Abstract
The genotype of the hepatitis C virus (HCV) is essential for determining treatment duration in clinical practice and for epidemiological and clinical studies. Currently, few genotyping assays that determine the HCV subtype are available. This report describes a microarray-based molecular technique for identifying the HCV genotype and subtype. It uses low-density hydrogel-based biochips containing genotype- and subtype-specific oligonucleotides based on the sequences of the NS5B region of the HCV genome. The biochip contains 120 oligonucleotides that identify genotypes 1 to 6 and 36 (1a, 1b, 1c, 1d, 1e, 2a, 2b, 2c, 2d, 2i, 2j, 2k, 2l, 2m, 3a, 3b, 3k, 4a, 4c, 4d, 4f, 4h, 4i, 4k, 4n, 4o, 4p, 4r, 4t, 5a, 6a, 6b, 6d, 6g, 6h, and 6k) subtypes. The procedure included amplification of a 380-nucleotide (nt) fragment of NS5B and its hybridization on the biochip. Tests on 345 HCV-positive samples showed that the assay agreed with NS5B sequencing 100% for the genotype and 99.7% for the subtype. The hybridization on the microarray and the NS5B sequence were in 100% agreement for identifying the most common subtypes, 1a, 1b, 4a, 4d, and 3a. This approach is a promising tool for HCV genotyping, especially for implementing the new anti-HCV drugs that require accurate identification of clinically relevant subtypes.
The hepatitis C virus (HCV) is a leading cause of chronic liver disease and increased risk of cirrhosis and hepatocellular carcinoma (51). More than 170 million people are infected with HCV worldwide (42). This enveloped, single-stranded positive-sense RNA virus is a member of the Flaviviridae family. The RNA genome contains a single large open reading frame composed of over 9,000 nucleotides (nt) encoding structural and nonstructural proteins (5). One of these proteins is an RNA-dependent RNA polymerase encoded by the so-called NS5B region. This error-prone enzyme lacks proofreading activity, which makes it responsible for the great genetic variability of HCV. Sequencing studies of HCV strains have identified 6 genotypes and more than 70 subtypes (43, 45).
The HCV genotype is considered to be the major baseline predictor of a sustained virological response (SVR) to antiviral therapy. Patients infected with HCV genotypes 2 and 3 are more sensitive to combination therapy with interferon and ribavirin than are those infected with genotype 1 (8, 11, 21). The available data on HCV genotype 4 suggest that its sensitivity to HCV treatment lies somewhere between those of genotypes 1 and 2/3 (17). The sensitivity of genotypes 5 and 6 could be similar to that of genotype 2 or 3 (1, 9, 19). The HCV subtype has recently been implicated as a potential predictor of SVR. One study of 597 difficult-to-treat patients found that subtypes 1b, 4a, and 4d were independently associated with SVR (16). The virological response to new anti-HCV agents could also be influenced by the HCV subtype (31, 42).
Several methods has been proposed for HCV genotyping (50), including commercially available techniques based on real-time PCR: the HCV genotyping analyte-specific reagent (ASR) assay (Abbott Molecular Inc., Des Plaines, IL) (23), semiautomated sequencing (the TruGene HCV 5′NC genotyping kit; Bayer HealthCare, Berkeley, CA) (10), and automated reverse hybridization (the Inno-LiPA HCV II assay; Innogenetics, Ghent, Belgium) (46, 49). Most HCV genotyping methods are based on analysis of the 5′ noncoding (NC) region of the HCV genome because the 5′ NC region is regularly amplified for HCV molecular diagnosis and quantification of the viral load. However, this highly conserved region is not suitable for accurately discriminating between subtypes and can lead to genotyping or subtyping errors (2, 3, 15, 39, 43). Hence, alternative genomic regions have been proposed for genotyping HCV, including the core fragment (35, 49) and the NS5B region (39). Sequencing and phylogenetic analysis of the NS5B region are presently considered to be the gold standard for HCV genotyping since they accurately identify the subtype and can be used to establish an epidemiological picture of circulating virus strains (27, 30, 39, 47). However, this method includes steps of purification of the amplified product, sequencing, and phylogenetic analysis that require the skill of laboratory personnel, a factor that can be a limitation to the wide use of the technique in routine clinical laboratories.
Therefore, an assay was developed that involves hybridization on an oligonucleotide microarray for identifying HCV genotypes and subtypes. It uses a low-density hydrogel-based microarray (biochip) that has been successfully used in many fields of molecular diagnostics (26, 28, 37). The microarray contains genotype- and subtype-specific oligonucleotides based on the corresponding sequences of the NS5B region. This report compares this approach to accurately identifying HCV genotype and subtype with direct NS5B sequencing.
MATERIALS AND METHODS
Collection of serum samples, HCV RNA isolation, and NS5B amplification.
All the HCV-positive patients attending Toulouse University Hospital between March 2007 and August 2008 for whom genotyping was requested were included in this study. A total of 345 samples from consecutive patients with HCV RNA concentrations of 1,622 to >10,000,000 IU/ml were included. The viral load in samples was quantified by the real-time RT-PCR Cobas AmpliPrep/Cobas TaqMan HCV test (CAP/CTM; Roche Diagnostic, Meylan, France) according to the manufacturer's instructions.
HCV RNA for genotyping was extracted with the Cobas AmpliPrep total nucleic acid isolation kit (TNAI) (Roche Diagnostics, Basel, Switzerland) following the manufacturer's instructions. Briefly, reverse transcription-PCR (RT-PCR) was performed using 10 μl of extracted RNA with primers Pr2r (5′-GGCGGAATTCCTGGTCATAGCCTCCGTGAA-3′) and Pr1f (5′-TATGAYACCCGCTGYTTTGACTC-3′) as previously described (39). PCR products were stored frozen for both NS5B sequencing and microarray genotyping.
Sequencing and phylogenetic analysis of the NS5B region.
