Abstract
The predominant cultivable microbiota from 20 refractory endodontic lesions (9 with abscesses and 11 without abscesses) were determined, and Propionibacterium acnes and Staphylococcus epidermidis were among the most predominant organisms. The number of species identified from lesions with abscesses (14.1 ± 2.6) was significantly greater (P < 0.001) than the number from lesions without abscesses (7.4 ± 5.9). Comparison of perioral isolates using repetitive extragenic palindromic PCR of the same species from the same subjects demonstrated that the endodontic and skin populations were significantly different. The P. acnes isolates were typed on the basis of recA gene sequence comparison, and only three types (types I, II, and III) were identified among 125 isolates examined. However, we found that type I (type IA and IB) isolates were primarily isolated from the skin, while types II and III were significantly more likely to be isolated from the endodontic lesions (P < 10−10). We found that the robustness of the recA phylotypes was not strong by comparing the partial gene sequences of six putative virulence determinants, PAmce, PAp60, PA-25957, PA-5541, PA-21293, and PA-4687. The resulting neighbor-joining trees were incongruent, and significant (phi test; P = 2.2 × 10−7) evidence of recombination was demonstrated, with significant phylogenetic heterogeneity being apparent within the clusters. P. acnes and S. epidermidis isolated from refractory endodontic infections, with or without periapical abscesses, are likely to be nosocomial infections.
Propionibacterium acnes and coagulase-negative staphylococci, including Staphylococcus epidermidis, have been identified among the microflora of endodontic infections (8, 12, 40, 50, 52, 55, 56, 63, 64), but their importance as endodontic pathogens has largely been ignored due to their nearly universal presence on the skin and the consequent likelihood of sample contamination. However, there is now considerable evidence that these organisms are increasingly isolated from human infections, and so their association with endodontic infections requires clarification. P. acnes, a non-spore-forming, Gram-positive anaerobic or aerotolerant rod, is a member of the resident microflora of the large intestine, conjunctiva, and external ear canal (10, 15) and accounts for approximately half of the total skin microbiota (59), predominating over other pilosebaceous flora (16, 39). Although traditionally considered to be relatively nonpathogenic, an increasing number of studies have implicated P. acnes as an opportunistic pathogen responsible for a wide range of infections and inflammatory conditions. In addition to its well-established role in the pathogenesis of acnes vulgaris (16, 34), it has also been linked to synovitis-acnes-pustulosis-hyperostosis-osteitis syndrome (44, 54), sarcoidosis (17), and prostate cancer (14). Recent studies have revealed trauma and surgery as the predisposing factors associated with numerous P. acnes infections, which include brain abscesses (36), osteomyelitis after lumbar puncture (1), discitis after surgery (23), spodylodiscitis following epidural catheterization (22, 25), postoperative mediastinitis (19, 58), endophthalmitis (7), and endocarditis (21). Furthermore, it is also emerging as an important pathogen in infections related to medical foreign-body implants, such as intraocular lenses (65), ventroperitoneal shunts, orthopedic implants (9, 28, 51), silicone implants (2), prosthetic heart valves (33, 42), and prosthetic joints (38, 61).
S. epidermidis is part of the human skin microflora, where, as a commensal organism, it usually exists in a benign relationship with the host. Although it is ranked first in nosocomial and implant-based infections, perhaps due to its ubiquity on the skin, it is more likely to contaminate devices at the time of insertion. As a recognized opportunistic pathogen, it is responsible for nosocomial infections of indwelling medical devices (43), such as peripheral or central intravenous catheters (CVCs), prosthetic joints, vascular grafts, and central nervous system shunts (46), and cardiac device infections, such as prosthetic valve endocarditis (PVE) (13), as well as ventricular assist device driveline-related infections (4). Clearly, a characteristic shared by both of these organisms is an association with the infection of prosthetic devices.
The treatment of endodontic infections involves the insertion of gutta-percha into the debrided and disinfected root canal and the restoration of the tooth. This treatment may fail, with the root canal becoming infected. A range of bacteria have been isolated from such infected sites, including P. acnes and S. epidermidis, but these have generally been considered contaminants. The objective of the present study was to confirm the endodontic origin of P. acnes and S. epidermidis isolates recovered from refractory endodontic infections. We have used DNA fingerprinting of the P. acnes and S. epidermidis isolates from endodontic lesions and samples from the perioral skin of the same individual using repetitive extragenic palindromic PCRs (REP-PCRs) (3). The recA phylotypes of the P. acnes isolates were determined, as we expected to find that the predominant phylotypes from the endodontic lesions would be types II and III, which are almost exclusively associated with infections of implanted prostheses, while types IA and IB are usually isolated from skin (26, 30, 31, 62). We also investigated the phylogenetic status and robustness of the recA phylotypes by comparing the intrastrain relationships of partial gene sequences of six putative virulence determinants, PAmce (mammalian cell entry gene), PAp60, PA-25957, PA-5541, PA-21293, and PA-4687 (20, 35). Here we have set out to determine if P. acnes and S. epidermidis isolated from refractory endodontic lesions are contaminants arising during the sample collection process or if they represent nosocomial infections.
MATERIALS AND METHODS
Sample collection.
Patients recruited into the study received treatment in the Endodontic Department of the Dental Institute at Guys' and St. Thomas's Hospital, King's College London. The selected teeth were single rooted with failed root canal treatment (refractory cases) with or without a periapical abscess. No communication between the abscess and the oral cavity or the skin surface was observed following clinical examination. All patients were medically stable and had no active, acute medical conditions, and they had not been prescribed antibiotics during the previous month. The sampled teeth had no root fracture or endoperiodontal lesion. All significant details about the history, clinical and radiographic signs, and symptoms about the involved tooth were recorded. The project was approved by the local ethics committee, and the patients gave their informed consent for their inclusion.
Twenty patients with refractory endodontic lesions (n = 9 with periapical abscesses and n = 11 without periapical abscesses) were investigated. After local anesthesia, the tooth to be treated was isolated with a rubber dam. The tooth and surrounding dam and clamp were cleaned with 30% (vol/vol) hydrogen peroxide and decontaminated with 2.5% sodium hypochlorite. After decontamination, the isolated tooth and surrounding dam were swabbed (LIP Limited, West Yorkshire, United Kingdom) to check for contamination.
The coronal restorations were removed, and the root filling was exposed. The coronal gutta-percha placed during the initial root canal treatment was removed using sterile Gates-Glidden drills, and the apical gutta-percha was removed with files. All of the material removed was transferred into 1 ml Tris-EDTA buffer (1.0 M Tris-HCl containing 0.1 M EDTA, pH 8.0, prepared in deionized water) (sample a). Radiographs were taken to ensure that all filling materials had been removed. An apex locator was used to determine the working length of the canal. Sterile saline solution was introduced into the canal, and the canal wall was filed up to the working length. The file samples were transferred into 1 ml Tris-EDTA buffer (sample b). The remaining root canal contents were absorbed onto paper points and transferred into 1 ml Tris-EDTA buffer (sample c). All samples were immediately transported on ice to the laboratory.
From 13 patients, perioral skin swab specimens (LIP Limited) were also taken for the isolation of P. acnes and S. epidermidis.
Microbial analysis of samples.
Each sample (samples a to c) was dispersed by vortexing with glass beads, diluted in fastidious anaerobic broth (Lab M, United Kingdom), and plated onto nonselective medium (fastidious anaerobic agar [FAA] supplemented with 5% [vol/vol] horse blood; Lab M) and also onto a range of selective media, including Rogosa agar (Oxoid, United Kingdom) (49) for lactobacilli, mupirocin-containing Trypticase phytone yeast (MTPY) agar (45) for bifidobacterium, Tryptone yeast cystine agar (TYC) agar (Lab M) (53) for streptococci, Veillonella agar (47, 48) and MacConkey agar (Oxoid, United Kingdom) for enteric bacteria, and CHROMagar (CHROMagar, France) for yeasts. The Rogosa, MTPY, and TYC agar media were incubated anaerobically for 3 days, the Veillonella agar was incubated anaerobically for 4 days, and FAA plates were incubated anaerobically for 7 days. The MacConkey agar and CHROMagar plates were incubated aerobically for 2 days at 37°C. After incubation, the colonies were counted and a predetermined number of colonies from each medium (n = 300 maximum per patient) were randomly selected for identification.
