Abstract
Rapid and effective methods for the isolation of Clostridium difficile from stool samples are desirable to obtain isolates for typing or to facilitate accurate diagnosis of C. difficile-associated diarrhea. We report on the evaluation of a prototype chromogenic medium (ID C. difficile prototype [IDCd]) for isolation of C. difficile. The chromogenic medium was compared using (i) 368 untreated stool samples that were also inoculated onto CLO medium, (ii) 339 stool samples that were subjected to alcohol shock and also inoculated onto five distinct selective agars, and (iii) standardized suspensions of 10 C. difficile ribotypes (untreated and alcohol treated) that were also inoculated onto five distinct selective agars. Two hundred thirty-six isolates of C. difficile were recovered from 368 untreated stool samples, and all but 1 of these strains (99.6%) were recovered on IDCd within 24 h, whereas 74.6% of isolates were recovered on CLO medium after 48 h. Of 339 alcohol-treated stool samples cultured onto IDCd and five other selective agars, C. difficile was recovered from 218 samples using a combination of all media. The use of IDCd allowed recovery of 96.3% of isolates within 24 h, whereas 51 to 83% of isolates were recovered within 24 h using the five other media. Finally, when they were challenged with pure cultures, all 10 ribotypes of C. difficile generated higher colony counts on IDCd irrespective of alcohol pretreatment or duration of incubation. We conclude that IDCd is an effective medium for isolation of C. difficile from stool samples within 24 h.
Clostridium difficile is the primary infectious cause of nosocomial diarrhea and is associated with recent administration of antibiotics. Renewed attention has been given to this pathogen due to the increase in both the incidence and the severity of Clostridium difficile infection (CDI) (17). Diagnosis of CDI is most commonly achieved by detection of toxins in stool samples using an immunoassay, although the positive predictive value of such assays can be unacceptably low in some populations (7). Methods for isolation of C. difficile using culture media are important in order to obtain isolates for typing, thus enabling outbreaks to be monitored and controlled (5, 6). More recently, routine culture of stool samples for C. difficile has been advocated as part of a diagnostic algorithm (10, 24, 26).
The first major advance in the design of selective culture media for C. difficile was the development of cycloserine-cefoxitin-fructose-egg yolk agar (CCFA) by George et al. (12). Various studies have challenged the optimal levels of cycloserine and cefoxitin (2, 18, 19), and reduced concentrations of 250 mg/liter cycloserine and 8 to 10 mg/liter cefoxitin are now widely used; however, the original formulation has been reaffirmed by others (20). Aspinall and Hutchinson recommended norfloxacin and moxalactam as alternative selective agents and reported a 20% increase in the yield of C. difficile from 832 stool samples when comparing their new medium with CCFA (1). Various additives have been proposed to promote germination of C. difficile spores and therefore enhance recovery, particularly from environmental samples. Bile salts such as cholic acid and sodium taurocholate may be particularly effective and have been used to supplement CCFA to allow enhanced recovery of C. difficile from surveillance cultures (3). Wilcox et al. adapted cefsulodin-cycloserine-egg yolk agar (CCEY) by omitting egg yolk and including lysozyme at 5 mg/liter (30). The use of CCEY plus lysozyme significantly increased the recovery of C. difficile from 197 environmental swab specimens compared with the recovery by use of CCEY alone. There are few comparisons of commercially available selective media for C. difficile. In one study, CCFA was compared with an Oxoid formulation that included cefoxitin at 8 mg/liter and cycloserine at 250 mg/liter, as well as neutral red, cholic acid, glucose, and 1% horse blood (4). In the same study, a medium from Becton Dickinson (BD) was also evaluated that contained the same level of selective agents and utilized mannitol plus a pH indicator to demonstrate fermentation by C. difficile. Ninety-four toxigenic isolates of C. difficile were isolated, with 90.4% being recovered on CCFA, 84% on the Oxoid formulation, and 42.6% on the BD formulation.
As C. difficile readily forms spores, various treatments such as heat shock and alcohol shock may be applied to specimens for culture to reduce or eradicate vegetative cells and hence limit the growth of contaminating flora. Alcohol shock in particular has been shown to increase the yield of C. difficile from stool samples in some studies (19, 23). Media containing chromogenic enzyme substrates to facilitate enhanced isolation of various bacterial pathogens have successfully been developed (21) but have not yet been evaluated for isolation of C. difficile. The aim of the present study was to evaluate a novel prototype chromogenic medium in comparison to commercially available media for detection of C. difficile.
(The part of this work comparing IDCd and CLO medium was presented at the 20th European Congress for Clinical Microbiology and Infectious Diseases, Vienna, Austria, 2010, abstr. P678.)
MATERIALS AND METHODS
Materials.
