Abstract
Two immunocompromised patients with 2009 H1N1 influenza pneumonia had viral shedding for over 5 weeks despite therapy with oseltamivir. Declining or persistently low cycle threshold values noted on serial qualitative real-time reverse transcriptase PCR (rRT-PCR) of respiratory specimens implied increasing viral load and probable drug resistance. Oseltamivir resistance was later confirmed by pyrosequencing.
CASE REPORTS
Case report 1.
A 52-year-old male with myelodysplastic syndrome underwent an allogeneic hematopoietic stem cell transplantation (HSCT) complicated by graft-versus-host disease (GVHD) and bronchiolitis obliterans of his lungs. On day 864 post-HSCT, he developed cough and pleuritic chest pain. Nodular cavitary infiltrates were identified by computed tomography scan (CT) of his thorax. Aspergillus fumigatus was isolated from bronchoalveolar lavage (BAL) fluid, and the BAL fluid Aspergillus galactomannan assay was positive. Voriconazole therapy was initiated for probable pulmonary aspergillosis. Rhinovirus was detected on PCR assay of BAL fluid. Mycobacterium avium complex was also isolated from BAL fluid and was considered to represent colonization.
On day 990 post-HSCT, in November 2009, he was hospitalized with fever, progressive dyspnea, and nonproductive cough. In addition to voriconazole, he was receiving methylprednisone, tacrolimus, sirolimus, imatinib mesylate, and extracorporeal photopheresis as therapy for GVHD. His oxygen saturation was 83% when he was breathing ambient air, and he had crackles at both lung bases. A computed tomography (CT) scan of the thorax demonstrated new bilateral lung infiltrates. The rapid influenza diagnostic test (RIDT) of the nasal wash was positive for influenza A virus, which was also isolated on viral culture. Oseltamivir (75 mg twice daily [BID] for 10 days) as well as empirical broad-spectrum antibiotic therapy was initiated. His symptoms improved within 7 days, and he was discharged to a rehabilitation facility. The patient was not retested for influenza virus at time of discharge.
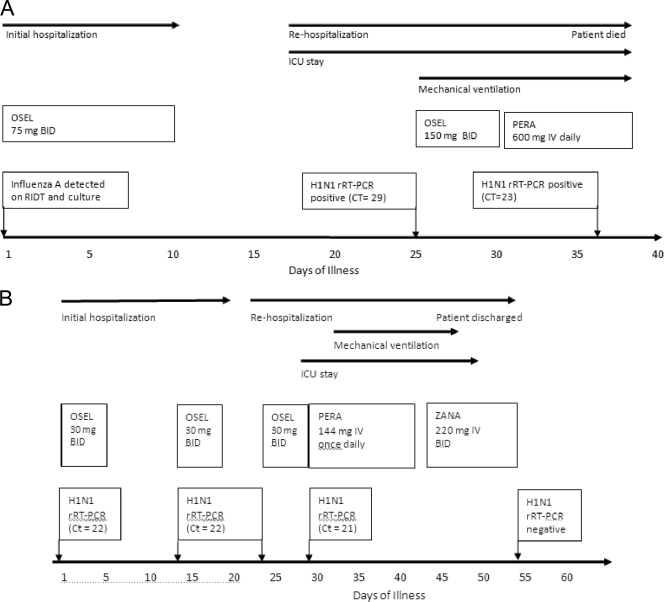
One week later he was admitted to the intensive care unit (ICU) with acute shortness of breath. A CT scan revealed interval improvement of the prior lung infiltrates. Empirical therapy with meropenem and vancomycin was begun for possible nosocomial lower respiratory tract infection. The methylprednisone dose was increased to 2 mg/kg/day for suspected worsening of bronchiolitis obliterans. On ICU day 8, he required mechanical ventilation. Bronchoscopy was performed; the BAL fluid tested positive for influenza A virus on RIDT, the novel 2009 H1N1 influenza virus (H1N1) was detected by real-time reverse transcriptase PCR (rRT-PCR), the cycle threshold (CT) was 29, and influenza A virus was isolated from viral culture. Therapy with high-dose oseltamavir (150 mg BID) was initiated, the steroid dose was tapered, and antibiotics were discontinued. Therapy was switched to intravenous (i.v.) peramivir on day 10. Aerosolized zanamivir could not be administered, as the patient was being mechanically ventilated; the i.v. formulation of zanamivir was not readily obtainable. On ICU day 22, bronchoscopy was repeated because of fever and blood-tinged purulent sputum. Numerous methicillin-resistant Staphylococcus aureus (MRSA) isolates were found in BAL fluid; i.v. vancomycin was initiated. The H1N1 rRT-PCR was positive (CT = 23), and influenza A virus was isolated from the BAL fluid. A rhinovirus PCR was positive in the BAL fluid 52 days after initial detection. The patient died after 24 days in the ICU (Fig. 1A).
FIG. 1.
Timelines of clinical course of H1N1 infection in patients 1 (A) and 2 (B).
