Abstract
Candida glabrata has emerged as the second most common etiologic agent, after Candida albicans, of superficial and invasive candidiasis in adults. Strain typing is essential for epidemiological investigation, but easy-to-use and reliable typing methods are still lacking. We report the use of a multilocus microsatellite typing method with a set of eight markers on a panel of 180 strains, including 136 blood isolates from hospitalized patients and 34 digestive tract isolates from nonhospitalized patients. A total of 44 different alleles were observed, generating 87 distinct genotypes. In addition to perfect reproducibility, typing ability, and stability, the method had a discriminatory power calculated at 0.97 when all 8 markers were associated, making it suitable for tracing strains. In addition, it is shown that digestive tract isolates differed from blood culture isolates by exhibiting a higher genotypic diversity associated with different allelic frequencies and preferentially did not group in clonal complexes (CCs). The demonstration of the occurrence of microevolution in digestive strains supports the idea that C. glabrata can be a persistent commensal of the human gut.
During the last decade, incidence of invasive candidiasis with non-albicans Candida species has increased (40, 46, 48). Candida glabrata is the most significant species that has emerged and now regularly ranks number two, after C. albicans, as the etiologic agent of superficial and invasive candidiasis occurring in adults (17, 27). Reasons for this change in species distribution remain uncertain but may be partially due to the natural resistance of C. glabrata to azole derivates, widely used since the 1980s (6, 10, 26). This lower susceptibility of C. glabrata to azoles (2, 23, 38) and delayed initiation of therapy due to delayed diagnosis (2, 20, 35) may explain in part why the prognosis for C. glabrata candidemia is worse than that for C. albicans candidemia (7, 18, 41, 51).
Currently, despite increasing clinical concern, the epidemiology of C. glabrata remains poorly known compared to that of C. albicans. While C. glabrata is considered a commensal of the human digestive tract, its natural reservoir is still uncertain. Possible transmission between patients has been suggested (43), and clusters of invasive infections have been reported (4, 36). An efficient and easy-to-use molecular typing method which would allow tracing strains would also allow better understanding of the spread of this species, notably in a hospital context. Up to now, the most reliable method for C. glabrata typing was Southern blotting with moderately repeated probes (3, 32). However, this method requires the use of radioactive elements, and the sequences of the probes are not available. Sequence-based methods such as multilocus sequence typing (MLST) and multilocus microsatellite analysis have been recently proposed (15, 19, 21), but in all cases one could hope for better discriminatory power.
A microsatellite-based typing method using a new set of eight markers has been set up for population genetic analysis of C. glabrata (14). In this work, we positively evaluate this method, which has a discriminatory power calculated at 0.97, for tracing C. glabrata strains. The results of typing a large panel of both blood culture and digestive tract isolates suggest (i) that C. glabrata can undergo microevolution within the gut, supporting a persistent commensal life cycle of C. glabrata, and (ii) that digestive and bloodstream isolates exhibit genetic diversity preferentially induced by polymorphism in some loci.
MATERIALS AND METHODS
Strains.
A panel of 180 strains was used for this study (see Table S1 at http://gepamy.free.fr/GEPAMY/Microsatellite_data.html). Clinical isolates were collected either from blood or digestive tract samples. All bloodstream isolates (n = 136) were collected during a prospective multicentric European study on nosocomial candidemia (47). Each strain was collected from a different patient. Digestive strains (n = 34) were collected from nonhospitalized patients. Five isolates were collected during a study comparing fecal yeast loads and species distributions in patients with proven viral or bacterial diarrhea with those in healthy subjects (41a). Twenty-six isolates were collected in the framework of a case control study analyzing the correlation between Candida digestive carriage, detected from mouth swab and stool sample cultures, and the presence of anti-Saccharomyces cerevisiae serum antibodies associated with Crohn's disease (CD) in subjects from northern Belgium and northeastern France (44). Three digestive tract isolates came from strain collections. Twenty-four digestive tract isolates were considered unrelated. In addition to these clinical strains, 10 strains originating from different countries or continents, either environmental or isolated from human beings, were typed in order to calculate the discriminatory power of our method. All 180 strains had their identification as C. glabrata confirmed using the commercial panel ID 32C test (bioMérieux, Marcy l'Etoile, France). A monoclonal subculture of each was stored at −80°C before testing.
Microsatellite typing.