A prototype chromogenic medium for C. difficile (ID C. difficile prototype [IDCd]) was provided as prepoured plates by bioMérieux, Craponne, France. CLO medium (reference no. 43431; bioMérieux, Basingstoke, United Kingdom) and BBL Clostridium difficile agar (reference no. 222228; Becton Dickinson, Oxford, United Kingdom) were purchased as prepoured plates from their respective manufacturers. Clostridium difficile agar base (reference no. CM0601; Oxoid, Basingstoke, United Kingdom) was prepared and sterilized according to the manufacturer's instructions, and each liter was supplemented at 50°C with 7% (vol/vol) defribrinated horse blood and two vials of selective supplement containing moxalactam-norfloxacin (reference no. SR0173; Oxoid). CCEY (reference no. BC2160; Bioconnections, Wetherby, United Kingdom) was prepared and sterilized according to the manufacturer's instructions, and each liter was supplemented at 50°C with 40 ml of egg yolk emulsion (reference no. S2073; Bioconnections), 10 ml of lysed horse blood, and two vials of selective supplement containing cefoxitin-cycloserine (reference no. S2073; Bioconnections). Finally, an identical batch of CCEY was prepared and supplemented as described above, except that egg yolk was excluded and replaced with 5 mg of lysozyme (reference no. 7651; Sigma, Poole, United Kingdom) per liter of agar, according to recommendations (30). Culture plates prepared in-house were lightly dried and stored at 4°C for a maximum of 1 week before use. Commercially supplied media were stored at 4°C and used before specified expiry dates. Prolyl-7-amido-4-methylcoumarin (reference no. 1-1290) was obtained from Bachem, (Saffron Walden, United Kingdom). The following strains, obtained from the National Collection of Type Cultures (NCTC), Colindale, United Kingdom, were selected to control culture media, atmospheric conditions, and identification protocols: C. difficile NCTC 11209, Clostridium bifermentans NCTC 506, Clostridium sordellii NCTC 8780, and Pseudomonas aeruginosa NCTC 10662. API strips were purchased from bioMérieux, Basingstoke, United Kingdom.
Comparison of the prototype chromogenic medium (IDCd) with CLO medium for isolation of C. difficile from stool samples.
Stool samples eligible for testing comprised any diarrheal samples for which testing for C. difficile toxin had been requested. On arrival at the laboratory, specimens were refrigerated and stored for up to 3 days before culture. On day 1, all samples were processed using the Vidas immunoassay for detection of C. difficile toxins A and B. This was to ensure that a predominance of positive samples could be selected for evaluation of the culture media. A total of 268 Vidas-positive samples and 100 Vidas-negative samples were selected for culture on the two media. Each specimen (1 ml or 1 g) was homogenized in 2 ml sterile distilled water to form an even suspension. Alcohol treatment was avoided so that the selectivity of the media could be assessed. A 10-μl aliquot of this suspension was inoculated onto IDCd and CLO medium and spread to obtain isolated bacterial colonies. All media were incubated in an anaerobic workstation at 37°C for a full 24-h period and then removed into air for interpretation and selection of colonies for identification. After a maximum of 30 min in air, the culture plates were reincubated anaerobically for a further 24 h.
All colonies were initially investigated by Gram stain, and any possible isolates of C. difficile were subcultured onto CCEY for further investigation (see below). Non-C. difficile isolates on CLO medium were investigated and assigned to presumptive genera by Gram stain and simple biochemical tests, e.g., a test for catalase. Non-C. difficile isolates forming black colonies on IDCd were fully identified to the genus or species level using appropriate API galleries.
Comparison of IDCd with five selective media for isolation of C. difficile from stool samples.
A total of 226 Vidas-positive samples and 113 Vidas-negative samples were selected for culture on the six media. These comprised IDCd, CLO medium, BBL medium, Oxoid medium, CCEY, and CCEY plus lysozyme (CCEY/L). Specimens were processed using the alcohol-shock method recommended by the United Kingdom Health Protection Agency. Each specimen (1 ml or 1 g) was homogenized in 1 ml of absolute alcohol, vortexed to form an even suspension, and left to stand at room temperature for 30 min. A 50-μl aliquot of this suspension was inoculated onto each of the six media and spread to obtain isolated bacterial colonies. All media were incubated at 37°C in an anaerobic workstation with an atmosphere comprising 80% nitrogen, 10% carbon dioxide, and 10% hydrogen. After incubation for a full 24-h period, the cultures were then removed into air for interpretation and selection of colonies for testing. After a maximum of 30 min in air, the culture plates were reincubated anaerobically for a further 24 h. All colonies, irrespective of appearance, were initially investigated by Gram stain, and any possible isolates of C. difficile were subcultured onto CCEY for further investigation (see below). Isolates that were proven not to be C. difficile were not formally identified further.
Identification, typing, and toxin testing of C. difficile.