The swabs taken from the prepared teeth prior to the removal of the restoration and the swabs from the perioral skin were plated directly onto FAA and incubated anaerobically for 7 days.
Identification of isolates.
All randomly selected isolates were subcultured on FAA and grown for 24 h. Bacterial genomic DNA was extracted by boiling 100 μl of a suspension of the cultured cells prepared in sterile distilled H2O for 10 min. The suspension was cooled on ice for 10 min and centrifuged at 13,000 × g for 2 min, and the supernatant containing the genomic DNA was stored at −20°C prior to analysis (38). A partial fragment of the 16S rRNA gene of all isolates was amplified using the following reaction mixture (final volume, 25 μl): either 0.3 μl of a 10-pmol/μl concentration each 27F-YMa, 27F-YMb, 27F-YMc, and 27F-YMd forward primers (Integrated DNA Technology, United Kingdom) (18), 0.7 μl of 1492R reverse primer (concentration, 10 pmol/μl), 22.5 μl of Reddymix buffer (Thermo Scientific, United Kingdom), and 0.6 μl DNA extract or 0.5 μl of 9F forward primer (concentration, 10 pmol/μl; MWG, United Kingdom), 0.5 μl of 907R reverse primer (concentration, 10 pmol/μl; MWG), 23 μl of Reddymix buffer (Thermo Scientific), and 1 μl DNA extract. The PCR products were cleaned with Microclean (Sigma, United Kingdom), in order to get ready for the sequence reaction. Amplicon sequencing was performed by using an ABI Prism BigDye Terminator sequencing kit (Applied Biosystems) with 30 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 2 min. Sequencing reaction products were run on an ABI 3730xl sequencer Applied Biosystems). All DNA sequences were analyzed, trimmed, and aligned using BioEdit software (version 7.0.0; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The partial gene sequences were identified by a BLAST search of the NCBI database (http://0-www.ncbi.nlm.nih.gov.ilsprod.ilb.neu.edu/BLAST/), the Human Oral Microbiome database (http://www.homd.org/), or the Ribosome Database Project database http://rdp.cme.msu.edu/). Phylogenetic trees were constructed by the neighbor-joining (NJ) method, based on 16S rRNA gene sequence comparisons, using the MEGA (version 4.1) program (http://www.megasoftware.net/). Most isolates were identified using these sequences, but if isolates were identified as Lactobacillus spp., Veillonella spp., or “Actinomyces naeslundii/A. viscosus,” these were more accurately identified by partial sequencing of the pheS (41), rpoB (5), and metG (24) genes, respectively.
Phylotyping of P. acnes isolates.
Endodontic and skin P. acnes isolates identified by partial 16S rRNA gene sequencing were typed by partial recA gene sequencing. The P. acnes recA gene was amplified using primer PAR-1 (positions −96 to −75; 5′-AGCTCGGTGGGGTTCTCTCATC-3′) and primer PAR-2 (positions +1105 to + 1083; 5′-GCTTCCTCATACCACTGGTCATC-3′), which generated a 1,201-bp amplicon (38). The reaction mixture comprised 0.5 μl of PAR-1 (concentration, 10 pmol/μl; Sigma), 0.5 μl of PAR-2 (concentration, 10 pmol/μl; Sigma), 23 μl of Reddymix (Thermo Scientific), and 1 μl DNA extract in a 25-μl final volume. The thermal cycling conditions included initial denaturation at 95°C for 3 min, denaturation at 95°C for 1 min, annealing at 55°C for 30 s, and extension at 72°C for 90 s, repeated for 35 cycles, and a final extension at 72°C for 10 min. The amplified products could be stored temporarily at 4°C. Amplified products were run on a 0.5% agarose gel and visualized under UV transillumination. Sequencing was performed as described above, and the P. acnes recA sequences were compared with GenBank sequences AY642055 (type IA), EU687255 (type IB), AY642061 (type II), and DQ672252 (type III). NJ trees were constructed using the Jukes-Cantor method with MEGA (version 4.1) software (www.megasoftware.net/).
Comparison of putative P. acnes virulence determinants.
A number of genes have been associated with the virulence of P. acnes (20, 35), including two invasion-associated proteins (PAmce and Pap60) and four predominant immunoreactive surface proteins (PA-25957, PA-5541, PA-21293 and PA-4687), the first two of which have homology with M-like protein of Streptococcus equi and have dermatan sulfate-binding activity, while the other proteins are similar to the product of the Corynebacterium diphtheriae htaA gene, which codes for an iron-regulated hemin binding protein. We used the previously reported primers to amplify and sequence internal fragments of PAmce and Pap60 (35) and designed primers to amplify and sequence fragments of each gene of these putative virulence determinants from a selection of P. acnes strains isolated in this study. PA5541 was amplified and sequenced using primers 5′-TCGACCTCAGCTTCAA-3′ and 5′-AGGCGCTTGACGATAT-3′, PA25957 was amplified and sequenced using primers 5′-CAATCGCAGCAATCAC-3′ and 5′-CTCAGTCTTTGCGATC-3′, PA21693 was amplified and sequenced using primers 5′-CCGAGTTCTATGGCAA-3′ and 5′-CCTGTTTGGTCATGGT-3′, while PA4687 was amplified and sequenced using primers 5′-GTATTGTTAGCCGTGC-3′ and 5′-ATCGTTTTGACCCTGC-3′. The amplicons sizes for the six genes used in the analyses were 783, 1,024, 491, 386, 464, and 473 bp, respectively.
The sequences of the same genes of the sequenced P. acnes strains (strains J165 and SK137 [type IA], KPA171202 and SK187 [type IB], and J139 [type II]) were downloaded from GenBank, and these were aligned with the sequences derived here using BioEdit and NJ trees for each gene and for the concatenated sequences of all the putative virulence genes that were constructed, using the Jukes-Cantor model, in MEGA (version 4.1). The concatenated sequences were tested for the presence of recombination using the phi test (11).
REP-PCR analysis of P. acnes and S. epidermidis.
Skin and endodontic P. acnes and S. epidermidis isolates from the same individual (n = 8 for S. epidermidis and n = 13 for P. acnes) were compared using REP-PCR (3). Primers REP1R-Dt (5′-555 NCG NCG NCA TCN GCC-3′) and REP2-Dt (5′-NCG NCT TAT CNG GCC TAC-3′) (Integrated DNA Technologies, Belgium) used in this study were described previously (3). The reaction mixture contained each of the four deoxynucleoside triphosphates (dNTPs; Thermo Scientific) at a concentration of 125 μM, 150 ng of each primer, 3.75 mM MgCl2 (Thermo Scientific), 2.5 U of Taq polymerase (Thermo Scientific), 20 mM (NH4)2SO4, 75 mM Tris-HCl (pH 9.0), 0.01% (wt/vol) Tween 80, and 3 μl of DNA solution made up to a final volume of 25 μl with sterile water. PCR amplification was performed in an automated thermocycler (Techne) with an initial denaturation (7 min at 95°C), followed by 32 cycles of denaturation (30 s at 94°C), annealing (1 min at 40°C), and extension (8 min at 65°C) and with a final extension (16 min at 65°C). The amplification products of REP-PCR (15 μl) were analyzed with a 2% Metasieve agarose gel (Flowgen, Staffordshire, England) containing Gel Red nucleic acid stain (10,000× in dimethyl sulfoxide; Cambridge Bioscience) and were separated electrophoretically on gels at 60 V for 5 h in 1× TBE (Tris-borate-EDTA) buffer. A molecular size marker (pGEM DNA markers; Promega) was included on all gels to facilitate comparison of tracks between gels. The gels were imaged (AlphaImager), and the resulting patterns were compared visually.
Biochemical characterization of P. acnes isolates.
