (1994) Phytochrome assembly in living cells of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 91, 12535–12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.J., Yu,Y. and Stillman,D.J. (1999) Distinct regions of the Swi5 and Ace2 transcription factors are required for specific gene activation. J. Biol. Chem., 274, 21029–21036. [DOI] [PubMed] [Google Scholar]
- Madden K. and Snyder,M. (1998) Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol., 52, 687–744. [DOI] [PubMed] [Google Scholar]
- Millward T., Cron,P. and Hemmings,B.A. (1995) Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proc. Natl Acad. Sci. USA, 92, 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J., Preuss,D., Moon,A., Wong,A., Drubin,D. and Botstein,D. (1994) Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol., 125, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr F., Becam,A.M. and Herbert,C.J. (1996) The sequence of 36.8 kb from the left arm of chromosome XIV reveals 24 complete open reading frames: 18 correspond to new genes, one of which encodes a protein similar to the human myotonic dystrophy kinase. Yeast, 12, 169–175. [DOI] [PubMed] [Google Scholar]
- O’Conallanin C., Doolin,M.T., Taggart,C., Thornton,F. and Butler,G. (1999) Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet., 262, 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Szostak,J.W. and Rothstein,R.J. (1983) Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol., 101, 228–245. [DOI] [PubMed] [Google Scholar]
- Peter M., Neiman,A.M., Park,H.O., van Lohuizen,M. and Herskowitz,I. (1996) Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J., 15, 7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Pompon D., Louerat,B., Bronine,A. and Urban,P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol., 272, 51–64. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits G.J., Kapteyn,J.C., van den Ende,H. and Klis,F.M. (1999) Cell wall dynamics in yeast. Curr. Opin. Microbiol., 2, 348–352. [DOI] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R.. Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D.J., Bankier,A.T., Seddon,A., Groenhout,E.G. and Nasmyth,K.A. (1988) Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J., 7, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Verde F., Wiley,D.J. and Nurse,P. (1998) Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl Acad. Sci. USA, 95, 7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Wissmann A., Ingles,J., McGhee,J.D. and Mains,P.E. (1997) Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev., 11, 409–422. [DOI] [PubMed] [Google Scholar]
- Yang S., Ayscough,K.R. and Drubin,D.G. (1997) A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J. Cell Biol., 136, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden O., Plamann,M., Ebbole,D.J. and Yanofsky,C. (1992) Cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J., 11, 2159–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]