Suspect colonies subcultured onto CCEY were confirmed to be C. difficile by their characteristic morphology, natural yellow-green colony fluorescence under long-wave UV light, lack of lecithinase activity (i.e., lack of opalescence surrounding colonies on CCEY), and ability to hydrolyze prolyl-7-amido-4-methylcoumarin. For the last test, substrate solution (1 mg/ml) was added directly to bacterial colonies on CCEY medium before incubation in air at 37°C for 30 min. Generation of blue fluorescence under long-wave UV light indicated a positive reaction and the presence of prolyl aminopeptidase. Further confirmation of species identity required generation of a PCR product in the PCR ribotyping assay, which was performed as described previously (28). For interest, any isolates of C. difficile recovered from Vidas-negative specimens were tested by PCR for the presence of the toxin B gene. This was performed using the Xpert C. difficile assay and the GeneXpert Dx system (both from Cepheid, Maurens-Scopont, France). The assay utilized primers for the toxin B gene (tcdB), binary toxin (cdt), and the tcdC deletion at nucleotide 117. Internal controls were included with each individual test cartridge. The test procedure was performed according to the manufacturer's instructions. Briefly, a subculture of the isolate was suspended in sterile, deionized water to a density equal to 0.5 McFarland unit. A swab was dipped into the prepared suspension and left to soak for 1 min. The swab was broken off into the sample reagent, and the suspension was vortexed at high speed for 10 s. The entire suspension was transferred into the sample chamber of the Xpert C. difficile cartridge. Reagents 1 and 2 were added to the appropriate chambers of the cartridge. The cartridge was programmed into and loaded onto the GeneXpert Dx system. Assay time was 45 min. PCR testing was not performed with C. difficile isolates from Vidas-positive stool samples. Such isolates were assumed to be toxigenic, although this was not proven.
Comparison of IDCd with five selective media for cultivation of pure strains of C. difficile.
Ten strains of C. difficile representing distinct ribotypes (ribotypes 001, 002, 005, 015, 016, 023, 027, 064, 078, and 106) were incubated anaerobically for 48 h on Columbia blood agar plates. Colonies of each were then suspended in 0.85% sodium chloride solution (saline) to a density equivalent to 2.0 McFarland units using a densitometer. This suspension was diluted 1/5,000 and 1/50,000 in saline, and 50 μl of each dilution was inoculated onto a culture plate. The inoculum was spread using a sterile loop to form a lawn. Each of the six different culture media described above was inoculated in this way. All media were incubated in an anaerobic workstation at 37°C for a full 24-h period and then removed into air so that colony counts could be performed. After a maximum of 30 min in air, culture plates were reincubated anaerobically for a further 24 h and further counts were performed. The experiment was performed in duplicate on separate occasions, and the colony counts were averaged. The above experiment was also repeated in duplicate using absolute alcohol in place of saline for preparation of the initial bacterial suspensions. After 30 min in alcohol, dilutions were performed in saline, cultured, and examined exactly as described above.
Statistical methods.
Student's t test was used to compare the mean colony counts obtained on different selective media. The various methods for isolation from clinical samples were compared with each other for statistical significance using McNemar's test. A P value of ≤0.05 was used to infer statistical significance.
RESULTS
Comparison of IDCd with CLO medium for isolation of C. difficile from stool samples.
Table 1 shows the performance of IDCd with 268 Vidas-positive stool samples. C. difficile was recovered from 230 specimens (86%) using a combination of both media, and all isolates were recovered on IDCd after 24 h of incubation. About 3% of isolates failed to generate black colonies after 24 h of incubation, and 1% remained colorless after 48 h of incubation. For calculation of sensitivity, these were regarded as undetected. Only 59% of the isolates could be recovered on CLO medium after 24 h of incubation, and even after 48 h of incubation, 24% of the isolates remained uncultured. The performance of CLO medium was significantly inferior to that of IDCd, even after 48 h of incubation (P < 0.001). Of the isolates that could be typed, it was found that 11 distinct ribotypes were isolated, including 001 (n = 33), 106 (n = 27), 016 (n = 22), 027 (n = 21), 015 (n = 12), and others (n < 10), including 002, 005, 023, 064, 078, and 105. Figure 1 shows the typical appearance of C. difficile on IDCd after 24 h of incubation.
TABLE 1.
Numbers of C. difficile isolates recovered on CLO medium and IDCd from 268 Vidas-positive stool samples
| Medium | No. of specimens positive by culture | % specimens from which isolates were recovered | Sensitivity (%) |
|---|---|---|---|
| Anya | 230 | ||
| CLO (24 h) | 136 | 59 | 59 |
| CLO (48 h) | 174 | 76 | 76 |
| IDCd (24 h) | 230 (224)b | 100 | 97 |
| IDCd (48 h) | 230 (227) | 100 | 99 |
Total number of isolates recovered.