Performance of the developed microarray was tested by comparing the results of hybridization with phylogenetic analysis of NS5B sequences, which is a standard method to identify HCV genotypes and subtypes. In fact, it is representative of phylogenetic analysis of the complete HCV genome (12). Two microliters of RT-PCR amplification mix was used for sequencing the NS5B region as previously described (39). The NS5B nucleotide sequences were aligned with CLUSTAL_X 1.83 software (48), and phylogenetic trees were created by the neighbor-joining (NJ) method. The reproducibility of the branching pattern was tested by bootstrap analysis (100 replicates). Genotypes and subtypes were determined when the bootstrap value was >70%. We used the TreeView 1.66 program to draw the phylogenetic trees (32).
Phylogenetic analyses were performed with the NS5B sequences from patients and 191 reference sequences available from the Los Alamos HCV database (14). These 191 reference sequences included genotype 1 (subtypes 1a, 1b, 1c, 1d, 1e, 1f, 1g, 1h, 1i, 1j, 1k, 1l, 1m, and 1 nontypeable), genotype 2 (subtypes 2a, 2b, 2c, 2d, 2e, 2f, 2g, 2h, 2i, 2j, 2k, 2l, 2m, 2o, 2p, 2q, 2r, and 2 nontypeable), genotype 3 (subtypes 3a, 3b, 3c, 3d, 3e, 3f, 3g, 3h, 3i, and 3k), genotype 4 (subtypes 4a, 4b, 4c, 4d, 4e, 4f, 4g, 4h, 4i, 4j, 4k, 4l, 4m, 4n, 4o, 4p, 4q, 4r, 4t, and 4 untypeable), genotype 5 (subtype 5a and 5 untypeable), and genotype 6 (subtypes 6a, 6b, 6c, 6d, 6f, 6g, 6h, 6i, 6j, 6k, 6l, 6o, 6p, 6q, 6t, and 6u). It allowed the determination of HCV genotype and subtypes of the samples subsequently tested by the microarray.
Oligonucleotide design.
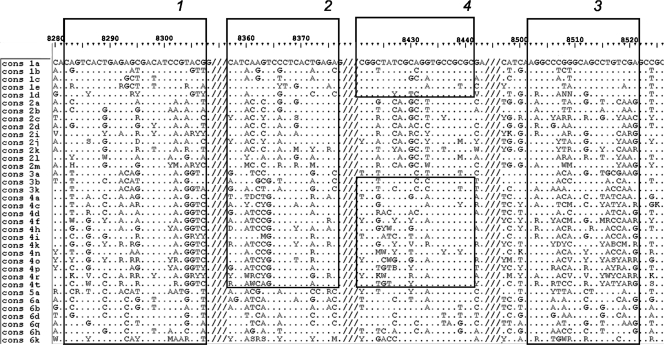
NS5B region nucleotide sequences were aligned using Bioedit software (Ibis Therapeutics, Carlsbad, CA). A total of 1,232 NS5B region sequences from GenBank and the Los Alamos HCV sequence database were aligned (14) (nt 8256 to 8616; numbering according to reference 5). This multiple alignment was used to generate unique consensus sequences for each genotype. Genotype-specific probes were then selected from within the corresponding consensus sequences. As the HCV genome is highly variable, several probes were designed for each genotype wherever possible so as to increase the reliability of the method. The sequences of the genotype-specific probes selected by this procedure were located in different segments of the NS5B region.
Next, consensus sequences were deduced for each subtype, and segments of the NS5B region that discriminated between the maximum number of subtypes within each genotype were selected (Fig. 1). The number of such segments was optimized for reliable identification of each subtype. Finally, probes for identifying subtypes were designed based on the sequences of the selected segments. This procedure did not exclude the possibility that individual probes could detect simultaneously two or more subtypes in different subtype-specific segments of the analyzed NS5B region.
FIG. 1.
Alignment of the subtype-specific consensus sequences of the NS5B region. The subtypes are indicated in the left-hand column. Residues identical to the consensus sequence of subtype 1a are indicated by dots. Numbering is from the first nucleotide of the H77 1a reference sequence (GenBank accession no. NC_004102). The positions of segments of the NS5B region used for selecting subtype-specific probes are boxed. The number of the segment corresponds to a group number and the designation of the subtype-specific oligonucleotide. Slashes indicate discontinuous sequences within the NS5B region.
The melting temperatures were calculated, and the secondary structures of the designed oligonucleotides were estimated with an Oligo analyzer (Integrated DNA Technologies). The lengths of the oligonucleotides were adjusted to maintain the range of melting temperatures within 2 to 3°C.
The sequences of oligonucleotides are listed in Table S1 in the supplemental material, and they are also available in a published patent application (D. Gryadunov, V. Mikhailovich, F. Nicot, M. Dubois, A. Zasedatelev, and J. Izopet, 2 February 2009, WO/2009/022939 2009, World Intellectual Property Organization).
Oligonucleotides for immobilization on the biochip and primers for amplification were synthesized and purified as described earlier (38). The molecular masses of oligonucleotides were measured with a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Compact MALDI 4; Kratos Analytical, Chestnut Ridge, NY) using sinapinic acid or 2-amino-5-nitropyridine as a matrix.
Microarray design.
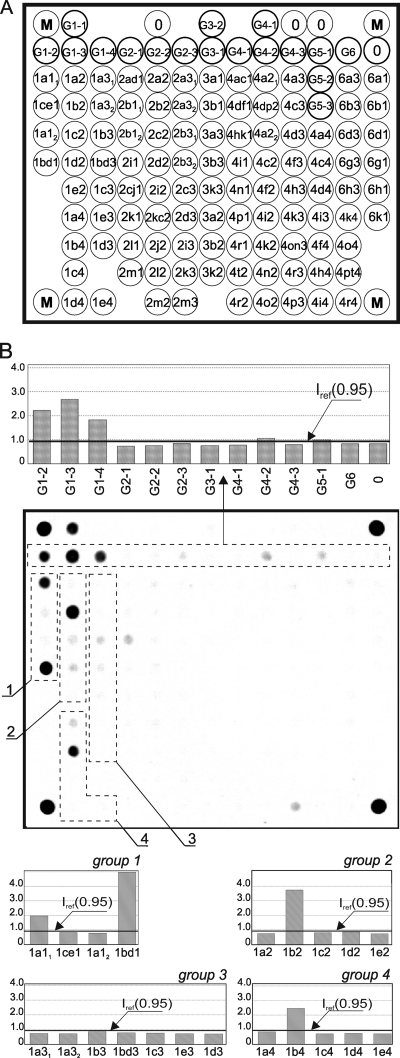
The diagnostic biochip comprised 120 immobilized oligonucleotides, four marker cells (M) for accurate positioning (image acquisition) by the processing software, and four elements of empty gel (0) needed to calculate the reference fluorescence intensity Iref (background). The arrangement of oligonucleotides immobilized on the biochip is shown in Fig. 2 A. The oligonucleotides labeled “G” that identified the genotype of the HCV sample were immobilized in the two top rows (all six genotypes). The probes immobilized in the lower rows identified the HCV subtypes.