All DNAs were extracted by a rapid method using a Chelex solution containing resin and heat shock as previously described (24). Molecular typing was done using a method previously described by some of us for a population genetic study of C. glabrata (14). It relies on length polymorphism analysis of 8 microsatellite markers found within the C. glabrata genome (http://cbi.labri.fr/Genolevures/elt/CAGL) using the Tandem Repeat Finder software from Gary Benson (8). Amplification with primers pairs previously described (14) was done using forward primers 5′ fluorolabeled with different dyes allowing sizing on the automated sequencer. PCR amplifications were performed in 15-μl reaction volumes containing 1× PCR buffer, 2.5 mM MgCl2, 0.5 μM each primer (Eurogentec, Liege, Belgium), 0.25 mM each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP) (BioLabs Inc.), and 0.6 U Taq Gold polymerase (Eurogentec, Liege, Belgium). The amplification program was performed as follows: 7 min at 94°C; 3 cycles of 30 s at 94°C, 30 s at 69°C, and 30 s at 72°C; 3 cycles of 30 s at 94°C, 30 s at 65°C, and 30 s at 72°C; 3 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C; 28 cycles of 30 s at 55°C and 30 s at 72°C; and a final extension of 15 min at 72°C. For each isolate, fluorolabeled amplified fragments were pooled and added to a molecular marker (Genescan HD400Rox; Applied Biosystems), enabling the determination of the fragment sizes after a run in an ABI 377 automatic sequencer (Applied Biosystems). Length polymorphism was evaluated by comparison with type strain CBS 138 (ATCC 2001), which was tested for each PCR and length analysis run.
Evaluation of the method.
Primer specificity was checked by testing closely related species belonging to the Nakaseomyces clade, namely, Candida nivariensis (CBS 9983), Candida bracarensis (CBS 10154), and Nakaseomyces (Kluyveromyces) delphensis (CBS 2170). To test genotype stability, we assessed the eight markers in the type strain CBS 138 (ATCC 2001) over 960 generations. Briefly, a culture was inoculated with a single clone, and every 3 days an aliquot was taken for DNA extraction and for reinoculation into fresh medium. This was performed 20 times successively. To test reproducibility, the type strain was tested for each PCR and length analysis run.
As recommended by the European Society of Clinical Microbiology and Infectious Diseases Study Group for Epidemiological Markers (ESGEM) guidelines (50), a subset of 50 unrelated strains (time of isolation or geographic origin) was selected for discriminatory power calculation, including the type strain CBS 138 (ATCC 2001) (see Table S1 at http://gepamy.free.fr/GEPAMY/Microsatellite_data.html). Isolates were collected from different patients in different hospitals and/or at different periods (difference between dates of isolation exceeded 1 year) or isolated from animals or plants. Forty isolates came from blood cultures. Discriminatory power was calculated using the Simpson index of diversity (D), given by the equation D = 1 − 1/[N(N − 1)]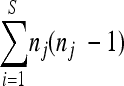 , where N is the number of strains tested, S is the number of different types, and nj is the number of strains exhibiting the j type (28). The value of D is between 0 and 1 and tends to 1 as the discriminatory power rises. In order to see whether scoring more loci was associated with increasing discriminatory power or whether one has reached a plateau, the number of different genotypes and the Simpson index were calculated from all combinations of 1 to 8 loci from the data set.
, where N is the number of strains tested, S is the number of different types, and nj is the number of strains exhibiting the j type (28). The value of D is between 0 and 1 and tends to 1 as the discriminatory power rises. In order to see whether scoring more loci was associated with increasing discriminatory power or whether one has reached a plateau, the number of different genotypes and the Simpson index were calculated from all combinations of 1 to 8 loci from the data set.
Genetic analysis.
Multilocus V1.3B software was used to describe the genetic diversity within the two groups of strains (i.e., collected from the digestive tract and blood): the number of different genotypes, frequency of the most prevalent genotype, and genotypic diversity, defined as the probability that two individuals taken at random have different genotypes. The last is defined as [n/(n − 1)](1 − 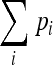 , where pi is the frequency of the ith genotype and n is the number of strains sampled (1). The value tends to 1 as the genotypic diversity increases. The software was also used to generate expected diversities by randomly shuffling (100 randomizations), independently for each locus, the alleles among the 24 unrelated digestive strains and a panel of 24 randomly chosen bloodstream strains.
, where pi is the frequency of the ith genotype and n is the number of strains sampled (1). The value tends to 1 as the genotypic diversity increases. The software was also used to generate expected diversities by randomly shuffling (100 randomizations), independently for each locus, the alleles among the 24 unrelated digestive strains and a panel of 24 randomly chosen bloodstream strains.