P. acnes isolates from skin (n = 29) and endodontic infections (n = 34) and reference strains P. acnes type IA (NCTC 737) and P. acnes type II (NCTC 10390) were characterized for their ability to ferment lactose, N-acetylglucosamine, erythritol, ribose, and sorbitol, according to a previously described method (6). The fermentation reactions were also performed in a Rapid ID 32A system, according to the manufacturer's instructions (bioMérieux United Kingdom, Limited). β-d-Fucosidase, β-N-acetylgalactosaminidase, sialidase, α-l-fucosidase, β-N-acetylglucosaminidase, α-glucosidase, β-glucosidase, β-galactosidase, α-arabinosidase, and β-galactosidase activities were determined using fluorogenic substrates, as described previously (6). The strains were also tested for the production of lipase (32), DNase, nonspecific proteinase (37), elastase (29), lecithinase (60), and hyaluronidase (57).
Statistical analysis.
Data distributions were compared using χ2 tests, means were compared using the Mann-Whitney U test in SPSSPC (version 16.0), and the analysis of the biochemical test results using canonical variates analysis was performed in the PAST-Palaeontological Statistics program (version 1.90; http://folk.uio.no/ohammer/past/).
RESULTS
No organisms were recovered from the samples taken from the sterilized tooth surfaces prior to removal of the restoration. The range of organisms cultured from the 20 endodontic lesions is shown in Tables 1 and 2. The lesions without abscesses (Table 1) were characterized by the presence primarily of Gram-positive facultative anaerobic organisms, and these organisms were also present in the lesions with abscesses. However, the cultivable flora of the lesions with abscesses was more complex, as these lesions also harbored obligate anaerobic Gram-positive and Gram-negative organisms. The mean number of taxa from the lesions with periapical abscesses (Table 2) was 14.1 ± 2.6, and that from lesions without periapical abscesses was 7.4 ± 5.9 (P < 0.001). P. acnes was recovered from 10 of the 11 endodontic lesions without periapical abscesses and from 8 of the 9 lesions with periapical abscesses (P > 0.1). Other propionibacteria isolated from the lesions included P. avidium, P. granulosum, P. propionicum, and Propionibacterium sp. strain FMA5. The median proportion of propionibacteria recovered from the file samples in the teeth without abscesses was 26.7% (range, not detected [ND] to 92%) compared to not detected (range, ND to 76.7%) in teeth with abscesses (P < 0.01). Staphylococci, including Staphylococcus caprae, S. condimenti, S. epidermidis, S. hominis, S. pasteuri and S. warneri, were isolated from the endodontic lesions, with S. epidermidis being isolated from 5/9 endodontic lesions with abscesses and from 9/11 of the lesions without abscesses (P > 0.1). The median proportion of staphylococci recovered from the file samples in the teeth without abscesses was 6.7% (range, ND to 73%) compared to not detected (range, ND to 13.3%) in teeth with abscesses (P < 0.01).
TABLE 1.
Distribution of all 51 bacterial taxa identified among 638 isolates recovered from 11 refractory endodontic lesions without periapical abscesses
| Organism | Presence in endodontic lesion (no periapical abscess) no.: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Gram-positive organisms | |||||||||||
| Obligate anaerobes | |||||||||||
| Clostridium sporogenes | − | + | − | − | − | − | − | − | − | − | − |
| Olsenella profusa | + | − | − | − | − | − | − | − | − | + | − |
| Facultative anaerobes | |||||||||||
| Abiotrophia defectiva | − | − | − | − | − | − | − | − | − | + | − |
| Actinomyces gerencseriae oral taxon 618 | − | − | − | − | − | − | − | − | + | − | − |
| Actinomyces massiliensis | − | + | − | − | − | − | − | − | − | − | − |
| Actinomyces meyeri | − | − | + | − | − | − | − | − | − | − | − |
| Actinomyces radicidentis | + | − | − | − | − | − | − | − | − | − | − |
| Actinomyces sp. oral taxon 169 clone AG004 | − | − | − | − | − | − | − | − | − | + | − |
| Actinomyces sp. oral clone CT047 | − | − | − | − | − | − | − | − | + | − | − |
| Bacillus subtilis | − | − | − | − | − | − | − | − | − | − | + |
| Bifidobacterium dentium | + | − | − | − | − | − | − | − | − | − | − |
| Enterococcus faecalis | − | − | − | − | − | + | + | − | − | − | − |
| Gemella haemolysans | − | − | − | − | + | − | − | − | + | + | + |
| Gemella sanguinis | − | − | − | − | − | − | − | − | − | + | − |
| Granulicatella elegans | − | − | − | − | − | − | − | − | − | + | − |
| Kocuria sp. oral taxon B56 | − | − | − | − | − | − | − | − | − | + | − |
| Lactobacillus fermentum | + | − | − | − | − | − | − | − | − | − | − |
| Lactobacillus paracasei | + | − | − | − | − | − | − | − | − | − | − |
| Lactobacillus rhamnosus | + | − | − | − | − | − | − | − | − | − | − |
| Lactobacillus salivarius | + | − | − | − | − | − | − | − | − | − | − |
| Micrococcus luteus strain EHFS1 | − | − | − | − | + | − | − | − | − | − | − |
| Micrococcus sp. oral taxon B64 | − | − | − | − | − | − | − | − | − | + | − |
| Propionibacterium acnes | + | − | + | + | + | + | + | + | + | + | + |
| Propionibacterium granulosum | + | − | − | − | − | − | − | − | − | − | − |
| Propionibacterium propionicum | − | − | − | − | − | − | − | − | − | + | − |
| Rothia dentocariosa | − | − | − | − | − | − | − | − | − | + | − |
| Rothia sp. oral taxon 188 | − | − | − | − | − | − | − | − | − | + | − |
| Staphylococcus capare | − | − | − | + | + | + | − | + | − | − | − |
| Staphylococcus condimenti | + | − | − | − | − | − | − | − | − | − | − |
| Staphylococcus epidermidis | + | + | + | + | + | + | − | + | − | + | + |
| Staphylococcus hominis | + | − | − | − | − | − | − | − | − | − | − |
| Staphylococcus pasteuri | + | − | − | − | − | − | − | − | − | − | − |
| Staphylococcus warneri | − | − | − | + | − | − | − | − | + | − | − |
| Streptococcus anginosus | − | + | − | − | − | − | − | − | − | − | − |
| Streptococcus cristatus | − | − | − | − | − | − | − | − | − | + | − |
| Streptococcus gordonii | − | − | − | − | − | − | − | − | − | + | − |
| Streptococcus mitis/S. oralis | − | + | − | − | + | − | − | − | − | + | − |
| Streptococcus mitis biovar 2 | − | + | − | − | + | − | − | − | − | − | − |
| Streptococcus mutans | + | − | − | − | − | + | − | − | − | − | − |
| Streptococcus sanguinis | − | + | − | − | + | − | − | − | + | − | − |
| Streptococcus sp. oral taxon C65 | − | − | − | − | − | − | − | − | − | + | − |
| Gram-negative organisms | |||||||||||
| Obligate anaerobes | |||||||||||
| Dialister invisus | − | − | − | − | + | − | − | − | − | − | − |
| Prevotella buccae | − | − | − | − | − | − | − | − | − | − | − |
| Prevotella loescheii | − | − | + | − | − | − | − | − | − | − | − |
| Prevotella sp. oral taxon 473 clone IK062 | − | − | − | − | + | − | − | − | − | − | − |
| Selenomonas noxia | − | − | + | − | − | − | − | − | − | − | − |
| Selenomonas sputigena | − | − | − | − | − | − | − | − | − | + | − |
| Tanerella forsythia | − | − | − | − | + | − | − | − | − | − | − |
| Veillonella parvula | − | − | − | − | + | − | − | − | − | − | − |
| Facultative anaerobe | |||||||||||
| Capnocytophaga gingivalis | − | + | − | − | − | − | − | − | − | − | − |
| Capnocytophaga ochracea | − | − | + | − | − | − | − | − | − | − | − |
| Haemophilus parainfluenza | − | − | − | − | + | − | − | − | − | ||
TABLE 2.