FIG. 2.
(A) Diagram of the biochip for hybridization. Elements with the letter G contain genotype-specific probes. Four probes (G1-1 to G1-4) are used to identify genotype 1, three (G2-1 to G2-3) are used to identify genotype 2, two (G3-1 to G3-2) are used to identify genotype 3, three (G4-1 to G4-3) are used to identify genotype 4, three (G5-1 to G5-3) are used to identify genotype 5, and one (G6) is used to identify genotype 6. The probes for identifying subtypes are named ixN, where i is the genotype number, x indicates the subtype, and N is the number of the group corresponding to a segment of the NS5B region. (B) Fluorescence hybridization pattern of biochip elements obtained by analyzing an HCV sample of genotype 1, subtype 1b. The group of genotype-specific probes (G2-1 to G6) and groups 1 to 4 of subtype-specific probes of genotype 1 are outlined with a broken line. The histogram of normalized fluorescence signals of row 2 elements containing genotype-specific oligonucleotides is shown above the hybridization pattern. The histograms of normalized fluorescence signals of elements comprising groups 1 to 4 of subtype-specific oligonucleotides belonging to genotype 1 are outlined below the fluorescence image. The calculated value of the mean signal of empty elements (Iref) is shown by a solid bold line on all histograms.
Four groups of oligonucleotides were designed to identify subtypes 1a, 1b, 1c, 1d, and 1e of genotype 1. Three groups of probes were designed to differentiate between subtypes 2a, 2b, 2c, 2d, 2i, 2j, 2k, 2l, and 2m of genotype 2. Three more groups of oligonucleotides identified the three subtypes of genotype 3—3a, 3b, and 3k. Finally, four groups of probes were included to identify subtypes 4a, 4c, 4d, 4f, 4h, 4i, 4k, 4n, 4o, 4p, 4r, and 4t of genotype 4. Genotype 5 has only one subtype, 5a; therefore, the three probes for identifying genotype 5 also identified subtype 5a. Subtypes 6a, 6b, 6d, 6g, 6h, and 6k of genotype 6 were identified using two groups of probes, each of which corresponded to a separate segment within the analyzed fragment of NS5B region (Fig. 1).
Biochip manufacture.
The biochips were manufactured as described earlier (38), with 35-μl hybridization chambers (Biochip-IMB, Ltd., Moscow, Russia). Each biochip contained semispherical gel elements 100 μm in diameter placed 300 μm apart. Quality control of large-scale microchip production was done by measuring the quantity of immobilized oligonucleotides in each gel element using TestChip software provided by Biochip-IMB, Ltd.
Amplification of the NS5B fragment for genotyping on the microarray.
The PCR amplification step was performed with 1 μl RT-PCR mixture using the primers Pr1f and Pr3r (5′-GCTAGTCATAGCCTCCGT-3′). The primer concentrations were 10 nM Pr1f and 100 nM Pr3r.
The reaction mixture (25 μl) contained 1.5 mM MgCl2; 10 mM KCl; 10 mM Tris-HCl, pH 8.3; 0.2 mM (each) dATP, dCTP, dGTP, and dUTP (Sileks, Russia); 0.04 mM fluorescently labeled dUTP (IMD-515-dUTP; Biochip-IMB, Ltd, Moscow, Russia); and 5 units Taq DNA polymerase (Sileks). Because of the difference in the concentrations of forward and reverse primers within each pair, the reaction yielded predominantly single-stranded fluorescently labeled product. PCR was performed as follows: 4 min at 95°C; 36 cycles of 20 s at 95°C, 20 s at 60°C, and 30 s at 72°C; and 5 min at 72°C.
Hybridization on the biochip and registration of the results.
Hybridization mixtures were prepared by adding 12 μl of PCR mixtures to 23 μl of 1.5 M guanidine thiocyanate (GuSCN), 0.075 M HEPES, pH 7.5, 7.5 mM EDTA. The biochip hybridization chamber was filled with the mixture, and the assembly was incubated for 14 to 16 h at 37°C. The chamber was then removed, and the microarray surface was washed three times (about 30 s each) with water at 37°C and air dried. The fluorescent pattern of biochips was registered using a fluorescence analyzer setup and specialized software (ImageWare; Biochip-IMB, Ltd.).
Interpretation of hybridization results. (i) Genotype identification (genotyping).
First, perfect hybridization duplexes were identified within the upper two rows containing oligonucleotides for identifying genotypes. Our statistics (33) indicated that the fluorescent intensity of perfect duplexes should be at least 2.0 times higher than the average background signal (Iref) with a standard deviation of 0.2. Thus, a 2.0-fold intensity difference was taken as the threshold value for selecting positive signals.
The intensities of selected positive signals corresponding to perfect duplexes were compared within each genotype-specific group. If the maximum signal Gimax in one group exceeded the maximum signal in the other groups by more than 1.5-fold, the analyzed specimen was considered to belong to the corresponding genotype.
If the ratio of the signals among Gimax observed within each individual group did not exceed 1.5, the genotype of the analyzed specimen could not be accurately determined and identification of subtype was not performed. The program stopped further processing when the signals within each genotype-specific group were below the threshold value and could not pass the initial selection.
(ii) Subtype identification (subtyping).
The subtype-specific oligonucleotides were combined in groups according to the selected segments of the NS5B region. Subtyping was performed strictly after the genotype had been successfully identified. It was crucial for the identification strategy to consider only the subtype-specific oligonucleotides corresponding to the specific genotype while excluding all other signals as irrelevant.