In order to describe the relationships among isolates at the microevolutionary level, we performed allelic profile-based comparisons using a minimum spanning tree (MSTree) analysis with BioNumerics v5.10 software (Applied-Maths, Sint Maartens-Latem, Belgium). MSTree analysis links profiles so that the sum of the distances (number of distinct alleles between two sequence types [STs]) is minimized (42). With this graphical representation, strains sharing the same allelic profile fall into the same circle, whose size is proportional to the number of strains with the profile. Founder genotypes, previously described (14) as any genotypes shared by at least 4 strains, were considered as such even if in the present study they do not cluster 4 strains. Single-locus variants (SLVs) were defined as genotypes with a single locus difference with the considered founder genotype. Clonal complexes (CCs) were defined as groups of strains including a founder genotype and its corresponding SLVs. When an SLV included more than four strains, its own SLVs, defined as successive SLVs, were included in the CC.
A dendrogram was also generated by the unweighted pair group method of average linkage (UPGMA) using the Phylip software package (version 3.69).
RESULTS
In order to validate this microsatellite-based typing method (14) as a reliable tool for tracing strains, we first evaluated its specificity, stability across generations, reproducibility, and discriminatory power based on the ESGEM guidelines (50). Considering the positive results, a molecular investigation of C. glabrata digestive tract colonization was initiated.
Evaluation of the method.
The primer pairs designed to amplify the 8 selected loci appeared specific for C. glabrata since no amplification was observed for C. bracarensis and Nakaseomyces (Kluyveromyces) delphensis and only marker 9 gave a positive PCR for C. nivariensis. In contrast, DNA from all C. glabrata strains tested was successfully amplified with all markers, giving a unique PCR product. Length changes associated with insertion/deletion within the flanking regions of the microsatellite loci were not observed after several alleles of each locus were sequenced (data not shown). Genomic stability of the tested regions was confirmed by demonstrating the lack of size modification in any of the assayed microsatellite markers over 960 generations of the type strain CBS 138. A perfect reproducibility was found, with identical results obtained for type strain CBS 138 across the runs.
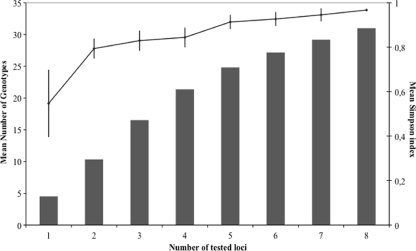
On the panel of 180 clinical strains, each marker appeared polymorphic, with 3, 4, 5, and 9 alleles for marker 2bis; markers 5 and 9; markers 2, 3, and 4; and markers 6 and 8, respectively. A total of 44 different alleles were thus observed, the combination of which generated 87 distinct genotypes in our strain collection. The discriminatory power, calculated using Simpson's index, for a subset of 50 unrelated strains reached 0.95 for a combination of 7 markers, and it was 0.97 for a combination of 8 markers (Fig. 1).
FIG. 1.
Discriminatory power, calculated with Simpson's index of diversity, and number of genotypes observed according to the various combinations of microsatellite loci. The line indicates the 0.95 threshold recommended by European Study Group on Epidemiological Markers guidelines (50).
Population structure.
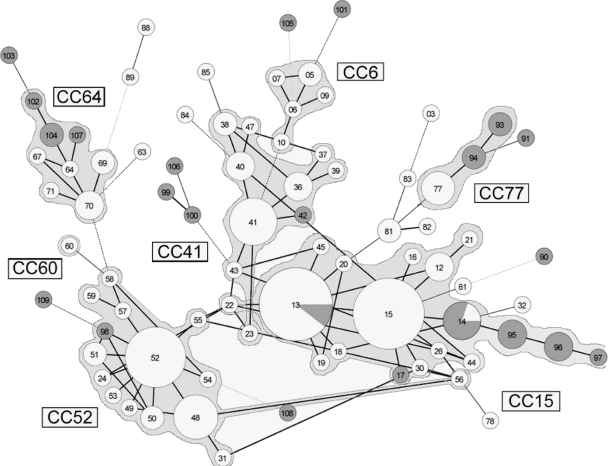
For the population structure study, we performed a MSTree analysis and generated a UPGMA tree based on genetic distances. Congruence between these two analyses was observed (Fig. 2; see Fig. S1 at http://gepamy.free.fr/GEPAMY/Microsatellite_data.html). To perform our analysis under more-stringent conditions, we reviewed the definition of clonal complexes in MSTree. Indeed, we used the high concordance observed between clades, defined using the MLST method described by Dodgson et al. (15, 16), and CCs (Table 1). Interestingly, CC64, which was not associated with a given MLST clade, corresponds in a recent paper by Lott et al. (33) to an MLST group clustering as many as 18.3% of the tested strains, supported by a 99% bootstrap value (100% in the Dodgson study), each with a synapomorphic NMT1 allele (NMT1-5). Conversely, we considered genotype RT60 to be included in a potential CC since the single strain we had exhibited allele 12 for the NMT1 locus (data not shown), characteristic of clade VII (15, 31, 33).
FIG. 2.
