Abstract
Vibrio cholerae O1 El Tor variant strains produced much more cholera toxin than did prototype El Tor strains. The amount of cholera toxin produced by El Tor variant strains both in vitro and in vivo was more or less equivalent to that produced by classical strains.
Vibrio cholerae O1 is classified into classical and El Tor biotypes. Among other genetic, biochemical, and physiological differences, each biotype has unique gene sequences encoding cholera toxin B subunit (CTB), that is, classical ctxB and El Tor ctxB. Besides these two prototype biotypes of V. cholerae O1, Nair et al. (9) in 2002 in Bangladesh isolated strains that possess phenotypic properties of both classical and El Tor biotypes carrying classical ctxB. The same group also isolated El Tor strains that had classical ctxB (10). For these new types of strains of V. cholerae O1, we have recently proposed the designations of hybrid and El Tor variants, respectively (13). Subsequent to the isolation of the El Tor variant in Bangladesh by Nair et al. (10), El Tor variant strains were isolated from several countries and areas in Asia and Africa (1, 11, 15-18). In Kolkata, India, we showed that El Tor variant strains appeared in 1990 and that a complete replacement of prototype El Tor strains by El Tor variant strains has occurred since 1995 (14).
In this study, we investigated the amount of cholera toxin (CT) produced both in vitro and in vivo by V. cholerae O1 El Tor variant strains isolated in Kolkata during a period from 1996 to 2007. It was found that El Tor variant strains produced a much larger amount of CT than did prototype El Tor strains and that the amount of CT produced by El Tor variant strains was more or less equivalent to that produced by classical strains.
V. cholerae O1 strains used in this study are listed in Table 1. AKI (3) and Syncase medium (2) were used for culturing the test strains. The rationale for selecting these media was that AKI preferentially supports the production of El Tor CT (3) while Syncase medium is reported to be the best medium supporting the production of CT by the classical biotype (2). Measurement of CT concentration produced by V. cholerae O1 strains was carried out as follows. Each strain was cultured either in AKI medium at 37°C for 20 h without shaking or in Syncase medium at 37°C for 20 h with shaking, and the optical density of the culture was measured at 600 nm (OD600). After centrifugation, the supernatants were collected and the concentration of CT (ng/ml/OD600) in the samples was measured by bead enzyme-linked immunosorbent assay (ELISA). The method of the bead ELISA employed was essentially that described by Oku et al. (12). In brief, a polystyrene bead (6.5 mm in diameter) was coated with anti-CT IgG and used as a solid phase. The coated bead was first incubated with the sample and then incubated with anti-CT IgG [F(ab′)]-horseradish peroxidase conjugate. Peroxidase activity was determined colorimetrically with 3,3′,5,5′-tetramethylbenzidine as the substrate. The absorbance at 450 nm (OD450) was linear between 0 and 0.5, representing CT concentrations of 0 to 20 ng/ml. The sample prepared as described above (the supernatant of the culture of the strain) was appropriately diluted so that the OD450 fell in the range of 0.1 to 0.5, and the amount of CT produced by the strain was expressed as ng/ml/OD600.
TABLE 1.
V. cholerae O1 strains used
| Biotype and straina |
|---|
| El Tor variant |
| AM157 (1996), 06-049 (2006), IDH60 (2007), BD200 (2002), 06-098 (2006), CRC220 (2000), AM168 (1996), DO2669 (1998), NLC96 (1999), CRC17 (2000), AM352 (1997), NLC41 (1999), NLC49 (1999), D26942 (1998), SC32 (2003), G27875 (2001), IDH32 (2007), SC216 (2003), NLC8 (1999) |
| El Tor |
| N16961, V100, V114, V113, VC60, M14716, V7, VC64, V54, V24, V32 |
| Classical |
| L362, GP15, GP8, GP148, GP147, 569B, GP145 |
Strains used are listed in the order of CT production (from high to low). The year of isolation is in parentheses.
The rabbit ileal loop test was carried out essentially as described by Koley et al. (7). Eight intestinal loops of about 10 cm, separated by uninoculated segments of 1 to 2 cm, were prepared in each animal. Test loops were inoculated with 1 ml of bacterial suspension containing approximately 109 cells. Negative-control loops were inoculated with 1 ml of phosphate-buffered saline. The loops were replaced in the peritoneal cavity, and the cavity was closed. After about 20 h the animal was sacrificed by intravenous injection of sodium pentobarbital and the loops were taken out. The volume of the accumulated fluid in ml and the length of the loop in cm were measured, and the extent of the fluid accumulation (FA) was expressed as ml/cm.