The signals within each group of subtype-specific probes were considered positive if their intensities were at least 2.0 times higher than the average background signal Iref. Positive signals within each group of subtype-specific probes that were at least 1.5 times stronger than other signals of the same group were selected for further processing. These signals were designated Sixj (where i is the genotype number, x is the symbol of a subtype according to the HCV subtype classification, and j is the number of the analyzed group of microarray elements). When two or more elements within the same group had signals differing from one another by less than 1.5-fold, then all such signals Sixj were selected as positive. As a result, a set of elements from the various groups ix1, iy1, ix2, iz2, ix3, etc., were detected whose signals were at least 1.5 times the rest of the signals in their groups. If the number of elements homologous to one subtype in such a set, for example, ix1 and ix3, or ix1 and ixy2, exceeded the number of elements corresponding to other subtypes by at least 1, the conclusion was that the analyzed specimen belonged to subtype x of genotype i.
When the elements of different groups in the set so obtained corresponded to a different subtype, for example, ix1, iy2, and iz3, or ix1 and iyz3, the sets of signals corresponding to individual subtypes were compared to each other. If the signal of an element corresponding to subtype x in one group was 3 or more times stronger than the strongest signals from the groups corresponding to other subtypes, the conclusion was that the analyzed specimen belonged to subtype x. If the ratio of signals Six1/Siy2 was 3 or less, we concluded that the subtype could not be determined. Similarly, if the probe specific for two subtypes was the strongest signal in the group, for example, ixy1, and there were no valid signals in other groups of the elements, the conclusion was that the subtype could not be determined. Finally, if the signals of subtype-specific groups of elements did not pass the primary signal filtration relative to Iref, the conclusion was that the subtype of the analyzed specimen could not be determined.
Statistical analysis.
The kappa coefficient was measured using Stata SE 9.2 (StataCorp LP, College Station, TX) to evaluate the concordance between the HCV subtypes determined by NS5B sequencing and the NS5B biochip assays. The overall proportions of HCV subtypes determined by NS5B sequencing and the NS5B biochip assays were checked using the chi-squared test. P values of <0.05 were considered significant.
RESULTS
Determining the genotype/subtype by biochip analysis of the NS5B region.
HCV genotyping based on analysis of the NS5B region was performed by hybridization on the biochip. The procedure consisted of three steps: (i) RT-PCR to amplify the NS5B region fragment, (ii) asymmetric PCR to obtain fluorescently labeled predominantly single-stranded DNA fragments, and (iii) hybridization of the labeled product on the biochip with gel elements carrying immobilized oligonucleotides.
Figure 2B shows an example of hybridization pattern and distribution of normalized signals of biochip elements resulting from analysis of a subtype 1b sample. As defined by the algorithm described in Materials and Methods, only signals in the G1 group containing genotype 1-specific probes were more than 2 times the threshold, with the maximum signal being produced by the G1-3 element (2.68). The signals in other groups containing genotype-specific probes were close to background (the deviation from Iref did not exceed 0.12). The conclusion was that this HCV sample belonged to genotype 1.
Further processing of the groups of elements containing specific probes for genotype 1 subtypes produced the following results. In group 1, the strongest signal was obtained from element 1bd1 (4.98). In group 2 it was 1b2 (3.72), and in group 4 it was 1b4 (2.46). Group 3 contained no elements with positive signals (see corresponding histogram in Fig. 2B). Consequently, this specimen was identified as subtype 1b.
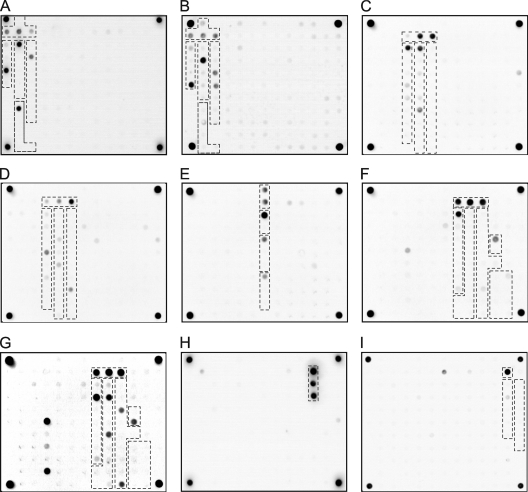
Additional examples of fluorescence patterns by hybridization with different HCV samples are shown in Fig. 3A to I. All the genotype-specific probes hybridized with the corresponding target genotypes without cross-reacting with the other genotypes. Analysis of some samples, for instance, 4d (Fig. 3G), resulted in cross-hybridization with oligonucleotides specific for subtypes of genotype 2. However, the data processing algorithm uses only the elements with subtype-specific probes of the genotype that was determined in the previous step regardless of the signals in other biochip elements. As a result, the subtypes for most samples were identified unambiguously.
FIG. 3.
Hybridization patterns obtained using HCV samples belonging to subtype 1a (A), 1b (B), 2a (C), 2i (D), 3a (E), 4a (F), 4d (G), 5a (H), and 6x (I). The groups of elements containing genotype- and subtype-specific oligonucleotides corresponding to the analyzed sample are contoured.
The ability of biochips to identify mixed HCV infections was examined. Specimens having 1a, 1b, 3a, and 4a subtypes were each adjusted to an equal HCV RNA concentration and subsequently mixed at different proportions as follows: 1a + 4a, 1b + 3a. The mixed infections were identified successfully as long as the amount of the minor species was no smaller than 20% (data not shown). The lower concentration of the minor subtype in a mixed sample led to decrease of signals in the corresponding groups of elements, and such a sample was identified as one that contained the dominant subtype only.
Analytical sensitivity and specificity.
The analytical sensitivity of this method was estimated by assaying 10-fold serial dilutions (with seronegative plasma) of a plasma standard containing 5.2 × 106 IU/ml of HCV subtype 1b. Four replicates were used for each dilution. The hybridization results obtained with the 2.0 × 102-IU/ml concentration of viral RNA were unambiguous.
The specificity of the procedure was tested using 24 seronegative plasma samples. All were identified as negative samples. With at least three replicates of each sample analyzed, the deviations of signals of genotype- and subtype-specific elements for identical samples remained within 20% of the average background signal (Iref). Therefore, there were no false-positive results.