Distribution of all 77 bacterial taxa identified among 1,004 isolates recovered from 9 refractory endodontic lesions with periapical abscesses
| Organism | Presence in endodontic lesion (with abscesses) no.: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| Gram-positive organisms | |||||||||
| Obligate anaerobes | |||||||||
| Atopobium species oral taxon 199 clone_C019 | + | − | − | − | − | − | − | − | − |
| Atopobium rimae | − | − | − | − | − | + | − | − | − |
| Eubacterium minutum | + | + | − | − | − | + | − | − | − |
| Eubacterium nodatum | − | + | − | − | − | − | − | − | − |
| Eubacterium infirmum | − | − | − | − | − | + | − | − | + |
| Eubacterium saburreum | − | − | − | − | + | − | − | − | − |
| Eubacterium sulci | − | − | − | − | − | − | + | − | − |
| Eubacterium yurii | − | − | − | − | − | − | + | − | − |
| Finegoldia magna | − | − | − | + | − | − | − | − | − |
| Lachnospiraceae (G-8) sp. clone JM048 | − | − | − | − | − | − | − | − | + |
| Mogibacterium diversum | − | − | − | − | − | − | + | − | + |
| Mogibacterium neglectum | − | − | − | − | − | − | + | − | − |
| Mogibacterium timidium | + | + | + | − | − | + | − | − | − |
| Olsenella profusa | − | − | − | − | − | + | − | − | − |
| Olsenella uli | − | + | + | − | − | + | − | − | − |
| Parvimonas micros | − | − | − | − | + | + | − | − | − |
| Peptostreptococcus stomatis | − | − | + | + | − | + | + | − | − |
| Pseudoramibacter alactolyticus | + | + | + | − | − | + | − | − | − |
| Slackia exigua | + | + | + | − | − | − | + | − | − |
| Solobacterium moorei | − | − | − | − | + | + | + | − | − |
| Uncultured Clostridium sp. clone MS027A1_C05 | − | − | − | + | − | − | − | − | − |
| Uncultured bacterium clone nbw335f06c1 | − | − | − | + | − | − | − | − | − |
| Facultative anaerobes | |||||||||
| Actinomyces israelii | − | − | − | − | − | − | − | + | − |
| Actinomyces sp. oral clone JA063 | − | − | − | + | − | − | − | − | − |
| Actinomyces sp. oral taxon 169 clone AG004 | − | − | − | − | + | − | − | + | − |
| Actinomyces sp. oral taxon 181 strain Hal1065 | − | − | − | − | − | − | − | − | + |
| Actinomyces strain B27SC | + | − | − | − | − | + | − | − | − |
| Actinomyces strain C29KA | − | + | − | − | − | − | − | − | − |
| Bifidobacterium dentium | − | − | + | − | − | − | − | − | − |
| Enterococcus faecalis | − | − | − | − | − | − | − | + | − |
| Gemella haemolysans | − | − | − | − | + | − | − | − | − |
| Granulicatella adiacens | − | − | − | − | − | − | − | + | − |
| Propionibacterium acnes | + | − | + | + | + | + | + | + | + |
| Propionibacterium avidium | + | − | − | − | + | − | − | − | − |
| Propionibacterium propionicum | − | − | − | − | + | − | − | − | − |
| Propionibacterium sp. strain FMA5 | − | − | + | − | − | − | − | − | − |
| Rothia dentocariosa | − | − | − | + | − | − | − | − | − |
| Staphylococcus capare | − | + | − | − | − | − | − | − | − |
| Staphylococcus epidermidis | + | + | − | − | + | − | + | − | + |
| Staphylococcus warneri | − | − | − | − | + | − | − | − | + |
| Streptococcus anginosus | − | − | − | − | − | + | + | − | − |
| Streptococcus australis | − | − | − | − | + | − | − | − | − |
| Streptococcus constellatus | − | + | + | − | − | − | − | − | − |
| Streptococcus genomospecies C8 | − | − | − | − | + | − | − | − | − |
| Streptococcus gordonii | − | − | + | − | − | − | − | − | − |
| Streptococcus infantis | − | − | − | + | − | − | − | − | − |
| Streptococcus mitis/S. oralis | + | − | − | − | − | − | − | − | − |
| Streptococcus mutans | − | − | + | − | − | − | − | − | − |
| Streptococcus sanguinis | − | − | − | − | + | − | − | − | + |
| Streptococcus sp. oral taxon 071 strain Hans H6 | − | − | − | − | + | − | − | + | − |
| Streptococcus sp. oral taxon 071 clone P4PA-13 | − | − | + | − | − | − | − | − | − |
| Gram-negative organisms | |||||||||
| Obligate anaerobes | |||||||||
| Anaeroglobus geminatus | − | − | − | − | − | − | + | − | − |
| Bacteroides thetaiotaomicron | − | − | − | + | − | − | − | − | − |
| Catonella morbi | − | − | − | − | − | − | − | − | + |
| Desulfovibrio fairfieldensis | − | − | − | + | − | − | − | − | − |
| Dialister invisus | + | + | − | − | − | − | + | − | − |
| Dialister pneumosintes | + | − | − | − | − | − | − | − | − |
| Dialister sp. oral clone BS095 | − | − | − | − | − | + | − | − | − |
| Fusobacterium naviforme | − | − | − | − | + | − | − | − | − |
| Fusobacterium nucleatum subsp. nucleatum | − | − | − | − | − | − | − | + | − |
| Fusobacterium nucleatum subsp. polymorphum | − | − | − | − | − | − | − | + | − |
| Fusobacterium nucleatum subsp. vincentii | − | + | − | − | + | − | + | + | − |
| Oribacterium sinus | − | − | − | − | − | − | − | − | + |
| Phascolarctobacterium faecium ACM3679 | − | − | − | + | − | − | − | − | − |
| Porphyromonas gingivalis | + | + | − | − | − | − | − | − | − |
| Prevotella baroniae | − | − | + | − | − | − | − | − | − |
| Prevotella buccae | − | + | − | − | − | − | − | − | − |
| Prevotella melaninogenica | − | − | − | − | − | − | − | − | + |
| Prevotella oralis | + | − | − | − | − | − | − | − | − |
| Prevotella sp. oral taxon 289 | − | − | − | − | − | − | + | − | − |
| Prevotella tannerae | + | − | − | − | − | − | − | − | − |
| Pyramidobacter piscolens | − | − | + | − | − | − | − | − | − |
| Selenomonas noxia | − | − | − | − | − | − | − | − | + |
| Tanerella forsythia | + | − | + | − | − | − | + | − | − |
| Veillonella dispar | − | + | − | − | − | − | − | + | − |
| Veillonella parvula | − | − | − | − | − | − | − | − | + |
| Facultative anaerobe | |||||||||
| Capnocytophaga sp. oral clone AA032 | − | − | − | − | − | + | − | − | − |
| Campylobacter gracilis | − | + | − | − | − | − | + | − | − |
| Campylobacter rectus | − | + | − | − | − | − | − | − | − |
| Lautropia mirabilis | − | − | − | + | − | − | − | − | − |
Phylotyping of P. acnes isolates.
Partial recA sequences were obtained for 65 skin and 60 endodontic P. acnes isolates; and on the basis of sequence alignment, 3 distinct phylogenetic lineages, type I, type II, and type III, were observed. The type I isolates segregated into two distinct groups, into which the sequences from known type IA and type IB sequences clustered. When the distribution of the different P. acnes types was considered (Table 3), the distribution of types recovered from the skin was significantly different from that of all P. acnes isolates recovered from all endodontic lesions (P < 10−10) and from that of the P. acnes isolates recovered only from patients from whom samples from skin were also taken (P = 2 × 10−8).
TABLE 3.
Comparison of distribution of P. acnes recA phylotypes from all endodontic lesions and from the 13 paired endodontic samples with the distribution of P. acnes phylotypes identified from the perioral samples
| Isolate | No. of isolates |
P value (χ2 test) | |||
|---|---|---|---|---|---|
| Type IA | Type IB | Type II | Type III | ||
| P. acnes from paired perioral lesions | 36 | 26 | 3 | 0 | |
| P. acnes from paired endodontic lesions | 7 | 5 | 25 | 2 | 4 × 10−8 |
| All endodontic P. acnes isolatesa | 9 | 12 | 35 | 4 | <10−10 |
n = 20.
Comparison of putative P. acnes virulence determinants.