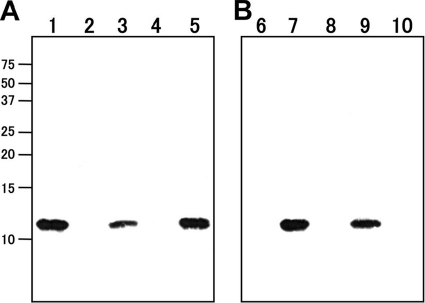
All 19 strains of V. cholerae O1 El Tor variant belonged to the El Tor biotype as evident from phenotypic traits such as resistance to 50 units of polymyxin B and a positive Voges-Proskauer test (19). All harbored El Tor biotype-specific alleles of tcpA and rstR when examined as described previously (5, 6). The ctxB gene of all strains was of classical type by mismatch amplification mutation assay (MAMA)-PCR carried out as described by Morita et al. (8). Further, the CTB produced by all strains was confirmed to be the classical type by Western blotting by using monoclonal antibody against either classical CTB or El Tor CTB, which was prepared by immunizing rats with a synthesized peptide (either NTQIYTLNDKC for El Tor CTB or NTQIHTLNDKC for classical CTB). Approximately 50 to 100 ng of CT (measured by bead ELISA) in the culture supernatant of each strain was analyzed. The results of the Western blotting of a representative strain (strain AM157) are shown in Fig. 1.
FIG. 1.
Results of Western blotting of the culture supernatant of a representative strain of El Tor variant biotype. Lanes 1 and 6, 100 ng of the purified classical CT; lanes 2 and 7, 100 ng of the purified El Tor CT; lanes 3 and 8, sample of El Tor variant strain AM157; lanes 4 and 9, sample of El Tor strain N16961; lanes 5 and 10, sample of classical strain L362. (A) Results with the monoclonal antibody against classical CTB. (B) Results with the monoclonal antibody against El Tor CTB. Numbers at left are molecular masses in kilodaltons (× 1,000).
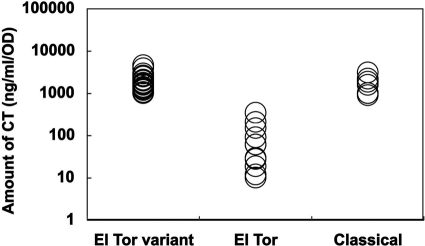
Figure 2 shows the distribution of the amounts of CT produced by strains examined. Each strain of El Tor variant, prototype El Tor, and classical biotype was cultured in 2 ml of AKI medium in a 10-ml test tube at 37°C for 20 h without shaking, and the supernatant of the culture was collected by centrifugation and was measured to determine the amount of CT by bead ELISA. It was found that most strains of El Tor variant produced much more CT than did most strains of prototype El Tor. All 19 El Tor variant strains produced more than 1,000 ng/ml/OD600 of CT, and among them 5 strains (AM157, 06-049, IDH60, BD200, and 06-098) produced more than 2,500 ng/ml/OD600, the highest (strain AM157) producing 4,656 ng/ml/OD600. The amount of CT produced varied but was not related to the year of isolation. Among 11 El Tor strains, 8 strains (V113, VC60, M14716, V7, VC64, V54, V24, and V32) produced less than 100 ng/ml/OD600, and among them 3 strains (V54, V24, and V32) produced less than 20 ng/ml/OD600. The rest of the strains (N16961, V100, and V114) produced more than 100 ng/ml/OD600, and the standard strain N16961 produced the largest amount (345 ng/ml/OD600). All 7 classical strains produced more than 900 ng/ml/OD600, and 2 of them (L362 and GP15) produced more than 2,000 ng/ml/OD600, the higher being L362 (3,028 ng/ml/OD600).
FIG. 2.
Amounts of CT produced by various biotypes of V. cholerae O1. Each circle represents an average of 4 determinations.
The amount of CT produced was measured during the growth of the strains in AKI medium with the representative strains of El Tor variant, prototype El Tor, and classical biotype, and it was found that the differences in the amounts of CT produced among these 3 biotypes were observed from the beginning of the growth (early logarithmic phase) till the late stationary phase (data not shown).
Table 2 shows the mean CT amounts produced by the strains of different biotypes with standard deviations. The amount of CT produced by El Tor variant strains was about 20 times more than that produced by prototype El Tor strains, and it was more or less equivalent to that produced by classical strains. A difference in the CT production between El Tor variant strains and prototype El Tor strains was statistically analyzed by Microsoft Excel 2004 for Mac, the P value (Student t test) being <0.05.