Comparison of biochip-based genotyping with NS5B sequencing.
Table 1 shows the results obtained by hybridization on the biochip and direct sequencing of NS5B segments. All (100%) of the 345 HCV RNA-positive sera analyzed were successfully genotyped by biochip hybridization. They included samples infected with all six HCV genotypes.
TABLE 1.
Comparison of HCV genotyping obtained by NS5B sequencing with that obtained by hybridization on the biochip
| NS5B sequencing | No. of isolates with HCV genotype and subtype assigned by hybridization on biochip |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1a | 1b | 1d | 1e | 2 | 2a | 2b | 2c | 2i | 2j | 2k | 2l | 3a | 4 | 4a | 4c | 4d | 4f | 4h | 4k | 4p | 4r | 5a | 6 | Total | |
| 1 | 1 | 1 | 1 | 3 | ||||||||||||||||||||||
| 1a | 103 | 103 | ||||||||||||||||||||||||
| 1b | 88 | 88 | ||||||||||||||||||||||||
| 1d | 1 | 1 | ||||||||||||||||||||||||
| 1e | 1 | 1 | ||||||||||||||||||||||||
| 2 | 3 | 1 | 4 | |||||||||||||||||||||||
| 2a | 1 | 4 | 5 | |||||||||||||||||||||||
| 2b | 5 | 5 | ||||||||||||||||||||||||
| 2c | 3 | 1 | 4 | |||||||||||||||||||||||
| 2i | 7 | 7 | ||||||||||||||||||||||||
| 2j | 1 | 1 | ||||||||||||||||||||||||
| 2k | 5 | 5 | ||||||||||||||||||||||||
| 2l | 1 | 1 | ||||||||||||||||||||||||
| 3a | 84 | 84 | ||||||||||||||||||||||||
| 4 | 0 | |||||||||||||||||||||||||
| 4a | 12 | 12 | ||||||||||||||||||||||||
| 4c | 1 | 1 | ||||||||||||||||||||||||
| 4d | 8 | 8 | ||||||||||||||||||||||||
| 4f | 1 | 1 | ||||||||||||||||||||||||
| 4h | 1 | 1 | ||||||||||||||||||||||||
| 4k | 1 | 1 | ||||||||||||||||||||||||
| 4p | 1 | 1 | ||||||||||||||||||||||||
| 4r | 2 | 2 | 4 | |||||||||||||||||||||||
| 5a | 3 | 3 | ||||||||||||||||||||||||
| 6 | 1 | 1 | ||||||||||||||||||||||||
| Total | 2 | 104 | 89 | 0 | 1 | 6 | 4 | 5 | 3 | 7 | 0 | 7 | 0 | 84 | 3 | 12 | 1 | 8 | 1 | 0 | 1 | 1 | 2 | 3 | 1 | 345 |
The samples included subtypes 1a, 1b, 1d, 1e, 2a, 2b, 2c, 2i, 2j, 2k, 2l, 3a, 4a, 4c, 4d, 4f, 4h, 4k, 4p, 4r, and 5a and samples of undetermined subtypes of genotypes 1, 2, 4, and 6, as determined by sequencing. The two methods were concordant for the subtypes of 329/330 samples (99.7%), with a kappa coefficient of 0.996 (P < 0.00001). One sample identified as 2c by NS5B sequencing was identified as 2k by NS5B biochip analysis. The NS5B sequencing method failed to determine the subtypes in 8 samples (2.3%), and the NS5B biochip methods failed in 12 samples (3.5%) (P = 0.36). Samples with an undetermined subtype by NS5B sequencing were identified as 1a, 1b, 2k, 4h, and 4r by NS5B biochip analysis. Samples with an undetermined subtype by NS5B biochip analysis were assigned to subtypes 1d, 2a, 2j, and 2l by NS5B sequencing. The subtypes of 5 samples were not determined by either method.
DISCUSSION
The gold standard for HCV genotyping remains PCR amplification followed by sequencing of one of the phylogenetically informative coding regions of the HCV genome, such as NS5B or core/E1, and comparison with the consensus sequences in GenBank or the Los Alamos hepatitis C virus databases (14). We have developed a novel microarray-based assay for identifying the HCV genotype and subtype and evaluated it in comparison with the phylogenetic analysis of the NS5B region as a reference method. The new NS5B microarray assay and NS5B sequencing were in almost complete agreement.
The assay relies on hybridization of a 380-nt NS5B fragment with oligonucleotides specific for HCV genotypes and subtypes immobilized on a biochip. The reliable identification of each individual genotype and subtype required the design of several oligonucleotides for each of them, in consequence of the variability of the NS5B region. The results were interpreted using an original algorithm that included preliminary processing of the hybridization signal intensities from the biochip elements and comparison of signals from elements within the sets of genotype-specific probes and then from sets of subtype-specific probes.
The new method enabled us to determine all six HCV genotypes with a sensitivity of approximately 2.0 × 102 IU/ml of HCV RNA. This analytical performance using biochip-based genotyping and subtyping is comparable to that of commercially available assays (50), including the new generation of line probe assays (49).
The new method was tested on 345 HCV-positive samples. The results were 100% concordant for the genotype and 99.7% concordant for the subtype with the results obtained by direct sequencing of the NS5B segment. The accuracy and reliability of the assay make it suitable for large-scale genotyping and subtyping projects.
Hybridization on the biochip correctly identified HCV isolates of subtypes 1a, 1b, 1e, 2a, 2b, 2c, 2i, 2k, 3a, 4a, 4c, 4d, 4f, 4k, 4p, 4r, and 5a. It failed to identify subtypes 1d, 2j, 2l, and 4h. This could be because there are fewer of these NS5B sequences in GenBank and other databases, which resulted in less accurate selection of subtype-specific probes. However, these subtypes are very infrequent in Europe—2.9% for 2l, 0.9% for 2j, and 1% for 4h (30, 47). However, the hybridization on the microarray and NS5B sequencing were in 100% agreement for identifying the most widespread and clinically relevant subtypes, such as 1a, 1b, 4a, 4d, and 3a. The only limitation of the study is that not many samples of HCV genotype 6 were tested because this is very rare in France.