Minimum spanning tree analysis of 170 C. glabrata strains based on allelic profiles at eight microsatellite loci. Each circle corresponds to a repeat type, the number of which is indicated inside the circle. The lines between circles indicate the similarity between profiles (bold, seven alleles in common; normal, six alleles; dotted, five alleles). Gray parts of circles indicate proportions of genotypes that contain digestive isolates. Cross-links corresponding to a single allelic mismatch are shown.
TABLE 1.
Correlation between subpopulations defined with an MLST scheme and the microsatellite typing method
| MLST cladea | MLST sequence type(s) | NMT1 allele by MLST | Microsatellite-based clonal complex |
|---|---|---|---|
| I | 3, 5 | 8 | 15 |
| II | 2, 8, 9, 11, 12 | 2 | 6 |
| IIIA | 18, 26, 79 | 3 | 41 |
| IIIB | 10, 15, 29, 80 | 3 | 41 |
| IV | 7, 30 | 4 | 77 |
| V | 6, 14 | 7 | 41 |
| VI | 16 | 11 | None (RT72, -73, -75) |
| VII | 36, 76 | 12 | None (RT60) |
| None* | 13, 19, 81 | 5 | 64 |
| None* | 22 | 6 | 52 |
Based on these data, we considered as forming a CC either founder genotypes or genotypes corresponding to an MLST clade and their related genotypes differing by single-locus variation. Seven distinct groups were thus delineated: CC52, CC15, CC64, CC6, CC77, CC41, and RT60 (Fig. 2). Two strains were assigned to CCs based on their NMT1 alleles (RT22 to CC15 and RT56 to CC52). No new CC was defined using this new data set, but diversity within CC52, CC64, CC15, and CC77 increased because some new repeat types represented SLVs: RT95, RT96, and RT97 represented successive SLVs of RT14 (CC15); RT98 represented an SLV of RT52 (CC52); RT94 and RT93 represented successive SLVs of RT77 (CC77); RT107 represented an SLV of RT64 (CC64); and RT104 and RT102 represented successive SLVs of RT64.
The inclusion of new unrelated strains thus supports the existence of the subpopulations defined as CCs and extends CC scope. Indeed 147 out of 170 (86.5%) clinical strains were included in CCs. Repartition values for strains from this European panel within the CCs were 36.5%, 20.6%, 12.9%, 7.7%, 4.7%, 3.5%, and 0.6% for CC15, CC52, CC41, CC64, CC77, CC6, and RT60, respectively. Considering the geographic origin of isolates, no difference in repeat type frequencies between countries or between northern and southern Europe (geographical limit at 46° north latitude) was observed. Since precise isolation time was not available for the digestive tract isolates collected in the framework of the Crohn's disease study and the period of isolation was short overall (1997 to 2003), time of isolation was not taken in consideration for the analysis.
Molecular investigation of C. glabrata digestive tract colonization.
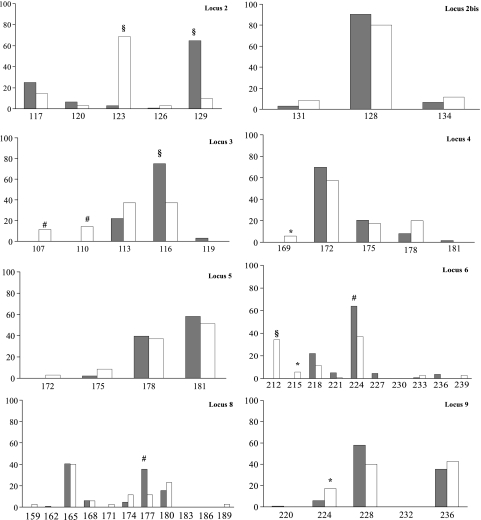
Microsatellite typing of the 34 available digestive tract isolates allowed the identification of 10 new alleles on 5 of the eight loci (locus 3, alleles 107 and 110; locus 4, allele 169; locus 5, allele 172; locus 6, alleles 212, 215, and 239; locus 8, alleles 159, 171, and 189) and 19 new genotypes (RT90 to RT108) in addition to the 41 alleles and 90 genotypes previously described (14) (see Table S1 at http://gepamy.free.fr/GEPAMY/Microsatellite_data.html). The MSTree's topology does not show any clustering of digestive tract strains, compared to bloodstream strains. By our current definition of CC, 6 of the 24 (25%) unrelated digestive tract strains (strains 154 [RT90], 151 [RT91], 162 [RT101], 173 [RT105], 174 [RT106], and 177 [RT108]) and 13 strains among the 136 (9.6%) bloodstream isolates (strains 60 [RT78], 137 [RT32], 138 [RT 61], 16 and 97 [RT81], 11 [RT82], 118 [RT83], 121 [RT3], 47 [RT84], 79 [RT85], 99 [RT63], 139 [RT89], and 50 [RT88]) did not group in any CC (Fisher's exact test; P < 0.05). In addition there was a higher number of strains with a unique genotype within the panel of digestive tract strains (16 among the unrelated 24) than among the isolates collected from blood culture (42 among 136) (Fisher's exact test; P < 0.002). Allelic frequencies varied according to the origin (bloodstream or digestive tract) of the strains for six of the eight microsatellite loci (loci 2, 3, 4, 6, 8, and 9) (Fig. 3). Genetic diversity was higher for the panel of 24 unrelated digestive tract strains (37 alleles generating 19 genotypes; genotypic diversity, 97.5%) than for a panel of 24 randomly selected unrelated bloodstream strains (27 alleles generating 15 genotypes; genotypic diversity, 93.1%). This was confirmed by comparing means of expected diversities estimated for both groups by randomly shuffling the alleles among strains (100 randomizations) independently for each locus (1-tailed Student t test; P < 0.01).