TABLE 2.
Comparison of the amounts of CT produced by strains of various biotypes of V. cholerae O1a
| Culture medium | CT concn (ng/ml/OD600) |
||
|---|---|---|---|
| El Tor variant | El Tor | Classical | |
| AKI | 2,044.1 ± 966.8 | 91.3 ± 104.6 | 1,664.4 ± 782.0 |
| Syncase | 81.3 ± 147.2 | 4.5 ± 3.7b | 114.7 ± 188.8 |
Strains examined were as listed in Table 1 unless indicated.
Only 5 strains of El Tor biotype (N16961, V113, VC64, VC60, and V24) grew in Syncase medium cultured at 37°C with shaking.
CT production by strains of El Tor variant, El Tor, and classical biotype was also examined when the strains were cultured in Syncase medium (2 ml in a 10-ml test tube) at 37°C for 20 h with shaking. As shown in Table 2, although the amount of CT produced in Syncase medium was much smaller than that produced in AKI medium, El Tor variant strains produced much more CT than did El Tor strains and produced an amount more or less equivalent to that produced by classical strains. The P value (Student t test) of the difference in the amounts produced between El Tor variant strains and prototype El Tor strains analyzed by Microsoft Excel 2004 for Mac was <0.05.
The ileal loop test was performed with a representative strain of El Tor variant (strain NLC41 producing 1,606 ng/ml/OD600 in AKI medium) together with representative strains of El Tor biotype (VC60 producing 60 ng/ml/OD600 in AKI medium) and classical biotype (L362 producing 3,028 ng/ml/OD600 in AKI medium). As shown in Table 3, the FA ratio of the El Tor variant NLC41 was almost the same as that of classical strain L362. On the other hand, El Tor strain VC60 did not cause measurable fluid accumulation. This is most probably because the number of inoculated cells was not high enough. The numbers of V. cholerae organisms in the accumulated fluid (CFU/ml) and the amounts of CT in the loop (ng/ml and ng/CFU) were also measured, showing that the El Tor variant strain grew better than did the classical strain in the loop; thus, the amount of CT in the loop inoculated with the El Tor variant strain was larger than that in the loop inoculated with the classical strain. Measurement of CFU/ml of the accumulated fluid of the prototype of El Tor strain was not possible as no fluid accumulation occurred.
TABLE 3.
Results of rabbit ileal loop testd
| Biotype | Strain | FA (ml/cm)a | CFU/mlb | CT (ng/ml)a | CT (ng/CFU) |
|---|---|---|---|---|---|
| El Tor variant | NLC41 | 0.90 ± 0.29 | 1.0 × 109 | 1,006 | 1.006 × 10−6 |
| El Tor | VC60 | 0 | —c | — | — |
| Classical | L362 | 0.83 ± 0.38 | 1.6 × 108 | 17.5 | 1.09 × 10−7 |
Averages of 4 determinations (2 loops each in 2 rabbits).
Averages of 2 determinations (2 loops of 1 representative rabbit).
—, not applicable as no fluid accumulation occurred.
Statistical analysis (Student t test) was performed by Microsoft Excel 2004 for Mac.
It is known that the clinical manifestation of cholera caused by classical strains is more severe than that caused by prototype El Tor strains (4). Although definite evidence to explain this is still not available, it has been hypothesized that a significant difference between the amounts of CT produced by these two biotype strains may reflect severity of clinical manifestation. If we were to accept the above hypothesis, a recent report by the World Health Organization (20) that the V. cholerae El Tor variant causes more severe episodes of cholera with higher case fatality rates might be explained by the results reported in this paper. However, Siddique et al. (16) reported that although El Tor variant strains appeared in 1998 in Bangladesh, the greater severity of cholera became evident only around 2006. Therefore, they concluded that it is not clear whether the observed higher proportion of severe dehydration is due to El Tor variants. Further study is needed to elucidate the role of CT produced by El Tor variant strains in the clinical manifestation of infection.
Acknowledgments
We thank Koichiro Yamamoto for his valuable discussion during this work. We also thank Debarati Ganguly for assistance in editing the manuscript.
This study was supported by the Japan Initiative for Global Research Network on Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 1 September 2010.