Numbers in parentheses indicate the number of isolates forming gray or black colonies.
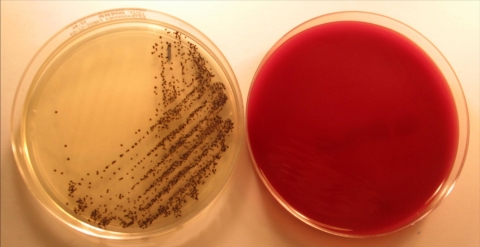
FIG. 1.
Culture of C. difficile from a stool sample after 24 h of incubation on IDCd medium (left) and CLO medium (right). On IDCd, C. difficile forms typical black colonies, whereas the CLO medium plate shows no growth.
C. difficile was recovered from six (6%) Vidas-negative stool samples using a combination of both media, and 5/6 isolates were recovered on IDCd after 24 h of incubation, with the sixth isolate being recovered on IDCd after 48 h. All six isolates formed black colonies on IDCd, but only two isolates were recovered using CLO medium (one after 24 h). All six isolates recovered from Vidas-negative stool samples were found to harbor the toxin B gene when they were tested by PCR.
When Vidas-positive stool samples were cultured onto IDCd, non-C. difficile isolates were recovered from about 10% of samples (26/268). Non-C. difficile isolates presenting as gray or black were recovered from about 8% of samples (22/268). Non-C. difficile isolates were much less common after only 24 h of incubation, with only four isolates forming black colonies from 268 toxin-positive samples (Table 2). The commonest false-positive species encountered was Clostridium clostridioforme, a spore-forming anaerobe typically presenting as a Gram-negative rod. This species was isolated from 4.5% of toxin-positive stool samples and from 22% of toxin-negative stool samples (Table 2).
TABLE 2.
Non-C. difficile isolates recovered on IDCd from 368 stool samples
| Organism | No. of isolates recovered |
|||||||
|---|---|---|---|---|---|---|---|---|
| Vidas-positive samples (n = 268) |
Vidas-negative samples (n = 100) |
|||||||
| Total |
Gray or black colonies |
Total |
Gray or black colonies |
|||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| Clostridium clostridioforme | 3 | 12 | 3 | 12 | 9 | 22 | 8 | 22 |
| Clostridium fallax | 1 | 1 | ||||||
| Clostridium perfringens | 1 | |||||||
| Enterococcus faecalis | 1 | 2 | 1 | 2 | 2 | 2 | ||
| Lactobacillus species | 4 | 2 | 2 | 2 | ||||
| Bacteroides distasonis | 1 | 1 | ||||||
| Bacteroides ovatus | 1 | 3 | 3 | 6 | 15 | 3 | 15 | |
| Bacteroides thetaiotaomicron | 1 | 1 | ||||||
| Bacteroides uniformis | 1 | 1 | ||||||
| Bacteroides vulgatus | 2 | 2 | 1 | 1 | 1 | |||
| Capnocytophaga sp. | 1 | 1 | ||||||
| Porphyromonas endontalis | 1 | 1 | ||||||
| Unidentified Gram-negative rod | 1 | 1 | 1 | 2 | ||||
| Total | 6 | 26 | 4 | 22 | 17 | 49 | 11 | 47 |
Non-C. difficile isolates were much more abundant (or much more easily recovered, due to lower yields of C. difficile) from toxin-negative samples, with 11 of 17 isolates forming gray or black colonies after 24 h of incubation (Table 2) and 47 of 49 isolates forming gray or black colonies after 48 h of incubation. Predominant species were Clostridium clostridioforme and Bacteroides spp. CLO medium was much less selective that IDCd, with non-C. difficile isolates being recovered from over half of all samples. Lactobacilli were the commonest species recovered (Table 3). Commensal bacteria frequently outnumbered C. difficile, and selection of C. difficile colonies was frequently compromised by the large amounts of other species present.
TABLE 3.
Non-C. difficile isolates recovered on CLO medium from 368 stool samples
| Organism | No. of isolates recovered |
|||
|---|---|---|---|---|
| Vidas-positive samples (n = 268) |
Vidas-negative samples (n = 100) |
|||
| 24 h | 48 h | 24 h | 48 h | |
| Lactobacillus species | 57 | 109 | 15 | 33 |
| Gram-negative rods | 14 | 25 | 10 | 25 |
| Gram positive cocci | 2 | 4 | 1 | 3 |
| Corynebacterium species | 2 | 4 | 2 | 2 |
| Clostridium perfringens | 2 | 2 | ||
| Yeast | 0 | 1 | ||
| Total | 77 | 144 | 28 | 64 |
Comparison of IDCd with five selective media for isolation of C. difficile from stool samples after alcohol-shock treatment.