FIG. 3.
Allelic frequencies within each of the eight microsatellite loci according to the origin of the isolates. Significant differences between blood culture (gray bars) and digestive tract (white bars) isolates by Fisher's exact test were observed (§, P < 0.001; #, P < 0.01; *, P < 0.05).
Among the six subjects from the CD study who had isolates collected from 2 different digestive tract sites (mouth and stool), one had isolates with identical genotype (isolates 147 and 148 [RT104]), three had isolates that differed by a single locus (isolates 155 [RT97] and 156 (RT96), isolates 169 [RT99] and 171 [RT100], and, isolates 167 [RT14] and 168 [RT95]), one had isolates which differed by two loci (isolates 160 [RT102] and 161 [RT103]), and another one had isolates which differed by three loci (isolates 170 [RT109] and 172 [RT98]). Among the four families for which isolates had been collected from different members, in one case isolates differed only by a single allele (isolates 149 [RT 93] and 150 [RT94]) and in the other cases, isolates did not share more than 5 alleles (see Table S1 at http://gepamy.free.fr/GEPAMY/Microsatellite_data.html).
DISCUSSION
C. glabrata has emerged as a serious cause of morbidity and mortality in patients with severe underlying disease such as cancer, in patients who have had recent surgery, and in patients admitted to intensive care units (27, 34, 47, 48). A reliable and discriminatory molecular typing method is urgently needed for investigating the spread of strains, notably in a hospital context. During the last decade, molecular typing methods have significantly evolved, offering the possibility of sequence analysis. MLST has emerged as a highly reliable and discriminatory method both for tracing strains and for establishing population structure in a number of Candida species (e.g., C. albicans, C. tropicalis, C. parapsilosis, and C. krusei) (13, 29, 45, 49). However, perhaps because of the highly clonal mode of reproduction of C. glabrata, MLST appears insufficient for tracing strains, despite allowing the definition of broad subpopulations (15, 31, 33). Microsatellite length polymorphism analysis represents an alternative method, which has been successfully applied to type pathogenic yeasts (5, 9, 11, 25). In two previous studies, microsatellite markers were used to differentiate strains of C. glabrata (19, 21). Levels of discriminatory power were 0.84 and 0.902, less than the 0.95 threshold generally considered for a typing method to be useful for epidemiological purposes (50), possibly due to the low number of loci tested (3 and 6, respectively). The current method has previously been used to analyze genetic population structure but has not been evaluated for tracing strains. Here, discriminatory power was calculated at 0.97. Typing ability and stability, two other important criteria for typing methods (50), were maximal. To our knowledge, this method is the first fully standardized, easy-to-implement method reaching such a level of discrimination to differentiate C. glabrata strains. Given its high reproducibility and portability, the method will allow comparison of strains and profiles from different studies through the MLVA-NET website (www.pasteur.fr/mlva) (22).
Application of the method to describe the biodiversity and genetic relationship of clinical isolates collected from the digestive tract gives new insights on the interaction between the human digestive tract and C. glabrata. As previously reported for C. albicans (12), the fact that isolates recovered from the same subject may have genotypes differing only by one allele is suggestive of microevolution within the gut. This supports the commonly accepted, but to our knowledge never demonstrated, idea that C. glabrata can be a persistent commensal of the human gut. Analyzing more sequential isolates over time would be necessary to definitively prove this hypothesis. The hypothesis is in opposition to the idea of transient digestive tract carriage, which could be due to ingestion of contaminated food, for example, a feature commonly found with other fungi like S. cerevisiae and Geotrichum candidum (30, 39). Demonstration of isolates with genotypes differing by only one allele in different members of a family also suggests intrafamilial transmission with further microevolution, as previously demonstrated for C. albicans (12). This is further supported by the fact that, in these cases, the considered genotypes (RT93 and RT94) are infrequent.