No mixed infections were encountered during the evaluation. Testing the analytical mixed samples revealed that the method is able to detect two different genotypes within the sample if the concentration of the minor genotype constitutes 20% or more of the total HCV RNAs.
Some recent studies have shown that HCV subtypes can predict the response to standard treatment regimens that include pegylated interferon and ribavirin. One French multicenter study of 597 treated patients showed that subtypes 1b, 4a, and 4d were independent predictors of SVR (16). A recent study also demonstrated that patients infected with HCV subtype 1b had a higher antiviral response than did patients infected with HCV subtype 1a (29). Another study of 1,532 patients infected with HCV genotype 4 showed that subtype 4a was more sensitive to anti-HCV treatment than was subtype 4d (36). Moreover, the development of new specific inhibitors of HCV enzymes whose antiviral responses and resistance profiles may be determined by the HCV subtype may require identification of the subtype prior to treatment (7, 20, 24). Several HCV inhibitors appear to act selectively against certain HCV genotype 1 subtypes, both in vitro and in vivo. Differences in the activities of NS3/4A protease inhibitors (telaprevir and boceprevir) against different subtypes have been reported. There is evidence that the selection of resistant variants and virus breakthrough is more frequent in patients infected with subtype 1b than in those harboring subtype 1a (13, 25, 40). The antiviral activities of nucleoside analogs of polymerase inhibitors are similar regardless of the HCV subtype, while nonnucleoside inhibitors are more active against subtype 1b than against subtype 1a (18, 31, 41). These findings suggest that the antiviral activity of new anti-HCV agents may also vary with the subtypes of genotypes other than 1. It is therefore essential to accurately discriminate between subtypes in order to tailor anti-HCV treatment schedules with HCV protease and polymerase inhibitors. There are few methods presently available other than direct sequencing of NS5B and core/E1 segments for identifying numerous subtypes. One of the commercially available methods, the INNO-LiPA v.2, discriminates better between subtypes 1a and 1b than does the previous version, INNO-LiPA v.1, but does not discriminate between subtypes 4a, 4c, and 4d (4, 49). Our new method correctly identified HCV subtypes 1a and 1b in more than 99% of samples; it also identified subtypes 4a and 4d.
All the experimental microarray-based methods for HCV genotyping use immobilized oligonucleotides from the 5′ untranslated region of the HCV genome. They can therefore identify only a small number of subtypes (1a, 1b, 2a/2b/2c, 3a, 3b, and 6a), although their determination of genotypes is reported to be almost 100% (6, 22, 34). In this work, use of probes complementary to subtype-specific sequences of the NS5B region enabled us to identify more than 20 HCV subtypes in the specimens tested. Other methods, such as real-time PCR, can identify a limited number of subtypes and genotypes (1a, 1b, 2a, 2b, 2c, 3, 4, 5, and 6) (23, 44). Only the clip sequencing method can, in theory, discriminate as many subtypes as can our procedure (35).
In conclusion, this new approach to analyzing the NS5B region of HCV based on hybridization with a low-density microarray is a promising tool for rapidly, sensitively, and accurately identifying viral genotype and subtype. It provides clinicians with the information needed for the choice of a correct individual treatment of hepatitis C. In addition, the performance of the new procedure and the range of identifiable genotypes and subtypes make it suitable for epidemiological surveys.
Supplementary Material
Acknowledgments
This work was supported by contract 02.522.11.2019 with the Federal Agency of Sciences and Innovations of the Russian Federation.
We are grateful to E. Kreindlin for manufacturing of microarrays, to S. Surzhikov and I. Grechishnikova for synthesis of oligonucleotides, to A. Chudinov for synthesis and selection of the fluorescent dyes, and to R. Urasov for help with the mathematical calculations and analysis of experimental data. We are especially grateful to A. Kolchinsky (Health Front Line, Ltd., Champaign, IL) for his assistance in the preparation of this paper. The English text was edited by Owen Parkes.
Footnotes
Published ahead of print on 15 September 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bonny, C., H. Fontaine, T. Poynard, C. Hezode, D. Larrey, P. Marcellin, M. Bourliere, J. P. Bronowicki, P. Merle, J. P. Zarski, T. Sapey, C. Guillemard, S. Ughetto, C. Henquell, C. Nicolas, C. Roche, K. Randl, G. Bommelaer, and A. Abergel. 2006. Effectiveness of interferon plus ribavirin combination in the treatment of naive patients with hepatitis C virus type 5. A French multicentre retrospective study. Aliment. Pharmacol. Ther. 24:593-600. [DOI] [PubMed] [Google Scholar]
- 2.Cantaloube, J. F., S. Laperche, P. Gallian, F. Bouchardeau, X. de Lamballerie, and P. de Micco. 2006. Analysis of the 5′ noncoding region versus the NS5b region in genotyping hepatitis C virus isolates from blood donors in France. J. Clin. Microbiol. 44:2051-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevaliez, S., M. Bouvier-Alias, R. Brillet, and J. M. Pawlotsky. 2009. Hepatitis C virus (HCV) genotype 1 subtype identification in new HCV drug development and future clinical practice. PLoS One 4:e8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costi, C., C. M. da Silva, N. N. Da Fre, T. Grandi, F. I. Hamester, A. Zaha, C. Niel, and M. L. Rossetti. 2009. Colorimetric microwell plate reverse-hybridization assay for detection and genotyping of hepatitis C virus. J. Virol. Methods 162:75-80. [DOI] [PubMed] [Google Scholar]