Nasal wash collected at the time of hospitalization and two subsequent BAL fluid specimens were sent to the Centers for Disease Control and Prevention (CDC) for antiviral resistance testing. Partial sequence analysis of the neuraminidase (NA) gene determined by pyrosequencing revealed oseltamivir-susceptible H1N1 virus in the initial nasal washing, but the virus isolated from BAL fluid after 25 and 36 days of initial H1N1 diagnosis had the H275Y mutation, indicative of resistance to oseltamivir (11). Confirmatory results of oseltamivir resistance became available several days after the patient died.
Case report 2.
A 3-year-old female receiving chemotherapy for acute myelogenous leukemia was hospitalized in October 2009 for neutropenic fever. She was diagnosed with a central line-associated bloodstream infection. A nasal swab obtained on day 1 of her illness tested positive for influenza A virus by RIDT and for H1N1 virus by rRT-PCR (CT = 22). She was treated with i.v. antibiotics for the bacteremia and received 5 days of oseltamivir therapy (30 mg BID). Due to neutropenia and persistently positive H1N1 rRT-PCR (CT = 22) from a repeat nasal wash, another 5-day course of oseltamivir was given and the patient was discharged home. The patient was not retested for influenza virus at the time of discharge.
On day 23 of her illness, she was rehospitalized for neutropenic fever, cough, rhinorrhea, and worsening respiratory distress. An H1N1 rRT-PCR (CT = 22) from nasal wash remained positive. Therapy with oseltamivir together with empirical broad-spectrum antibiotics was initiated. She was transferred to the ICU on day 28 for worsening respiratory failure. Bronchoscopy was performed, and H1N1 virus was detected in the BAL fluid by rRT-PCR (CT = 21). Therapy with i.v. peramivir was begun at a dose of 12 mg/kg/day. Her respiratory failure worsened; she required intubation and high-frequency oscillatory ventilation. On day 43 of illness, therapy with i.v. zanamivir at a dose of 20 mg/kg every 12 h, obtained via Emergency Investigational New Drug (EIND) approval through the Food and Drug Administration, was started. The patient's respiratory status gradually improved, and she was extubated on day 47. Viral respiratory cultures and rRT-PCR for H1N1 virus obtained on day 53 of her illness were negative (Fig. 1B).
The CDC confirmed the presence of the H275Y mutation in the H1N1 virus isolated from the BAL fluid obtained on day 29 of illness, while the patient was on oseltamivir therapy.
HSCT recipients and cancer patients are at risk for severe disease caused by H1N1 virus and are targeted for treatment with either oseltamivir or zanamivir (3). So far, 1.2% of the H1N1 isolates tested at the CDC are resistant to oseltamivir (4).
The currently available rRT-PCR assays for H1N1 are qualitative and do not provide an estimate of the viral burden. In rRT-PCR, the crossing threshold is an empirically derived value that separates fluorescence due to amplification of the target from background fluorescence (9). The PCR cycle number or cycle threshold (CT), at which the fluorescence exceeds the crossing threshold, can be correlated with the number of copies of the target sequence present originally in the reaction mixture. Assays based on rRT-PCR usually have between 35 and 45 cycles of amplification. Samples that contain high copy numbers of target nucleic acid are detected sooner and will have low CT values (usually around 20). However, those with very low numbers of the target are detected later, with CT values above 30 (9).
Our two severely immunocompromised patients with H1N1 pneumonia had prolonged viral shedding despite oseltamivir therapy and subsequently developed resistance to the drug. Declining CT values in our adult patient indicated an increasing H1N1 viral load, suggestive of the emergence of resistance while on therapy. Similarly, oseltamivir resistance in the child was suggested by the persistently low CT values indicative of high viral loads despite oseltamivir therapy.
Antiviral resistance test results are not readily available to guide modification of therapy. The current rRT-PCR assays for H1N1 virus are not quantitative; however, the CT value of the rRT-PCR test may serve as a surrogate marker of viral load. Each three-cycle decrease in CT value is considered almost equal to a 10-fold increase in viral load (8).
Moreover, in our adult patient with multiple potential copathogens, namely, A. fumigatus, MRSA, M. avium complex, and rhinovirus, the increasing viral load as indicated by the declining CT value suggested that H1N1 virus was an important contributor to his respiratory failure. CT values may also be useful in guiding the duration of infection control measures in patients with prolonged viral shedding. However, respiratory secretions are not standardized and CT values in this context need further validation.
In immunocompetent patients treated with oseltamivir, peak shedding of H1N1 in respiratory secretions occurred with the onset of symptoms, and no virus was detected after 8 days and 5 days of symptom onset using rRT-PCR and viral culture, respectively (10). In contrast, our two cases highlight the prolonged shedding (36 and 29 days) of the H1N1 virus that can occur in severely immunocompromised patients despite oseltamivir therapy. Similarly, the CDC has reported two HSCT recipients treated with oseltamivir with prolonged shedding of H1N1 and subsequent emergence of oseltamivir resistance as indicated by the presence of the H275Y mutation (2). Since only partial sequencing of the NA gene was performed on our patient isolates, it is not known if any additional mutations were present.