Some observations point to differences between strains during digestive tract carriage and the pathogenic process: (i) differences in allelic frequencies, (ii) higher proportions of digestive tract isolates corresponding to unique genotypes, and (iii) because of these higher proportions, higher proportions of digestive tract isolates than of bloodstream isolates which did not group in CCs. These data give rise to a number of comments. One hypothesis could be that the strains circulating within hospital centers differ from those found in the community. Previous work has shown a lack of influence of the city where the strains had been isolated, given that in most cases there was a single institution per city (14). However, endemicity of strains with peculiar traits allowing their maintenance in favorable conditions or due to selective pressure within university centers cannot be ruled out. To our knowledge, a genotypic comparative survey has never been performed either for C. glabrata or C. albicans, and a specific survey focused on the possible distinction of community versus nosocomial strains is warranted. The fact that most often genotypes of digestive tract isolates did not group in any CC raises the question of their propensity for epidemiologic spread and capacity to adapt to the bloodstream conditions. Correlation between genotypes and isolation at particular body sites has been suggested for C. albicans by Odds et al. (37), who showed significant differences in the proportions of isolates from blood, commensal carriage, and superficial infections among the five most important clades. However, we are currently unable to make a hypothesis to link these observations and the function of the chromosomal regions where our microsatellites are located. Thus, our data suggest that the genetic evolution of digestive tract isolates may be different, both in terms of diversity and loci responsible for this polymorphism, from that of isolates collected in pathogenic conditions. Further basic work to correlate these observations with virulence is warranted.
In summary, our results support the usefulness of the herein-developed molecular typing method for tracing C. glabrata strains. Analysis of digestive tract isolates supports the idea that C. glabrata is a persistent colonizer of the human digestive tract, where it appears to undergo microevolution. As for C. albicans (12), the digestive tract could be a reservoir from which intrafamilial transmission can occur. Further studies should allow completion and exploitation of these preliminary data on genetic diversity and evolution of C. glabrata in the context of the human digestive tract.
Acknowledgments
This work was supported in part by ACI no. MIC0314 and GDR 2354 from the CNRS. The platform Genotyping of Pathogens and Public Health receives financial support from Institut Pasteur and the Institut de Veille Sanitaire.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Agapov, P., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1:101-102. [Google Scholar]
- 2.Arendrup, M. C., K. Fuursted, B. Gahrn-Hansen, H. C. Schonheyder, J. D. Knudsen, I. M. Jensen, B. Bruun, J. J. Christensen, and H. K. Johansen. 2008. Semi-national surveillance of fungaemia in Denmark 2004-2006: increasing incidence of fungaemia and numbers of isolates with reduced azole susceptibility. Clin. Microbiol. Infect. 14:487-494. [DOI] [PubMed] [Google Scholar]
- 3.Arif, S., T. Barkham, E. G. Power, and S. A. Howell. 1996. Techniques for investigation of an apparent outbreak of infections with Candida glabrata. J. Clin. Microbiol. 34:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asmundsdottir, L. R., H. Erlendsdottir, G. Haraldsson, H. Guo, J. Xu, and M. Gottfredsson. 2008. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin. Infect. Dis. 47:e17-e24. [DOI] [PubMed] [Google Scholar]
- 5.Bart-Delabesse, E., J. Sarfati, J. P. Debeaupuis, W. van Leeuwen, A. van Belkum, S. Bretagne, and J. P. Latge. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassetti, M., E. Righi, A. Costa, R. Fasce, M. P. Molinari, R. Rosso, F. B. Pallavicini, and C. Viscoli. 2006. Epidemiological trends in nosocomial candidemia in intensive care. BMC Infect. Dis. 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Abraham, R., N. Keller, N. Teodorovitch, A. Barzilai, R. Harel, Z. Barzilay, and G. Paret. 2004. Predictors of adverse outcome from candidal infection in a tertiary care hospital. J. Infect. 49:317-323. [DOI] [PubMed] [Google Scholar]