A total of 226 Vidas-positive stool samples were subjected to alcohol shock and cultured onto the six selective media. From these, 199 isolates of C. difficile were recovered using a combination of all media. A total of 10 ribotypes were recovered, including 001 (n = 35), 016 (n = 23), 027 (n = 12), 015 (n = 10), and others (n < 10), including 002, 005, 023, 064, 078, and 106.
All isolates were recovered on IDCd medium, and 197 (99%) were present after only 24 h of incubation. Recovery was significantly better on IDCd than on any other medium after 24 h of incubation (P < 0.001). On IDCd, 94.5% of all C. difficile isolates were recovered as gray/black colonies after 24 h of incubation, and 98% presented as gray/black colonies after 48 h of incubation. Recovery of C. difficile on the other five media was good after 48 h of incubation (sensitivity, 96 to 98%) but was more variable after only 24 h, ranging from 53% for CCEY/L to 87% for CCEY (Table 4). Both BBL medium and CCEY/L were inferior to all other test media after 24 h of incubation (P < 0.001).
TABLE 4.
Recovery of C. difficile and other species from 226 Vidas-positive stool samples using six selective mediaa
| Incubation length and parameter | Result |
|||||
|---|---|---|---|---|---|---|
| IDCd | BBL medium | Oxoid medium | CLO medium | CCEY | CCEY/L | |
| 24 h | ||||||
| No. of C. difficile isolates | 197 (188)b | 129 | 171 | 167 | 174 | 106 |
| % of total | 99 (94.5) | 65 | 86 | 84 | 87 | 53 |
| No. of isolates of other species | 2 (2) | 0 | 2 | 1 | 0 | 0 |
| 48 h | ||||||
| No. of C. difficile isolates | 199 (195) | 194 | 196 | 193 | 195 | 191 |
| % of total | 100 (98) | 97 | 98 | 97 | 98 | 96 |
| No. of isolates of other species | 5 (5) | 10 | 23 | 9 | 1 | 1 |
A total of 199 isolates were recovered within 48 h using a combination of all media.
Numbers in parentheses indicate the number or percentage of isolates forming gray or black colonies.
A total of 19 isolates of C. difficile were recovered from 113 Vidas-negative stool samples cultured following alcohol shock using a combination of all six media. Only 6 of these 19 isolates were positive by PCR for the toxin B gene. Seventeen isolates were recovered on CLO medium after 48 h of incubation, with fewer isolates being recovered on the other media. IDCd showed the best recovery after 24 h of incubation (Table 5). Despite the use of alcohol shock, other flora were recovered on all media, and IDCd was the least selective with Vidas-negative stool samples, from which 34 isolates of other species generated gray or black colonies after 48 h of incubation (Table 5).
TABLE 5.
Recovery of C. difficile from 113 Vidas-negative stool samples using six selective mediaa
| Incubation length and parameter | Result |
|||||
|---|---|---|---|---|---|---|
| IDCd | BBL medium | Oxoid medium | CLO medium | CCEY | CCEY/L | |
| 24 h | ||||||
| No. of C. difficile isolates | 13 (13)b | 7 | 10 | 12 | 9 | 5 |
| % of total | 68 (68) | 37 | 53 | 63 | 47 | 26 |
| No. of isolates of other species | 12 (12) | 1 | 2 | 0 | 0 | 0 |
| 48 h | ||||||
| No. of C. difficile | 16 (16) | 14 | 16 | 17 | 16 | 15 |
| % of total | 84 (84) | 74 | 84 | 89 | 84 | 79 |
| No. of isolates of other species | 34 (34) | 26 | 9 | 18 | 6 | 9 |
A total of 19 isolates were recovered within 48 h using a combination of all media.
Numbers in parentheses indicate number or percentage of isolates forming gray or black colonies.
Comparison of IDCd with five selective media for cultivation of pure strains of C. difficile.
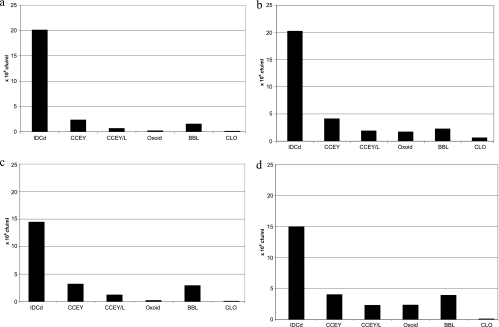
Figure 2 a to d shows the average colony counts of 10 distinct ribotypes of C. difficile on six selective agars. All 10 ribotypes consistently showed a higher count on IDCd medium than on any other agar, irrespective of incubation time or the use of alcohol-shock treatment. The mean count of the 10 ribotypes on IDCd was significantly higher than that on any other medium under any conditions (P < 0.005). For example, the mean count for the 10 ribotypes on IDCd was at least 3.7 times higher than that achieved on CCEY under any conditions and over 30 times higher than that achieved on CLO medium under any conditions. It was notable that there was very little increase in colony count on IDCd over the second period of incubation (<1% increase for cells in saline suspension and 3.5% increase for suspensions in alcohol), on the basis of the average figures for the 10 ribotypes. This contrasted with 684% and 921% increases in colony numbers during the second period of incubation on Oxoid medium for saline-treated and alcohol-treated cells, respectively. After 48 h of incubation, mean colony counts on CCEY and BBL medium were significantly higher than the mean colony count on CLO medium in untreated suspensions (P < 0.05), and mean colony counts from alcohol suspensions were significantly lower on CLO medium than on any other medium used (P < 0.005).