REFERENCES
- 1.Ansaruzzaman, M., N. A. Bhuiyan, G. B. Nair, D. A. Sack, M. Lucas, J. L. Deen, J. Ampuero, C. L. Chaignat, and the Mozambique Cholera Vaccine Demonstration Project Coordination Group. 2004. Cholera in Mozambique, variant of Vibrio cholerae. Emerg. Infect. Dis. 10:2057-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkelstein, R. A., P. Atthasampunna, M. Chulasamaya, and P. Charunmethee. 1966. Pathogenesis of experimental cholera: biologic activities of purified procholeragen A. J. Immunol. 96:440-449. [PubMed] [Google Scholar]
- 3.Iwanaga, M., and K. Yamamoto. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keasler, S. P., and R. H. Hall. 1993. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341:1661. [DOI] [PubMed] [Google Scholar]
- 6.Kimsey, H. H., G. B. Nair, A. Ghosh, and M. K. Waldor. 1998. Diverse CTX phage and evolution of new pathogenic Vibrio cholerae. Lancet 352:457-458. [DOI] [PubMed] [Google Scholar]
- 7.Koley, H., R. Mitra, A. Basu, A. K. Mukhopadhyay, P. K. Saha, B. S. Ramakrishna, S. Krishnan, Y. Takeda, and G. B. Nair. 1999. Response of wild-type mutants of Vibrio cholerae O1 possessing different combinations of virulence genes in the ligated rabbit ileal loop and in Ussing chambers: evidence for the presence of additional secretogen. J. Med. Microbiol. 48:51-57. [DOI] [PubMed] [Google Scholar]
- 8.Morita, M., M. Ohnishi, M. E. Arakawa, N. A. Bhuiyan, S. Nusrin, M. Alam, A. K. Siddique, F. Quadri, H. Izumiya, G. B. Nair, and H. Watanabe. 2008. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol. Immunol. 52:314-317. [DOI] [PubMed] [Google Scholar]
- 9.Nair, G. B., S. M. Faruque, N. A. Bhuiyan, M. Kamruzzaman, A. K. Siddique, and D. A. Sack. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair, G. B., F. Qadri, J. Holmgren, A. M. Svennerholm. A. Safa, N. A. Bhuiyan, Q. S. Ahmad, S. M. Faruque, A. S. Faruque, Y. Takeda, and D. A. Sack. 2006. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J. Clin. Microbiol. 44:4211-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen, B. M., J. H. Lee, N. T. Cuong, S. Y. Choi, N. T. Hien, D. D. Anh, H. R. Lee, M. Ansaruzzaman, H. P. Endtz, J. Chun, A. L. Lopez, C. Czerkinsky, J. D. Clemens, and D. W. Kim. 2009. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J. Clin. Microbiol. 47:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oku, Y., Y. Uesaka, T. Hirayama, and Y. Takeda. 1988. Development of a highly sensitive bead-ELISA to detect bacterial protein toxins. Microbiol. Immunol. 32:807-816. [DOI] [PubMed] [Google Scholar]
- 13.Raychoudhuri, A., A. K. Mukhopadhyay, T. Ramamurthy, R. K. Nandy, Y. Takeda, and G. B. Nair. 2008. Biotyping of Vibrio cholerae O1: time to redefine the scheme. Indian J. Med. Res. 128:695-698. [PubMed] [Google Scholar]
- 14.Raychoudhuri, A., T. Patra, K. Ghosh, T. Ramamurthy, R. K. Nandy, Y. Takeda, G. B. Nair, and A. K. Mukhopadhyay. 2009. Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 15:131-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safa, A., J. Sultana, P. D. Cam, J. C. Mwansa, and R. Y. C. Kong. 2008. Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg. Infect. Dis. 14:987-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddique, A. K., G. B. Nair, M. Alam, D. A. Sack, A. Huq, A. Nizam, I. M. Longini, Jr., F. Qadri, S. M. Faruque, R. R. Colwell, S. Ahmed, A. Iqbal, N. A. Bhuiyan, and R. B. Sack. 2010. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol. Infect. 138:347-352. [DOI] [PubMed] [Google Scholar]
- 17.Sithivong, N., H. Izumiya, K. Munnalath, T. Phouthavane, K. Chomlasak, L. Sisavath, A. Vongdouangchanh, P. Vongprachanh, H. Watanabe, and M. Ohnishi. 2010. Cholera outbreak, Laos, 2007. Emerg. Infect. Dis. 16:745-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taneja, N., A. Mishra, G. Sangar, G. Singh, and M. Sharma. 2009. Outbreaks caused by new variants of Vibrio cholerae O1 El Tor, India. Emerg. Infect. Dis. 15:352-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. 1987. Manual for laboratory investigations of acute enteric infections. World Health Organization, Geneva, Switzerland.
- 20.World Health Organization. 2008. Cholera 2007. Wkly. Epidemiol. Rec. 83:269-284. [PubMed] [Google Scholar]