- 8.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beretta, S., J. P. Fulgencio, A. Enache-Angoulvant, C. Bernard, S. El Metaoua, T. Ancelle, M. Denis, and C. Hennequin. 2006. Application of microsatellite typing for the investigation of a cluster of cases of Candida albicans candidaemia. Clin. Microbiol. Infect. 12:674-676. [DOI] [PubMed] [Google Scholar]
- 10.Blot, S., R. Janssens, G. Claeys, E. Hoste, F. Buyle, J. J. De Waele, R. Peleman, D. Vogelaers, and K. Vandewoude. 2006. Effect of fluconazole consumption on long-term trends in candidal ecology. J. Antimicrob. Chemother. 58:474-477. [DOI] [PubMed] [Google Scholar]
- 11.Botterel, F., C. Desterke, C. Costa, and S. Bretagne. 2001. Analysis of microsatellite markers of Candida albicans used for rapid typing. J. Clin. Microbiol. 39:4076-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bougnoux, M. E., D. Diogo, N. Francois, B. Sendid, S. Veirmeire, J. F. Colombel, C. Bouchier, H. Van Kruiningen, C. d'Enfert, and D. Poulain. 2006. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J. Clin. Microbiol. 44:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bougnoux, M. E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brisse, S., C. Pannier, A. Angoulvant, T. de Meeus, L. Diancourt, O. Faure, H. Muller, J. Peman, M. A. Viviani, R. Grillot, B. Dujon, C. Fairhead, and C. Hennequin. 2009. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell 8:287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodgson, A. R., C. Pujol, D. W. Denning, D. R. Soll, and A. J. Fox. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 41:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodgson, A. R., C. Pujol, M. A. Pfaller, D. W. Denning, and D. R. Soll. 2005. Evidence for recombination in Candida glabrata. Fungal Genet. Biol. 42:233-243. [DOI] [PubMed] [Google Scholar]
- 17.Dongari-Bagtzoglou, A., P. Dwivedi, E. Ioannidou, M. Shaqman, D. Hull, and J. Burleson. 2009. Oral Candida infection and colonization in solid organ transplant recipients. Oral Microbiol. Immunol. 24:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fidel, P. L., Jr., J. A. Vazquez, and J. D. Sobel. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foulet, F., N. Nicolas, O. Eloy, F. Botterel, J. C. Gantier, J. M. Costa, and S. Bretagne. 2005. Microsatellite marker analysis as a typing system for Candida glabrata. J. Clin. Microbiol. 43:4574-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricker-Hidalgo, H., B. Lebeau, H. Pelloux, and R. Grillot. 2004. Use of the BACTEC 9240 system with Mycosis-IC/F blood culture bottles for detection of fungemia. J. Clin. Microbiol. 42:1855-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grenouillet, F., L. Millon, J. M. Bart, S. Roussel, I. Biot, E. Didier, A. S. Ong, and R. Piarroux. 2007. Multiple-locus variable-number tandem-repeat analysis for rapid typing of Candida glabrata. J. Clin. Microbiol. 45:3781-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guigon, G., J. Cheval, R. Cahuzac, and S. Brisse. 2008. MLVA-NET—a standardised web database for bacterial genotyping and surveillance. Euro Surveill. 13(19):pii=18863. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=18863. [PubMed] [Google Scholar]
- 23.Hachem, R., H. Hanna, D. Kontoyiannis, Y. Jiang, and I. Raad. 2008. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 112:2493-2499. [DOI] [PubMed] [Google Scholar]
- 24.Hennequin, C., E. Abachin, F. Symoens, V. Lavarde, G. Reboux, N. Nolard, and P. Berche. 1999. Identification of Fusarium species involved in human infections by 28S rRNA gene sequencing. J. Clin. Microbiol. 37:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hennequin, C., A. Thierry, G. F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hope, W., A. Morton, and D. P. Eisen. 2002. Increase in prevalence of nosocomial non-Candida albicans candidaemia and the association of Candida krusei with fluconazole use. J. Hosp. Infect. 50:56-65. [DOI] [PubMed] [Google Scholar]
- 27.Horn, D. L., D. Neofytos, E. J. Anaissie, J. A. Fishman, W. J. Steinbach, A. J. Olyaei, K. A. Marr, M. A. Pfaller, C. H. Chang, and K. M. Webster. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48:1695-1703. [DOI] [PubMed] [Google Scholar]
- 28.Hunter, P. R. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen, M. D., N. A. Gow, M. C. Maiden, D. J. Shaw, and F. C. Odds. 2007. Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J. Clin. Microbiol. 45:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacques, N., and S. Casaregola. 2008. Safety assessment of dairy microorganisms: the hemiascomycetous yeasts. Int. J. Food Microbiol. 126:321-326. [DOI] [PubMed] [Google Scholar]