FIG. 2.
Average colony count for 10 distinct ribotypes of C. difficile on six selective agars under different conditions. (a) No alcohol treatment, 24 h of incubation; (b) no alcohol treatment, 48 h of incubation; (c) inoculum treated with alcohol, 24 h of incubation; (d) inoculum treated with alcohol, 48 h of incubation.
DISCUSSION
For several reasons, there is renewed interest in culture media for the isolation of C. difficile. One reason is the emergence of so-called hypervirulent strains that cause outbreaks of infection associated with an increased severity of disease and significant mortality (14, 17). In order to track the spread of such strains, it is usually necessary to isolate them by culture and perform molecular typing. Also, due to the well-recognized limitations of immunoassays, there is a desire by some laboratories to have access to a test with high sensitivity and specificity that may be used as part of a diagnostic algorithm (10, 25, 26). The fecal cytotoxin assay for direct detection of toxin in stool samples using cell lines and specific neutralization was recognized as the “gold standard” for diagnosis of CDI. However, there is clear evidence that culture followed by demonstration of toxin production by isolates (toxigenic culture) is a more sensitive assay for detection of toxigenic C. difficile than the fecal cytotoxin assay (8, 13, 15, 16, 22, 27). One study reported that 29 patients with proven pseudomembranous colitis tested negative in the fecal cytotoxin assay; however, 9 of the 29 samples were submitted for culture and all 9 contained toxigenic C. difficile (16). In one large 7-year study, toxigenic culture resulted in the diagnosis of 355 cases of CDI that would have been missed using the fecal cytotoxin assay alone (8). For these reasons, cytotoxigenic culture is now widely recognized as the gold standard (9). It is worth emphasizing that culture of C. difficile alone has little positive predictive value for diagnosis of CDI without subsequent demonstration of the isolate's ability to produce cytotoxin. This can be directly demonstrated by testing culture supernatants on cell lines or by using immunoassays or may be inferred by PCR (8, 11, 29).
We have compared a prototype chromogenic medium for C. difficile with five selective agars, including four brands from leading suppliers in the United Kingdom. Samples were preselected on the basis of the results obtained with the Vidas immunoassay. This was purely to ensure that a predominance of positive samples was used to compare the different culture media. The most notable feature of IDCd was its ability to induce high colony counts of C. difficile within 24 h of incubation. In this respect it was significantly better than the comparators when it was challenged with pure cultures, presumably due to a superior ability to stimulate germination. It is well recognized that higher colony counts can be obtained by the inclusion of suitable germinants. For example, a recent study compared CLO medium with Columbia blood agar supplemented with 0.1% taurocholate plus cycloserine-cefoxitin. The authors challenged both media with 130 stool samples and found that colony counts of C. difficile were, on average, 30 times higher on the medium containing taurocholate (24). On IDCd, when media were challenged with pure cultures, average colony counts were at least 30 times higher than those obtained on CLO medium and at least 100 times higher when alcohol-treated suspensions were used. One advantage of enhanced germination is that colonies readily form within 24 h of incubation. By combining all of the results of this study, it can be seen that 454 of 707 stool samples were found to contain C. difficile and 445 (98%) were recovered on IDCd within 24 h of incubation. Given this high recovery rate, it can be argued that IDCd should not be incubated beyond 24 h, as the specificity of the medium decreases as other flora is increasingly isolated. It should be noted that when alcohol shock was used, there was no significant difference between IDCd and CCEY, Oxoid medium, and BBL medium after 48 h of incubation. One limitation of the study is that after 24 h of incubation plates were removed into air for up to 30 min to allow effective investigation of colonies that may have become overgrown at 48 h. It is therefore conceivable that the yield of C. difficile might have been higher on any medium after 48 h, if anaerobic incubation had not been interrupted.
Wilcox et al. (30) reported that the incorporation of lysozyme into CCEY medium was advantageous for the recovery of C. difficile from environmental samples. Although no benefit has been reported with stool samples, it was surprising to see an apparent detrimental effect of lysozyme on the recovery of C. difficile from alcohol-treated stool samples, and this was particularly noticeable after 24 h of incubation (P < 0.001).