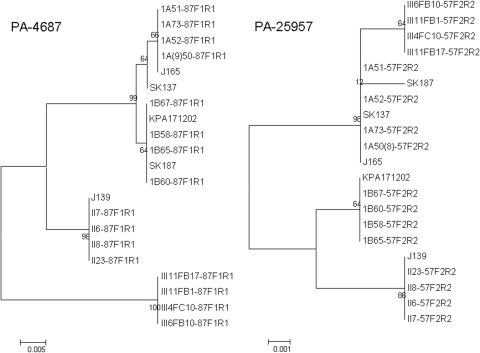
Individual NJ trees were constructed for each of the partial putative virulence gene sequences determined for the strains isolated in this study, along with the corresponding sequences derived from each of the 5 sequenced P. acnes genomes. In PA-4687, we found that each of the type III isolates had evidence of a 5-nucleotide (nt) deletion, such that the gene could no longer be transcribed; these nucleotides were removed from the sequences of the other phylotypes to enable the construction of the NJ trees. The NJ trees for PA-25957 and PA-4687 are shown in Fig. 1 (the remaining trees are shown in Fig. 1S in the supplemental material). From these it is apparent that the type II and III isolates formed distinct clusters with each gene. All the type I sequences for PA-5541, PA-4687, PAmce, and PA-21693 formed either single clusters or clustered together but clustered as subclusters supported by bootstrap values of ≤65. The type IA and IB strains formed distinct discrete clusters with the partial sequences of PA-25957 and Pap60, except that SK187 (type IB) was in the type IA cluster, while the type IB and II strains were on the same lineage with Pap60. The trees for the different genes were clearly incongruent (Fig. 1).
FIG. 1.
Neighbor-joining trees comparing the taxonomic relationships of representatives of P. acnes types IA, IB, II, and III determined by analysis of partial sequences of PA-4687 and PA-25957. Bootstrap values are indicated at corresponding nodes. J165 and SK137 are type IA, SK187 and KPA171202 are type IB, and J139 is type II by recA sequencing. The phylotype of the isolates recovered in this study is denoted by the first letters of the strain name (1A [for type IA], 1B [for type IB], II, or III). Bars, 0.005 or 0.001 substitution per site.
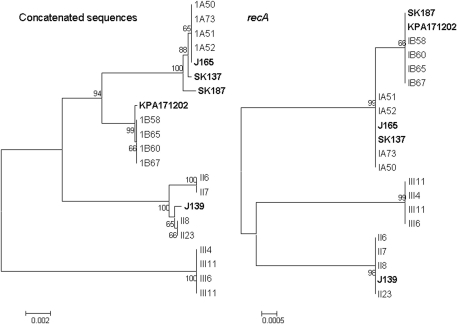
We next constructed an NJ tree using the concatenated sequences of the putative virulence determinants in order to determine if the recA-based determination of phylotype was replicated in this analysis. These data are shown in Fig. 2. In the tree derived from the recA sequences, the 4 lineages are well discriminated. However, in the trees derived from the concatenated sequence data (3,616 bp), the type III isolates formed a distinct lineage, as did the type II isolates, in which two subbranches supported by significant bootstrap values of >95 were evident. The type IB isolates formed a single lineage composed of all isolates except SK187, which was found in the type IA lineage. The type IA lineage appeared to be diverse, with SK187 and SK137 forming branches distinct from the other 5 isolates in this lineage. There was statistical evidence of recombination when the concatenated sequences were tested (phi test, P = 2.2 × 10−7).
FIG. 2.
Neighbor-joining trees comparing the taxonomic relationships of representatives of P. acnes types IA, IB, II, and III determined by analysis of a partial recA sequence and the concatenated partial sequences of 6 putative virulence determinants. Bootstrap values are indicated at the corresponding nodes. J165 and SK137 type are IA, SK187 and KPA171202 are type IB, and J139 is type II by recA sequencing. The phylotype of the isolates recovered in this study is denoted by the first letters of the strain name (1A [for type IA], 1B [for type IB], II, or III). Bars, 0.002 or 0.0005 substitution per site.
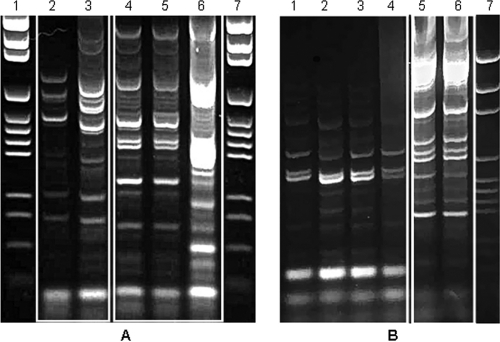
REP-PCR comparison of paired isolates.
When the endodontic and skin P. acnes isolates from the same individuals (n = 13) were compared using REP-PCR, it was found that all skin P. acnes isolates were distinct from all the endodontic isolates except in 3 patients, for whom a REP-PCR pattern was common to some of the P. acnes isolates from both the skin and the endodontic lesion (χ2 = 56.05; P < 0.001). A typical comparison of the REP-PCR patterns of skin and endodontic isolates from the same person is shown in Fig. 3A. Similarly, in the case of S. epidermidis, when the REP-PCR patterns of endodontic and skin isolates from the same individual (n = 8) were compared, all skin isolates were clearly distinct from the endodontic ones (χ2 = 37.73; P < 0.001). A REP-PCR pattern comparing skin and endodontic S. epidermidis isolates from the same patient is shown in Fig. 3B.
FIG. 3.
REP-PCR patterns obtained from the P. acnes isolates of patient 6 (A) and the S. epidermidis isolates of patient 13 (B). (A) Lanes 1 and 7, molecular size marker (pGEM DNA markers; Promega); lanes 2 and 3, skin isolates; lanes 4 to 6, isolates from the endodontic sample. (B) Lane 7, molecular size marker; lanes 5 and 6, isolates from the endodontic sample; lanes 1 to 4, skin isolates.
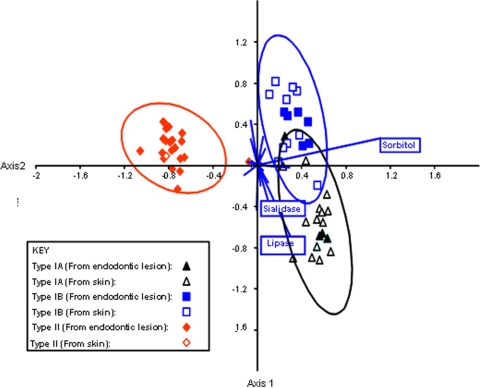
Phenotypic comparison of P. acnes phylotypes.
The phenotypic properties of the P. acnes phylotypes are shown in Table 4. The phenotypic properties which differentiate between P. acnes types IA, IB, and II were investigated using canonical variates analysis (Fig. 4). Type III strains were not investigated due to the small number of isolates. The ability to ferment sorbitol and to produce sialidase and lipase activities differentiated between types IA, IB, and II.
TABLE 4.
Biochemical test results for 65 P. acnes isolates, including reference strains P. acnes type IA (NCTC 737) and P. acnes type II (NCTC 10390)a
| Phenotypic test | % of P. acnes isolates |
|||
|---|---|---|---|---|
| Type IA (n = 20) | Type IB (n = 19) | Type II (n = 22) | Type III (n = 4) | |
| Lactose | 5 | 0 | 0 | 0 |
| N-Acetylglucosamine | 55 | 53 | 95 | 100 |
| Erythritol | 5 | 47 | 9 | 0 |
| Ribose | 95 | 89 | 100 | 100 |
| Sorbitol | 100 | 100 | 5 | 25 |
| Urease | 5 | 0 | 0 | 0 |
| Arginine dihydrolase | 95 | 100 | 95 | 100 |
| β-Galactosidase | 75 | 84 | 64 | 25 |
| α-Glucosidase | 15 | 84 | 77 | 50 |
| β-Glucoronidase | 5 | 16 | 0 | 0 |
| β-N-Acetyl-glucosaminidase | 100 | 100 | 95 | 100 |
| Mannose fermentation | 100 | 100 | 86 | 75 |
| Raffose fermentation | 0 | 5 | 0 | 0 |
| Nitrate reduction | 95 | 100 | 91 | 75 |
| Indole production | 95 | 79 | 59 | 50 |
| Arginine arylamidase | 40 | 37 | 59 | 25 |
| Leucyl glycine arylamidase | 10 | 11 | 18 | 0 |
| Phenylalanine | 0 | 0 | 5 | 0 |
| Leucine arylamidase | 0 | 0 | 5 | 0 |
| Pyroglutamic acid arylamidase | 5 | 11 | 9 | 0 |
| Alanine arylamidase | 100 | 95 | 82 | 50 |
| Serine arylamidase | 95 | 84 | 100 | 50 |
| β-N-Acetyl-galatosaminidase | 70 | 26 | 59 | 0 |
| Sialidase | 100 | 37 | 14 | 25 |
| Lipase | 95 | 26 | 0 | 25 |
Major differential characteristics distinguishing between types IA, IB, and II are highlighted in boldface. Data from type III strains were not included in the analysis due to the small number of isolates tested. All isolates were positive for glycine arylamidase activity and negative for β-fucosidase, α-fucosidase, β-glucosidase, α-galactosidase, α-arabinosidase, DNase, nonspecific proteinase, elastase, lecithinase, and hyaluronidase activities.