- 31.Lin, C. Y., Y. C. Chen, H. J. Lo, K. W. Chen, and S. Y. Li. 2007. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 45:2452-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockhart, S. R., S. Joly, C. Pujol, J. D. Sobel, M. A. Pfaller, and D. R. Soll. 1997. Development and verification of fingerprinting probes for Candida glabrata. Microbiology 143(Pt. 12):3733-3746. [DOI] [PubMed] [Google Scholar]
- 33.Lott, T. J., J. P. Frade, and S. R. Lockhart. 2010. Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. surveillance studies. Eukaryot. Cell 9:619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malani, A., J. Hmoud, L. Chiu, P. L. Carver, A. Bielaczyc, and C. A. Kauffman. 2005. Candida glabrata fungemia: experience in a tertiary care center. Clin. Infect. Dis. 41:975-981. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, M. H., V. Letscher-Bru, B. Jaulhac, J. Waller, and E. Candolfi. 2004. Comparison of Mycosis IC/F and Plus Aerobic/F media for diagnosis of fungemia by the Bactec 9240 system. J. Clin. Microbiol. 42:773-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nedret Koc, A., S. Kocagoz, F. Erdem, and Z. Gunduz. 2002. Outbreak of nosocomial fungemia caused by Candida glabrata. Mycoses 45:470-475. [DOI] [PubMed] [Google Scholar]
- 37.Odds, F. C., M. E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S. Y. Li, A. Tavanti, M. C. Maiden, N. A. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Geographic variation in the susceptibilities of invasive isolates of Candida glabrata to seven systemically active antifungal agents: a global assessment from the ARTEMIS Antifungal Surveillance Program conducted in 2001 and 2002. J. Clin. Microbiol. 42:3142-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pottier, I., S. Gente, J. P. Vernoux, and M. Gueguen. 2008. Safety assessment of dairy microorganisms: Geotrichum candidum. Int. J. Food Microbiol. 126:327-332. [DOI] [PubMed] [Google Scholar]
- 40.Presterl, E., F. Daxbock, W. Graninger, and B. Willinger. 2007. Changing pattern of candidaemia 2001-2006 and use of antifungal therapy at the University Hospital of Vienna, Austria. Clin. Microbiol. Infect. 13:1072-1076. [DOI] [PubMed] [Google Scholar]
- 41.Rennert, G., H. S. Rennert, S. Pitlik, R. Finkelstein, and R. Kitzes-Cohen. 2000. Epidemiology of candidemia—a nationwide survey in Israel. Infection 28:26-29. [DOI] [PubMed] [Google Scholar]
- 41a.Rimek, D., and R. Kappe. 2007. Fecal yeast loads in patients with bacterial and viral diarrhea and the impact of soluble fecal substances on the growth of Candida, poster P116. Abstr. 3rd Trends in Medical Mycology, Turin, Italy, 28 to 31 October, 2007.
- 42.Schouls, L. M., H. G. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwab, U., F. Chernomas, L. Larcom, and J. Weems. 1997. Molecular typing and fluconazole susceptibility of urinary Candida glabrata isolates from hospitalized patients. Diagn. Microbiol. Infect. Dis. 29:11-17. [DOI] [PubMed] [Google Scholar]
- 44.Standaert-Vitse, A., B. Sendid, M. Joossens, N. Francois, P. Vandewalle-El Khoury, J. Branche, H. Van Kruiningen, T. Jouault, P. Rutgeerts, C. Gower-Rousseau, C. Libersa, C. Neut, F. Broly, M. Chamaillard, S. Vermeire, D. Poulain, and J. F. Colombel. 2009. Candida albicans colonization and ASCA in familial Crohn's disease. Am. J. Gastroenterol. 104:1745-1753. [DOI] [PubMed] [Google Scholar]
- 45.Tavanti, A., A. D. Davidson, E. M. Johnson, M. C. Maiden, D. J. Shaw, N. A. Gow, and F. C. Odds. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tortorano, A. M., C. Kibbler, J. Peman, H. Bernhardt, L. Klingspor, and R. Grillot. 2006. Candidaemia in Europe: epidemiology and resistance. Int. J. Antimicrob. Agents 27:359-366. [DOI] [PubMed] [Google Scholar]
- 47.Tortorano, A. M., J. Peman, H. Bernhardt, L. Klingspor, C. C. Kibbler, O. Faure, E. Biraghi, E. Canton, K. Zimmermann, S. Seaton, and R. Grillot. 2004. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 23:317-322. [DOI] [PubMed] [Google Scholar]
- 48.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 49.van Asbeck, E. C., K. V. Clemons, A. N. Markham, and D. A. Stevens. 2009. Correlation of restriction fragment length polymorphism genotyping with internal transcribed spacer sequence, randomly amplified polymorphic DNA and multilocus sequence groupings for Candida parapsilosis. Mycoses 52:493-498. [DOI] [PubMed] [Google Scholar]
- 50.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1-46. [DOI] [PubMed] [Google Scholar]
- 51.Velasco, E., and R. Bigni. 2008. A prospective cohort study evaluating the prognostic impact of clinical characteristics and comorbid conditions of hospitalized adult and pediatric cancer patients with candidemia. Eur. J. Clin. Microbiol. Infect. Dis. 27:1071-1078. [DOI] [PubMed] [Google Scholar]