IDCd is the first chromogenic medium for C. difficile and contains an enzyme substrate that is hydrolyzed to generate black colonies. The nature of the chromogenic system is proprietary and undisclosed by the manufacturer. By combining all study results, it can be seen that 96.6% of C. difficile isolates recovered on IDCd formed gray/black colonies after 24 h and 98.4% were gray or black after 48 h on IDCd. The few isolates of C. difficile forming colorless colonies on IDCd (7/451 isolates; 1.6%) were regarded as undetected, but in practice they were readily distinguishable as C. difficile, due to a characteristic flat irregular colony. The C. difficile isolates forming colorless colonies were ribotype 023 (n = 3), ribotype 001 (n = 1), and untypeable (n = 3). The main advantage of the chromogenic reaction was the formation of black colonies that contrasted sharply with the clear background agar, enabling easy detection of C. difficile. As most other species that were recovered also produced black colonies, there is scope for improvement in the specificity of the agar by either improved inhibition or differentiation of competing flora. In conclusion, IDCd offers effective isolation of C. difficile within only 24 h with or without the use of alcohol-shock treatment. Further studies in different geographical locations are warranted to further assess the suitability of this medium for routine diagnostic use. Studies to examine the compatibility of immunoassays for confirmation of toxin production by isolated colonies would also be worthwhile.
Acknowledgments
We are grateful to bioMérieux, La Balme les Grottes, France, for providing funding and media to support this study. We are also grateful to the North East Strategic Health Authority, United Kingdom, for providing funding for ribotyping of Clostridium difficile isolates.
J.D.P. has received financial support for research or consultancy from suppliers of chromogenic culture media, including bioMérieux and Becton Dickinson. D.H. and S.O. are employed by bioMérieux and have received financial remuneration for patent applications and registrations. The other authors have no relevant disclosures.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Aspinall, S. T., and D. N. Hutchinson. 1992. New selective medium for isolating Clostridium difficile from faeces. J. Clin. Pathol. 45:812-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartley, S. L., and V. R. Dowell, Jr. 1991. Comparison of media for the isolation of Clostridium difficile from fecal specimens. Lab. Med. 22:335-338. [Google Scholar]
- 3.Bliss, D. Z., S. Johnson, C. R. Clabots, K. Savik, and D. N. Gerding. 1997. Comparison of cycloserine-cefoxitin-fructose agar (CCFA) and taurocholate-CCFA for recovery of Clostridium difficile during surveillance of hospitalized patients. Diagn. Microbiol. Infect. Dis. 29:1-4. [DOI] [PubMed] [Google Scholar]
- 4.Bloedt, K., M. Riecker, S. Poppert, and N. Wellinghausen. 2009. Evaluation of new selective culture media and a rapid fluorescence in situ hybridization assay for identification of Clostridium difficile from stool samples. J. Med. Microbiol. 58:874-877. [DOI] [PubMed] [Google Scholar]
- 5.Brazier, J. S. 1998. The diagnosis of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):29-40. [DOI] [PubMed] [Google Scholar]
- 6.Brazier, J. S., and B. I. Duerden. 1998. Guidelines for optimal surveillance of Clostridium difficile infection in hospitals. Commun. Dis. Public Health 1:229-230. [PubMed] [Google Scholar]
- 7.Crobach, M. J., O. M. Dekkers, M. H. Wilcox, and E. J. Kuijper. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin. Microbiol. Infect. 15:1053-1066. [DOI] [PubMed] [Google Scholar]
- 8.Delmée, M., J. Van Broeck, A. Simon, M. Janssens, and V. Avesani. 2005. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: a plea for culture. J. Med. Microbiol. 54:187-191. [DOI] [PubMed] [Google Scholar]
- 9.Eastwood, K., P. Else, A. Charlett, and M. Wilcox. 2009. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J. Clin. Microbiol. 47:3211-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenner, L., A. F. Widmer, G. Goy, S. Rudin, and R. Frei. 2008. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J. Clin. Microbiol. 46:328-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García, A., J. L. Pérez, A. Pulido, J. Niubó, P. Pérez, and R. Martín. 2000. Evaluation of four rapid methods for the investigation of the toxigenic capacity of Clostridium difficile strains isolated in a selective medium. Enferm. Infecc. Microbiol. Clin. 18:109-112. [PubMed] [Google Scholar]