If absorption of oral oseltamivir is compromised, i.v. peramivir, an NA inhibitor, is recommended for therapy (1). H1N1 isolates that are oseltamivir resistant due to the H275Y mutation have reduced in vitro susceptibility to peramavir as well (7). Zanamivir is the recommended drug for the treatment of oseltamivir-resistant H1N1 infection. If inhaled zanamavir is not tolerated or is contraindicated, as for patients on mechanical ventilation, then i.v. zanamavir may be obtainable through an EIND application (5, 6). The poor response to i.v. peramivir in our patients suggests that empirical therapy with zanamavir should be initiated if persistently low or declining CT values of the rRT-PCR test for H1N1 virus are seen while the patient is on oseltamivir therapy, pending results of antiviral resistance. Moreover, these two cases highlight the importance of having access to rapid antiviral susceptibility testing, as is the case for bacterial isolates, for the prompt detection of resistance and to guide the selection of appropriate antiviral therapy.
For H1N1 infection poorly responsive to therapy, clinicians often request serial rRT-PCR tests of respiratory secretions to determine clearance of the virus. CT values of rRT-PCR tests are not routinely made available to clinicians; it is therefore important for physicians to contact the laboratory to obtain these CT values. Low CT values indicate high viral load, and declining CT values suggest increasing viral load. Declining or persistently low CT values during therapy, as in our patients, correlated with clinical worsening and the development of oseltamivir resistance long before confirmatory resistance testing results were available. Likewise, rising CT values corresponding to clinical improvement of oseltamivir-resistant H1N1 infection treated with zanamivir have been reported (5, 6). Such data can also help discern the pathogenic role of H1N1 virus in immunosuppressed patients with multiple potential copathogens.
Acknowledgments
None of the authors have any conflict of interest.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Birnkrant, D., and E. Cox. 2009. The emergency use authorization of peramivir for treatment of 2009 H1N1 influenza. N. Engl. J. Med. 361(23):2204-2207. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2009. Oseltamavir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients—Seattle, Washington, 2009. MMWR Morb. Mortal. Wkly. Rep. 58(32):893-896. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 19 April 2010, accession date. Updated interim recommendations: special considerations for clinicians regarding 2009 H1N1 influenza in severely immunosuppressed patients. http://www.cdc.gov/h1n1flu/immunosuppression/index.htm.
- 4.Centers for Disease Control and Prevention. 19 April 2010, accession date. Seasonal influenza weekly report. http://www.cdc.gov/flu/weekly/.
- 5.Gaur, A. H., B. Bagga, S. Barman, R. Hayden, A. Lamptey, J. M. Hoffman, D. Bhojwani, P. M. Flynn, E. Tuomanen, and R. Webby. 2010. Intravenous zanamivir for oseltamivir-resistant 2009 H1N1 influenza. N. Engl. J. Med. 362(1):88-89. [DOI] [PubMed] [Google Scholar]
- 6.Kidd, I. M., J. Down, E. Nastouli, R. Shulman, P. R. Grant, D. C. Howell, and M. Singer. 2009. H1N1 pneumonitis treated with intravenous zanamivir. Lancet 374:1036. [DOI] [PubMed] [Google Scholar]
- 7.Okomo-Adhiambo, M., H. T. Nguyen, K. Sleeman, T. G. Sheu, V. M. Deyde, R. J. Garten, X. Xu, M. W. Shaw, A. I. Klimov, and L. V. Gubareva. 2010. Host cell selection of influenza neuraminidase variants: implications for drug resistance monitoring in A(H1N1) viruses. Antiviral Res. 85:381-388. [DOI] [PubMed] [Google Scholar]
- 8.Picard, C., M. Silvy, and J. Gabert. 2006. Overview of real-time RT-PCR strategies for quantification of gene rearrangements in myeloid malignancies, p. 27-68. In H. Iland, M. Hertzberg, and P. Marlton (ed.), Myeloid leukemia: methods and protocols. Humana Press, Totawa, NJ. [DOI] [PubMed]
- 9.Smith, C. J. 2005. Quantitative real-time PCR, p. 141-142. In M. A. Osborn and C. J. Smith (ed.), Molecular microbial ecology. Taylor & Francis Group, New York, NY.
- 10.To, K. K., K. H. Chan, I. W. Li, T. Y. Tsang, H. Tse, J. F. Chan, I. F. Hung, S. T. Lai, C. W. Leung, Y. W. Kwan, Y. L. Lau, T. K. Ng, V. C. Cheng, J. S. Peiris, and K. Y. Yuen. 2010. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J. Med. Virol. 82(1):1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. 19 April 2010, accession date. CDC pyrosequencing assay to detect H275Y mutation in the neuraminidase of novel A (H1N1) viruses. http://www.who.int/csr/resources/publications/swineflu/NA_DetailedPyrosequencing_20090513.pdf.