FIG. 4.
Phenotypic differences between P. acnes type IA, type IB, and type II. The chart is a canonical variance analysis scatter plot of isolates along the first two canonical axes, showing maximal separation between the three groups. The null hypothesis of no phenotypic difference between the three groups was tested by multivariate analysis of a variance (P < 0.01, Bonferroni-corrected P values for comparison of pairs of groups) and nonparametric analysis of similarities (P < 0.001, Euclidean distance measure). The biplot (blue lines) displays the contribution of individual tests to the separation; the three most important tests are indicated.
DISCUSSION
We have used a culture-based approach to investigate the microbiota of refractory root canal infections. With a culture-based approach, it is likely that, overall, fewer taxa than are detectable with 16S rRNA cloning or cloning combined with culture approaches may be identified. Thus, in a study of 5 primary endodontic lesions in four patients, a mean of 20.2 taxa (range, 7 to 29) was identified using a combined cloning and cultural approach (40), whereas a mean of 14.1 taxa was identified from the refractory lesions with abscesses. We investigated refractory lesions which had previously been treated, so DNA derived from dead bacteria may be present, which might lead to an overestimation of the diversity of the microbiota if a 16S rRNA cloning approach was used. However, as the purpose of this study was not just to catalogue the taxa present at a site but to investigate the genotypic and phenotypic properties of P. acnes and S. epidermidis, it was necessary to culture the microbiota and to isolate these species.
Previous studies of the endodontic microbiota (2, 9, 28, 33, 38, 42, 51, 61) have recovered P. acnes and various Staphylococcus species, including S. epidermidis, but these organisms are generally regarded as contaminants since they are skin commensals. In the present study, we have used a decontamination protocol to remove contaminating organisms from the tooth surface prior to entering the diseased tooth, removed the gutta-percha points, sampled the infected root dentine bordering the root canal, and isolated P. acnes and S. epidermidis. The same species were also isolated from the perioral area and compared using REP-PCR DNA fingerprinting. The endodontic and skin isolates of both P. acnes and S. epidermis are distinct and significantly different. These observations confirm the endodontic origin of P. acnes and S. epidermidis and the rejection of the hypothesis that these species recovered from endodontic samples are contaminants. While we did not investigate further the other Propionibacterium or Staphylococcus species, it must also be presumed that these are also infecting organisms. Foreign-body implants are one of the predisposing factors associated with numerous P. acnes infections (1, 14, 22, 23). P. acnes infections are also associated with the presence of foreign bodies in the root canals and may have been introduced into the root canals at the time of obturation of the canal with the gutta-percha or may have infected the canal subsequently, after failure. Similarly, S. epidermidis is associated with infections of prostheses, and it is also significant that S. caprae, which was isolated from 25% of patients and which is routinely isolated from goats and may be used in cheese ripening, is also associated with infections in patients with an indwelling catheter or foreign materials, such as cardiac prostheses (27). This supports the hypothesis that the gutta-percha in the root canal may represent a foreign body susceptible to colonization by the same range of organisms that infect foreign materials or prostheses.
This hypothesis was supported when the recA phylotypes of the isolates were considered. We confirmed the previous observations, as nucleotide sequencing of the recA housekeeping gene of P. acnes isolates from both endodontic lesions and samples from perioral skin revealed only 3 different phylogenetic lineages (37, 38). In agreement with these observations, we found that P. acnes type I was the predominant phylotype on skin, whereas types II and III have rarely been recovered from samples from skin (37). In the present study, P. acnes type II and type III were the prevalent phylotypes in the endodontic infections, and the distribution of type II was similar to that reported for type II isolates from failed prosthetic hip implants (38), orthopedic implants (51), and radical prostatectomy specimens from subjects with prostate cancer (14). Type III strains have previously been recovered only from spine intervertabral disc material and from a prosthetic hip joint removed during revision arthroplasty. Here we failed to isolate any type III strains from the skin, but 4 isolates were identified from the infected root canals of 3 patients. The natural habitat of types II and III is not known, but these phylotypes apparently exhibit a tropism for inert foreign surfaces, or at least an ability to bind to such materials. Although P. acnes isolates of the type II phylotype were the most prevalent endodontic isolates in our study, the virulence determinants have not been identified and none of the differential phenotypic characteristics investigated here appear to be relevant; and all isolates tested, irrespective of their phylotype, possessed genes coding for each of the 6 putative virulence determinants. However, all of the type III isolates have a 5-nt deletion in PA-4687, involved in iron transport, suggesting that this gene did not yield a functional product, which was supported by our inability to demonstrate mRNA for this gene in the reverse transcription-PCR experiments. The role of these genes in bacterial virulence remains unclear.
The recA typing of P. acnes isolates appears to be robust, as all isolates fell neatly into the three phylotypes. The significance of this typing is not clear, but it does provide a classification for P. acnes isolates which matches their site of isolation. Extensive DNA-DNA homology testing of P. acnes isolates suggests that the species is homogeneous (30), although it cannot be known if strains equivalent to types II and III were tested. To test the robustness of the recA typing scheme, we determined internal sequences of six putative virulence factors and used the concatenated sequences of these partial gene sequences in phylogenetic analyses. From previous investigations, it was apparent that the relationships found with recA sequencing were not always mirrored when the sequences of other P. acnes genes were compared. Thus, in a small study of 3 P. acnes strains, the sequences of CAMP factors 1, 2, 4, and 5 exhibited the same phylogenetic relationship between the type IA, IB, and II strains as that determined previously with recA; however, for CAMP factor 3, the type IA sequence had a closer relationship to the type II sequence, with the type IB sequence being more distinct (62). From the present analysis of the concatenated sequences, it is clear that types II and III remain distinct from the type I lineage. The type III strains are identical, despite being isolated from 3 different, unrelated patients; but within the type II lineage, two distinct subclusters were evident, and this was supported by significant bootstrap values. The type I lineage shows greater rearrangement; notably, SK187 (type IB) is found within the type IA subcluster. Within both the type IA and IB subclusters, there is evidence of significant branching, with KPA17202, SK187, and SK137 being on discrete branches. These data demonstrate that while recA phylotyping permits facile classification of P. acnes strains, the use of more partial or complete gene sequences to establish phylogenetic relationships between strains gives rise to a more complicated population structure than was initially apparent. The reasons for the disparity between the recA typing and the relationships evident when the concatenated sequences were used to type the strains may be intraspecies recombination, for which evidence was obtained in the analyses presented here. This is also supported by the observation that not all the individual gene trees were congruent, with types IB and II being in the same lineage with Pap60 and all type I and II strains being on the same lineage with PA-5541, yet type II strains were on discrete lineages with each of the other four genes. It may be that the type II and III lineages have evolved, presumably from type I, to be better able to cause infections associated with foreign-body implants.