- 12.George, W. L., V. L. Sutter, D. Citron, and S. M. Finegold. 1979. Selective and differential medium for isolation of Clostridium difficile. J. Clin. Microbiol. 9:214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerding, D. N. 2007. New definitions will help, but cultures are critical for resolving unanswered questions about Clostridium difficile. Infect. Control Hosp. Epidemiol. 28:113-115. [DOI] [PubMed] [Google Scholar]
- 14.Goorhuis, A., D. Bakker, J. Corver, S. B. Debast, C. Harmanus, D. W. Notermans, A. A. Bergwerff, F. W. Dekker, and E. J. Kuijper. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47:1162-1170. [DOI] [PubMed] [Google Scholar]
- 15.Jaksic, A. S., G. R. Nimmo, and B. W. Dwyer. 2009. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: microbiologists (should) do it with culture. Pathology 41:187-188. [DOI] [PubMed] [Google Scholar]
- 16.Johal, S. S., J. Hammond, K. Solomon, P. D. James, and Y. R. Mahida. 2004. Clostridium difficile associated diarrhoea in hospitalised patients: onset in the community and hospital and role of flexible sigmoidoscopy. Gut 53:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuijper, E. J., F. Barbut, J. S. Brazier, N. Kleinkauf, T. Eckmanns, M. L. Lambert, D. Drudy, F. Fitzpatrick, C. Wiuff, D. J. Brown, J. E. Coia, H. Pituch, P. Reichert, J. Even, J. Mossong, A. F. Widmer, K. E. Olsen, F. Allerberger, D. W. Notermans, M. Delmée, B. Coignard, M. Wilcox, B. Patel, R. Frei, E. Nagy, E. Bouza, M. Marin, T. Akerlund, A. Virolainen-Julkunen, O. Lyytikäinen, S. Kotila, A. Ingebretsen, B. Smyth, P. Rooney, I. R. Poxton, and D. L. Monnet. 2008. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 13(31):pii=18942. [PubMed] [Google Scholar]
- 18.Levett, P. N. 1985. Effect of antibiotic concentration in a selective medium on the isolation of Clostridium difficile from faecal specimens. J. Clin. Pathol. 38:233-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marler, L. M., J. A. Siders, L. C. Wolters, Y. Pettigrew, B. L. Skitt, and S. D. Allen. 1992. Comparison of five cultural procedures for isolation of Clostridium difficile from stools. J. Clin. Microbiol. 30:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mundy, L. S., C. J. Shanholtzer, K. E. Willard, D. N. Gerding, and L. R. Peterson. 1995. Laboratory detection of Clostridium difficile. A comparison of media and incubation systems. Am. J. Clin. Pathol. 103:52-56. [DOI] [PubMed] [Google Scholar]
- 21.Perry, J. D., and A. M. Freydière. 2007. The application of chromogenic media in clinical microbiology. J. Appl. Microbiol. 103:2046-2055. [DOI] [PubMed] [Google Scholar]
- 22.Reller, M. E., C. A. Lema, T. M. Perl, M. Cai, T. L. Ross, K. A. Speck, and K. C. Carroll. 2007. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 45:3601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley, T. V., J. S. Brazier, H. Hassan, K. Williams, and K. D. Phillips. 1987. Comparison of alcohol shock enrichment and selective enrichment for the isolation of Clostridium difficile. Epidemiol. Infect. 99:355-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousseau, C., I. Poilane, F. Diakite, L. Feghoul, P. Cruaud, and A. Collignon. 2010. Comparison of three Clostridium difficile culture media: interest of enhancing spore germination media? Pathol. Biol. 58:58-61. [DOI] [PubMed] [Google Scholar]
- 25.Sharp, S. E., W. M. Ivie, M. R. Buckles, D. M. Coover, J. C. Pohl, and P. A. Hatcher. 2009. A simple 3-step algorithm for improved laboratory detection of Clostridium difficile toxin without the need for tissue culture cytotoxicity neutralization assays. Diagn. Microbiol. Infect. Dis. 64:344-346. [DOI] [PubMed] [Google Scholar]
- 26.Shin, B. M., E. Y. Kuak, E. J. Lee, and J. G. Songer. 2009. Algorithm combining toxin immunoassay and stool culture for diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 47:2952-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staneck, J. L., L. S. Weckbach, S. D. Allen, J. A. Siders, P. H. Gilligan, G. Coppitt, J. A. Kraft, and D. H. Willis. 1996. Multicenter evaluation of four methods for Clostridium difficile detection: ImmunoCard C. difficile, cytotoxin assay, culture, and latex agglutination. J. Clin. Microbiol. 34:2718-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stubbs, S. L., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thonnard, J., F. Carreer, V. Avesani, and M. Delmée. 1996. Toxin A detection on Clostridium difficile colonies from 24-h cultures. Clin. Microbiol. Infect. 2:50-54. [DOI] [PubMed] [Google Scholar]
- 30.Wilcox, M. H., W. N. Fawley, and P. Parnell. 2000. Value of lysozyme agar incorporation and alkaline thioglycollate exposure for the environmental recovery of Clostridium difficile. J. Hosp. Infect. 44:65-69. [DOI] [PubMed] [Google Scholar]