- 7.Erhardt, A., K. Deterding, Y. Benhamou, M. Reiser, X. Forns, S. Pol, J. L. Calleja, S. Ross, H. C. Spangenberg, J. Garcia-Samaniego, M. Fuchs, J. Enriquez, J. Wiegand, J. Stern, K. Wu, G. Kukolj, M. Marquis, P. Beaulieu, G. Nehmiz, and J. Steffgen. 2009. Safety, pharmacokinetics and antiviral effect of BILB 1941, a novel hepatitis C virus RNA polymerase inhibitor, after 5 days oral treatment. Antivir. Ther. 14:23-32. [PubMed] [Google Scholar]
- 8.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 9.Fung, J., C. L. Lai, I. Hung, J. Young, C. Cheng, D. Wong, and M. F. Yuen. 2008. Chronic hepatitis C virus genotype 6 infection: response to pegylated interferon and ribavirin. J. Infect. Dis. 198:808-812. [DOI] [PubMed] [Google Scholar]
- 10.Germer, J. J., D. W. Majewski, M. Rosser, A. Thompson, P. S. Mitchell, T. F. Smith, S. Elagin, and J. D. Yao. 2003. Evaluation of the TRUGENE HCV 5′NC genotyping kit with the new GeneLibrarian module 3.1.2 for genotyping of hepatitis C virus from clinical specimens. J. Clin. Microbiol. 41:4855-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadziyannis, S. J., and J. S. Koskinas. 2004. Differences in epidemiology, liver disease and treatment response among HCV genotypes. Hepatol. Res. 29:129-135. [DOI] [PubMed] [Google Scholar]
- 12.Hraber, P. T., W. Fischer, W. J. Bruno, T. Leitner, and C. Kuiken. 2006. Comparative analysis of hepatitis C virus phylogenies from coding and non-coding regions: the 5′ untranslated region (UTR) fails to classify subtypes. Virol. J. 3:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieffer, T. L., C. Sarrazin, J. S. Miller, M. W. Welker, N. Forestier, H. W. Reesink, A. D. Kwong, and S. Zeuzem. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631-639. [DOI] [PubMed] [Google Scholar]
- 14.Kuiken, C., P. Hraber, J. Thurmond, and K. Yusim. 2008. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 36:D512-D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laperche, S., F. Lunel, J. Izopet, S. Alain, P. Deny, G. Duverlie, C. Gaudy, J. M. Pawlotsky, J. C. Plantier, B. Pozzetto, V. Thibault, F. Tosetti, and J. J. Lefrere. 2005. Comparison of hepatitis C virus NS5b and 5′ noncoding gene sequencing methods in a multicenter study. J. Clin. Microbiol. 43:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legrand-Abravanel, F., P. Colson, H. Leguillou-Guillemette, L. Alric, I. Ravaux, F. Lunel-Fabiani, M. Bouviers-Alias, P. Trimoulet, M. L. Chaix, C. Hezode, J. Foucher, H. Fontaine, A. M. Roque-Afonso, M. Gassin, E. Schvoerer, C. Gaudy, B. Roche, M. Doffoel, L. D'Alteroche, S. Vallet, Y. Baazia, B. Pozzetto, V. Thibault, J. B. Nousbaum, D. Roulot, H. Coppere, T. Poinard, C. Payan, and J. Izopet. 2009. Influence of the HCV subtype on the virological response to pegylated interferon and ribavirin therapy. J. Med. Virol. 81:2029-2035. [DOI] [PubMed] [Google Scholar]
- 17.Legrand-Abravanel, F., F. Nicot, A. Boulestin, K. Sandres-Saune, J. P. Vinel, L. Alric, and J. Izopet. 2005. Pegylated interferon and ribavirin therapy for chronic hepatitis C virus genotype 4 infection. J. Med. Virol. 77:66-69. [DOI] [PubMed] [Google Scholar]
- 18.Legrand-Abravanel, F., F. Nicot, and J. Izopet. 2010. New NS5B polymerase inhibitors for hepatitis C. Expert Opin. Invest. Drugs 19:963-975. [DOI] [PubMed] [Google Scholar]
- 19.Legrand-Abravanel, F., K. Sandres-Saune, K. Barange, L. Alric, J. Moreau, P. Desmorat, J. P. Vinel, and J. Izopet. 2004. Hepatitis C virus genotype 5: epidemiological characteristics and sensitivity to combination therapy with interferon-alpha plus ribavirin. J. Infect. Dis. 189:1397-1400. [DOI] [PubMed] [Google Scholar]
- 20.Liang, Y., H. Ishida, O. Lenz, T. I. Lin, O. Nyanguile, K. Simmen, R. B. Pyles, N. Bourne, M. Yi, K. Li, and S. M. Lemon. 2008. Antiviral suppression vs restoration of RIG-I signaling by hepatitis C protease and polymerase inhibitors. Gastroenterology 135:1710-1718. [DOI] [PubMed] [Google Scholar]
- 21.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao, H., H. Zhang, J. Zhao, Z. Lu, G. Jin, S. Gu, H. Wang, and Y. Wang. 2010. Clinical evaluation of a colorimetric oligonucleotide chip for genotyping hepatitis C virus. Clin. Biochem. 43:214-219. [DOI] [PubMed] [Google Scholar]
- 23.Martro, E., V. Gonzalez, A. J. Buckton, V. Saludes, G. Fernandez, L. Matas, R. Planas, and V. Ausina. 2008. Evaluation of a new assay in comparison with reverse hybridization and sequencing methods for hepatitis C virus genotyping targeting both 5′ noncoding and nonstructural 5b genomic regions. J. Clin. Microbiol. 46:192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCown, M. F., S. Rajyaguru, S. Kular, N. Cammack, and I. Najera. 2009. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob. Agents Chemother. 53:2129-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHutchison, J. G., G. T. Everson, S. C. Gordon, I. M. Jacobson, M. Sulkowski, R. Kauffman, L. McNair, J. Alam, and A. J. Muir. 2009. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360:1827-1838. [DOI] [PubMed] [Google Scholar]