The gutta-percha used as root canal filling material is marketed unsterilized and may also be contaminated during root canal filling procedures, as may be the paper points used to dry root canals prior to filling of the root canal. The results of the present study suggest that S. epidermidis and P. acnes isolates from the refractory lesions might be the result of nosocomial infections occurring at the time of the root canal treatment. Root canal treatments of infected teeth can be undertaken using a single- or a multivisit approach. The nosocomial nature of some endodontic infections would support the single-visit technique, as this would reduce the likelihood of such infections occurring.
Supplementary Material
Acknowledgments
We acknowledge support from the Higher Education Commission of Pakistan; from the Dental Institute, King's College London; and from the Department of Health via the National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre Award to Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust.
Footnotes
Published ahead of print on 25 August 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Abolnik, I. Z., J. V. Eaton, and D. J. Sexton. 1995. Propionibacterium acnes vertebral osteomyelitis following lumbar puncture: case report and review. Clin. Infect. Dis. 21:694-695. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, C. Y., C. Y. Ko, E. A. Wagar, R. S. Wong, and W. W. Shaw. 1996. Microbial evaluation: 139 implants removed from symptomatic patients. Plast. Reconstr. Surg. 98:1225-1229. [DOI] [PubMed] [Google Scholar]
- 3.Alam, S., S. R. Brailsford, R. A. Whiley, and D. Beighton. 1999. PCR-based methods for genotyping viridans group streptococci. J. Clin. Microbiol. 37:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrecubieta, C., F. A. Toba, M. von Bayern, H. Akashi, M. C. Deng, Y. Naka, and F. D. Lowy. 2009. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog. 5:e1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beighton, D., S. C. Gilbert, D. Clark, M. Mantzourani, M. Al-Haboubi, F. Ali, E. Ransome, N. Hodson, M. Fenlon, L. Zoitopoulos, and J. Gallagher. 2008. Isolation and identification of Bifidobacteriaceae from human saliva. Appl. Environ. Microbiol. 74:6457-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beighton, D., J. M. Hardie, and R. A. Whiley. 1991. A scheme for the identification of viridans streptococci. J. Med. Microbiol. 35:367-372. [DOI] [PubMed] [Google Scholar]
- 7.Benz, M. S., I. U. Scott, H. W. Flynn, Jr., N. Unonius, and D. Miller. 2004. Endophthalmitis isolates and antibiotic sensitivities: a 6-year review of culture-proven cases. Am. J. Ophthalmol. 137:38-42. [DOI] [PubMed] [Google Scholar]
- 8.Brito, L. C., F. R. Teles, R. P. Teles, E. C. Franca, A. P. Ribeiro-Sobrinho, A. D. Haffajee, and S. S. Socransky. 2007. Use of multiple-displacement amplification and checkerboard DNA-DNA hybridization to examine the microbiota of endodontic infections. J. Clin. Microbiol. 45:3039-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brook, I., and E. H. Frazier. 1999. Aerobic and anaerobic microbiology of surgical-site infection following spinal fusion. J. Clin. Microbiol. 37:841-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brook, I., and E. H. Frazier. 1991. Infections caused by Propionibacterium species. Rev. Infect. Dis. 13:819-822. [DOI] [PubMed] [Google Scholar]
- 11.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavez de Paz, L. E., A. Molander, and G. Dahlen. 2004. Gram-positive rods prevailing in teeth with apical periodontitis undergoing root canal treatment. Int. Endod. J. 37:579-587. [DOI] [PubMed] [Google Scholar]
- 13.Chu, V. H., J. M. Miro, B. Hoen, C. H. Cabell, P. A. Pappas, P. Jones, M. E. Stryjewski, I. Anguera, S. Braun, P. Munoz, P. Commerford, P. Tornos, J. Francis, M. Oyonarte, C. Selton-Suty, A. J. Morris, G. Habib, B. Almirante, D. J. Sexton, G. R. Corey, and V. G. Fowler, Jr. 2009. Coagulase-negative staphylococcal prosthetic valve endocarditis—a contemporary update based on the International Collaboration on Endocarditis: prospective cohort study. Heart 95:570-576. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, R. J., B. A. Shannon, J. E. McNeal, T. Shannon, and K. L. Garrett. 2005. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J. Urol. 173:1969-1974. [DOI] [PubMed] [Google Scholar]
- 15.Doyle, A., B. Beigi, A. Early, A. Blake, P. Eustace, and R. Hone. 1995. Adherence of bacteria to intraocular lenses: a prospective study. Br. J. Ophthalmol. 79:347-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eady, E. A., and E. Ingham. 1994. Propionibacterium acnes—friend or foe? Rev. Med. Microbiol. 5:163-173. [Google Scholar]
- 17.Eishi, Y., M. Suga, I. Ishige, D. Kobayashi, T. Yamada, T. Takemura, T. Takizawa, M. Koike, S. Kudoh, U. Costabel, J. Guzman, G. Rizzato, M. Gambacorta, R. du Bois, A. G. Nicholson, O. P. Sharma, and M. Ando. 2002. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J. Clin. Microbiol. 40:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, J. A., C. I. Reich, S. Sharma, J. S. Weisbaum, B. A. Wilson, and G. J. Olsen. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74:2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friberg, O., R. Svedjeholm, J. Kallman, and B. Soderquist. 2007. Incidence, microbiological findings, and clinical presentation of sternal wound infections after cardiac surgery with and without local gentamicin prophylaxis. Eur. J. Clin. Microbiol. Infect. Dis. 26:91-97. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa, A., K. Uchida, Y. Ishige, I. Ishige, I. Kobayashi, T. Takemura, T. Yokoyama, K. Iwai, K. Watanabe, S. Shimizu, N. Ishida, Y. Suzuki, T. Suzuki, T. Yamada, T. Ito, and Y. Eishi. 2009. Characterization of Propionibacterium acnes isolates from sarcoid and non-sarcoid tissues with special reference to cell invasiveness, serotype, and trigger factor gene polymorphism. Microb. Pathog. 46:80-87. [DOI] [PubMed] [Google Scholar]
- 21.Gunthard, H., A. Hany, M. Turina, and J. Wust. 1994. Propionibacterium acnes as a cause of aggressive aortic valve endocarditis and importance of tissue grinding: case report and review. J. Clin. Microbiol. 32:3043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halkic, N., C. Blanc, M. E. Corthesy, and J. M. Corpataux. 2001. Lumbar spondylodiscitis after epidural anaesthesia at a distant site. Anaesthesia 56:602-603. [PubMed] [Google Scholar]
- 23.Harris, A. E., C. Hennicke, K. Byers, and W. C. Welch. 2005. Postoperative discitis due to Propionibacterium acnes: a case report and review of the literature. Surg. Neurol. 63:538-541. [DOI] [PubMed] [Google Scholar]
- 24.Henssge, U., T. Do, D. R. Radford, S. C. Gilbert, D. Clark, and D. Beighton. 2009. Emended description of Actinomyces naeslundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov., previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int. J. Syst. Evol. Microbiol. 59:509-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Palazon, J., J. P. Puertas-Garcia, J. F. Martinez-Lage, and J. A. Tortosa. 2003. Lumbar spondylodiscitis caused by Propionibacterium acnes after epidural obstetric analgesia. Anesth. Analg. 96:1486-1488. [DOI] [PubMed] [Google Scholar]
- 26.Higaki, S., T. Kitagawa, M. Kagoura, M. Morohashi, and T. Yamagishi. 2000. Correlation between Propionibacterium acnes biotypes, lipase activity and rash degree in acne patients. J. Dermatol. 27:519-522. [DOI] [PubMed] [Google Scholar]
- 27.Irlinger, F. 2008. Safety assessment of dairy microorganisms: coagulase-negative staphylococci. Int. J. Food Microbiol. 126:302-310. [DOI] [PubMed] [Google Scholar]
- 28.Jakab, E., R. Zbinden, J. Gubler, C. Ruef, A. von Graevenitz, and M. Krause. 1996. Severe infections caused by Propionibacterium acnes: an underestimated pathogen in late postoperative infections. Yale J. Biol. Med. 69:477-482. [PMC free article] [PubMed] [Google Scholar]
- 29.Janda, J. M. 1986. Elastolytic activity among staphylococci. J. Clin. Microbiol. 24:945-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, J. L., and C. S. Cummins. 1972. Cell wall composition and deoxyribonucleic acid similarities among the anaerobic coryneforms, classical propionibacteria, and strains of Arachnia propionica. J. Bacteriol. 109:1047-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishishita, M., T. Ushijima, Y. Ozaki, and Y. Ito. 1979. Biotyping of Propionibacterium acnes isolated from normal human facial skin. Appl. Environ. Microbiol. 38:585-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouker, G., and K. E. Jaeger. 1987. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 53:211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazar, J. M., and D. S. Schulman. 1992. Propionibacterium acnes prosthetic valve endocarditis: a case of severe aortic insufficiency. Clin. Cardiol. 15:299-300. [DOI] [PubMed] [Google Scholar]
- 34.Leyden, J. J. 2001. The evolving role of Propionibacterium acnes in acne. Semin. Cutan. Med. Surg. 20:139-143. [DOI] [PubMed] [Google Scholar]
- 35.Lodes, M. J., H. Secrist, D. R. Benson, S. Jen, K. D. Shanebeck, J. Guderian, J. F. Maisonneuve, A. Bhatia, D. Persing, S. Patrick, and Y. A. Skeiky. 2006. Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology 152:3667-3681. [DOI] [PubMed] [Google Scholar]
- 36.Mathisen, G. E., R. D. Meyer, W. L. George, and D. M. Citron. 1984. Brain abscess and cerebritis. Rev. Infect. Dis. 6(Suppl. 1):101-106. [DOI] [PubMed] [Google Scholar]
- 37.McDowell, A., A. L. Perry, P. A. Lambert, and S. Patrick. 2008. A new phylogenetic group of Propionibacterium acnes. J. Med. Microbiol. 57:218-224. [DOI] [PubMed] [Google Scholar]
- 38.McDowell, A., S. Valanne, G. Ramage, M. M. Tunney, J. V. Glenn, G. C. McLorinan, A. Bhatia, J. F. Maisonneuve, M. Lodes, D. H. Persing, and S. Patrick. 2005. Propionibacterium acnes types I and II represent phylogenetically distinct groups. J. Clin. Microbiol. 43:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGinley, K. J., G. F. Webster, and J. J. Leyden. 1978. Regional variations of cutaneous propionibacteria. Appl. Environ. Microbiol. 35:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munson, M. A., T. Pitt-Ford, B. Chong, A. Weightman, and W. G. Wade. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J. Dent. Res. 81:761-766. [DOI] [PubMed] [Google Scholar]
- 41.Naser, S. M., P. Dawyndt, B. Hoste, D. Gevers, K. Vandemeulebroecke, I. Cleenwerck, M. Vancanneyt, and J. Swings. 2007. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 57:2777-2789. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill, T. M., R. Hone, and S. Blake. 1988. Prosthetic valve endocarditis caused by Propionibacterium acnes. Br. Med. J. (Clin. Res. ed.) 296:1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto, M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry, A. L., and P. A. Lambert. 2006. Propionibacterium acnes. Lett. Appl. Microbiol. 42:185-188. [DOI] [PubMed] [Google Scholar]
- 45.Rada, V., and J. Petr. 2000. A new selective medium for the isolation of glucose non-fermenting bifidobacteria from hen caeca. J. Microbiol. Methods 43:127-132. [DOI] [PubMed] [Google Scholar]
- 46.Rogers, K. L., P. D. Fey, and M. E. Rupp. 2009. Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23:73-98. [DOI] [PubMed] [Google Scholar]
- 47.Rogosa, M. 1956. A selective medium for the isolation and enumeration of the veillonella from the oral cavity. J. Bacteriol. 72:533-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogosa, M., R. J. Fitzgerald, M. E. Mackintosh, and A. J. Beaman. 1958. Improved medium for selective isolation of Veillonella. J. Bacteriol. 76:455-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogosa, M., J. A. Mitchell, and R. F. Wiseman. 1951. A selective medium for the isolation and enumeration of oral lactobacilli. J. Dent. Res. 30:682-689. [DOI] [PubMed] [Google Scholar]
- 50.Rolph, H. J., A. Lennon, M. P. Riggio, W. P. Saunders, D. MacKenzie, L. Coldero, and J. Bagg. 2001. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 39:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sampedro, M. F., K. E. Piper, A. McDowell, S. Patrick, J. N. Mandrekar, M. S. Rouse, J. M. Steckelberg, and R. Patel. 2009. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn. Microbiol. Infect. Dis. 64:138-145. [DOI] [PubMed] [Google Scholar]
- 52.Sassone, L., R. Fidel, L. Figueiredo, S. Fidel, M. Faveri, and M. Feres. 2007. Evaluation of the microbiota of primary endodontic infections using checkerboard DNA-DNA hybridization. Oral Microbiol. Immunol. 22:390-397. [DOI] [PubMed] [Google Scholar]
- 53.Schaeken, M. J., J. S. van der Hoeven, and H. C. Franken. 1986. Comparative recovery of Streptococcus mutans on five isolation media, including a new simple selective medium. J. Dent. Res. 65:906-908. [DOI] [PubMed] [Google Scholar]
- 54.Schaeverbeke, T., L. Lequen, B. de Barbeyrac, L. Labbe, C. M. Bebear, Y. Morrier, B. Bannwarth, C. Bebear, and J. Dehais. 1998. Propionibacterium acnes isolated from synovial tissue and fluid in a patient with oligoarthritis associated with acne and pustulosis. Arthritis Rheum. 41:1889-1893. [DOI] [PubMed] [Google Scholar]
- 55.Sunde, P. T., I. Olsen, G. J. Debelian, and L. Tronstad. 2002. Microbiota of periapical lesions refractory to endodontic therapy. J. Endod. 28:304-310. [DOI] [PubMed] [Google Scholar]
- 56.Sundqvist, G., D. Figdor, S. Persson, and U. Sjogren. 1998. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 85:86-93. [DOI] [PubMed] [Google Scholar]
- 57.Tam, Y. C., and E. C. Chan. 1985. Purification and characterization of hyaluronidase from oral Peptostreptococcus species. Infect. Immun. 47:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tammelin, A., A. Hambraeus, and E. Stahle. 2002. Mediastinitis after cardiac surgery: improvement of bacteriological diagnosis by use of multiple tissue samples and strain typing. J. Clin. Microbiol. 40:2936-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tancrede, C. 1992. Role of human microflora in health and disease. Eur. J. Clin. Microbiol. Infect. Dis. 11:1012-1015. [DOI] [PubMed] [Google Scholar]
- 60.Thaler, J. O., B. Duvic, A. Givaudan, and N. Boemare. 1998. Isolation and entomotoxic properties of the Xenorhabdus nematophilus F1 lecithinase. Appl. Environ. Microbiol. 64:2367-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tunney, M. M., S. Patrick, M. D. Curran, G. Ramage, D. Hanna, J. R. Nixon, S. P. Gorman, R. I. Davis, and N. Anderson. 1999. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J. Clin. Microbiol. 37:3281-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valanne, S., A. McDowell, G. Ramage, M. M. Tunney, G. G. Einarsson, S. O'Hagan, G. B. Wisdom, D. Fairley, A. Bhatia, J. F. Maisonneuve, M. Lodes, D. H. Persing, and S. Patrick. 2005. CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiology 151:1369-1379. [DOI] [PubMed] [Google Scholar]
- 63.Vianna, M. E., H. P. Horz, G. Conrads, M. Feres, and B. P. Gomes. 2008. Comparative analysis of endodontic pathogens using checkerboard hybridization in relation to culture. Oral Microbiol. Immunol. 23:282-290. [DOI] [PubMed] [Google Scholar]
- 64.Vianna, M. E., H. P. Horz, B. P. Gomes, and G. Conrads. 2005. Microarrays complement culture methods for identification of bacteria in endodontic infections. Oral Microbiol. Immunol. 20:253-258. [DOI] [PubMed] [Google Scholar]
- 65.Winward, K. E., S. C. Pflugfelder, H. W. Flynn, Jr., T. J. Roussel, and J. L. Davis. 1993. Postoperative Propionibacterium endophthalmitis. Treatment strategies and long-term results. Ophthalmology 100:447-451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.