- 26.Mikhailovich, V., D. Gryadunov, A. Kolchinsky, A. A. Makarov, and A. Zasedatelev. 2008. DNA microarrays in the clinic: infectious diseases. Bioessays 30:673-682. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, D. G., B. Willems, M. Deschenes, N. Hilzenrat, R. Mousseau, and S. Sabbah. 2007. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J. Clin. Microbiol. 45:1102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasedkina, T. V., N. A. Guseva, O. A. Gra, O. N. Mityaeva, A. V. Chudinov, and A. S. Zasedatelev. 2009. Diagnostic microarrays in hematologic oncology: applications of high- and low-density arrays. Mol. Diagn. Ther. 13:91-102. [DOI] [PubMed] [Google Scholar]
- 29.Nicot, F., L. Alric, K. Barange, S. Métivier, J. M. Dramard, J. M. Combis, B. Castan, J. J. Meurisse, J. L. Payen, D. Garipuy, H. Desmorat, J. M. Peron, S. Thebault, T. Morin, C. Renou, P. Barel, B. Guerin, Y. Imbert, S. Sire, K. Sauné, E. Chatelut, and J. Izopet. Influence of HCV genotype 1 subtypes on the virus response to peg interferon alpha-2a plus ribavirin therapy. J. Med. Virol., in press. [DOI] [PubMed]
- 30.Nicot, F., F. Legrand-Abravanel, K. Sandres-Saune, A. Boulestin, M. Dubois, L. Alric, J. P. Vinel, C. Pasquier, and J. Izopet. 2005. Heterogeneity of hepatitis C virus genotype 4 strains circulating in south-western France. J. Gen. Virol. 86:107-114. [DOI] [PubMed] [Google Scholar]
- 31.Nyanguile, O., B. Devogelaere, L. Vijgen, W. Van den Broeck, F. Pauwels, M. D. Cummings, H. L. De Bondt, A. M. Vos, J. M. Berke, O. Lenz, G. Vandercruyssen, K. Vermeiren, W. Mostmans, P. Dehertogh, F. Delouvroy, S. Vendeville, K. VanDyck, K. Dockx, E. Cleiren, P. Raboisson, K. A. Simmen, and G. C. Fanning. 2010. 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus. J. Virol. 84:2923-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 33.Pan'kov, S. V., V. R. Chechetkin, O. G. Somova, O. V. Antonova, O. V. Moiseeva, D. V. Prokopenko, R. A. Yurasov, D. A. Gryadunov, and A. V. Chudinov. 2009. Kinetic effects on signal normalization in oligonucleotide microchips with labeled immobilized probes. J. Biomol. Struct. Dyn. 27:235-244. [DOI] [PubMed] [Google Scholar]
- 34.Park, J. C., J. M. Kim, O. J. Kwon, K. R. Lee, Y. G. Chai, and H. B. Oh. 2010. Development and clinical evaluation of a microarray for hepatitis C virus genotyping. J. Virol. Methods 163:269-275. [DOI] [PubMed] [Google Scholar]
- 35.Ross, R. S., S. Viazov, B. Wolters, and M. Roggendorf. 2008. Towards a better resolution of hepatitis C virus variants: CLIP sequencing of an HCV core fragment and automated assignment of genotypes and subtypes. J. Virol. Methods 148:25-33. [DOI] [PubMed] [Google Scholar]
- 36.Roulot, D., V. Bourcier, V. Grando, P. Deny, Y. Baazia, H. Fontaine, F. Bailly, L. Castera, V. De Ledinghen, P. Marcellin, R. Poupon, M. Bourliere, J. P. Zarski, and F. Roudot-Thoraval. 2007. Epidemiological characteristics and response to peginterferon plus ribavirin treatment of hepatitis C virus genotype 4 infection. J. Viral Hepat. 14:460-467. [DOI] [PubMed] [Google Scholar]
- 37.Rubina, A. Y., A. Kolchinsky, A. A. Makarov, and A. S. Zasedatelev. 2008. Why 3-D? Gel-based microarrays in proteomics. Proteomics 8:817-831. [DOI] [PubMed] [Google Scholar]
- 38.Rubina, A. Y., S. V. Pan'kov, E. I. Dementieva, D. N. Pen'kov, A. V. Butygin, V. A. Vasiliskov, A. V. Chudinov, A. L. Mikheikin, V. M. Mikhailovich, and A. D. Mirzabekov. 2004. Hydrogel drop microchips with immobilized DNA: properties and methods for large-scale production. Anal. Biochem. 325:92-106. [DOI] [PubMed] [Google Scholar]
- 39.Sandres-Saune, K., P. Deny, C. Pasquier, V. Thibaut, G. Duverlie, and J. Izopet. 2003. Determining hepatitis C genotype by analyzing the sequence of the NS5b region. J. Virol. Methods 109:187-193. [DOI] [PubMed] [Google Scholar]
- 40.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelka, U. Muh, M. Welker, D. Wincheringer, Y. Zhou, H. M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]
- 41.Sarrazin, C., and S. Zeuzem. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447-462. [DOI] [PubMed] [Google Scholar]
- 42.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 43.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 44.Sohn, Y. H., S. Y. Ko, M. H. Kim, and H. B. Oh. 2010. Performance evaluation of the Abbott RealTime HCV Genotype II for hepatitis C virus genotyping. Clin. Chem. Lab. Med. 48:469-474. [DOI] [PubMed] [Google Scholar]
- 45.Stuyver, L., W. van Arnhem, A. Wyseur, F. Hernandez, E. Delaporte, and G. Maertens. 1994. Classification of hepatitis C viruses based on phylogenetic analysis of the envelope 1 and nonstructural 5B regions and identification of five additional subtypes. Proc. Natl. Acad. Sci. U. S. A. 91:10134-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, F., F. Nicot, K. Sandres-Saune, M. Dubois, F. Legrand-Abravanel, L. Alric, J. M. Peron, C. Pasquier, and J. Izopet. 2007. Genetic diversity of HCV genotype 2 strains in south western France. J. Med. Virol. 79:26-34. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verbeeck, J., M. J. Stanley, J. Shieh, L. Celis, E. Huyck, E. Wollants, J. Morimoto, A. Farrior, E. Sablon, M. Jankowski-Hennig, C. Schaper, P. Johnson, M. Van Ranst, and M. Van Brussel. 2008. Evaluation of Versant hepatitis C virus genotype assay (LiPA) 2.0. J. Clin. Microbiol. 46:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weck, K. 2005. Molecular methods of hepatitis C genotyping. Expert Rev. Mol. Diagn. 5:507-520. [DOI] [PubMed] [Google Scholar]
- 51.Younossi, Z., J. Kallman, and J. Kincaid. 2007. The effects of HCV infection and management on health-related quality of life. Hepatology 45:806-816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.