Abstract
Previous studies have confirmed the association of the acid producers Streptococcus mutans and Lactobacillus spp. with childhood caries, but they also suggested these microorganisms are not sufficient to explain all cases of caries. In addition, health-associated bacterial community profiles are not well understood, including the importance of base production and acid catabolism in pH homeostasis. The bacterial community composition in health and in severe caries of the young permanent dentition was compared using Sanger sequencing of the ribosomal 16S rRNA genes. Lactobacillus species were dominant in severe caries, and levels rose significantly as caries progressed from initial to deep lesions. S. mutans was often observed at high levels in the early stages of caries but also in some healthy subjects and was not statistically significantly associated with caries progression in the overall model. Lactobacillus or S. mutans was found either at low levels or not present in several samples. Other potential acid producers observed at high levels in these subjects included strains of Selenomonas, Neisseria, and Streptococcus mitis. Propionibacterium FMA5 was significantly associated with caries progression but was not found at high levels. An overall loss of community diversity occurred as caries progressed, and species that significantly decreased included the Streptococcus mitis-S. pneumoniae-S. infantis group, Corynebacterium matruchotii, Streptococcus gordonii, Streptococcus cristatus, Capnocytophaga gingivalis, Eubacterium IR009, Campylobacter rectus, and Lachnospiraceae sp. C1. The relationship of acid-base metabolism to 16S rRNA gene-based species assignments appears to be complex, and metagenomic approaches that would allow functional profiling of entire genomes will be helpful in elucidating the microbial pathogenesis of caries.
Dental caries is the most common chronic disease of childhood, affecting nearly three-fourths of all children by the age of 17 years (50). The majority of children experience mild caries in the permanent dentition that is easily managed, but nearly 20% of children suffer more aggressive caries (19) that is destructive and often recurrent. The cariogenicity of Streptococcus mutans and Lactobacillus species in tooth-associated biofilms has long been established based on culture studies (51), but this approach has provided a limited ability to study the role of other species present in biofilm communities. Recently DNA-based methods have been used to study early childhood caries (5, 11, 23), caries of the primary and permanent teeth in children and young adults (1), root caries in the elderly (42), and advanced dentin lesions (8, 10, 36). Taken together these studies have confirmed the association of S. mutans and Lactobacillus species with childhood caries, but they also suggest that these species are not sufficient to explain all cases of caries. In addition, health-associated bacterial community profiles are not well understood, including the importance of species that produce basic compounds that lower pH and species that metabolize lactic acid to lower-pKa acids.
The purpose of the present study was to compare bacterial community profiles associated with severe dental caries and health in the young permanent dentition. This was accomplished by using an open-ended molecular approach, 16S rRNA gene cloning, and Sanger sequencing. A previously established clinical model was used (5) in which samples representing the full range from early- to late-stage caries were collected to reconstruct the natural history of caries. These samples were compared within individuals and to samples from healthy control subjects. The data indicated that caries has a heterogeneous etiology, with multiple profiles of acid-producing species observed. An overall loss of community diversity occurred as caries progressed, and species that are part of a health-associated ecosystem were identified.
MATERIALS AND METHODS
Clinical methods. (i) Subject recruitment.
Subjects with dental caries and a dentally healthy control group were recruited at the Nationwide Children's Hospital Dental Clinic in Columbus, OH. General exclusionary criteria for either group included (i) age greater than 16 years, (ii) indications for infective endocarditis prophylaxis, (iii) antibiotic use in the past 30 days, and (iv) professional cleaning in the past 30 days. Only one child per family was included in each group. The inclusion requirement for the caries group was the presence of at least three permanent teeth with cavitated, multisurface lesions involving a smooth surface and with at least one of these three teeth with a vital pulp. Age-, race-, and gender-matched healthy control subjects who were caries-free and had no existing restorations were also recruited. Institutional Review Board approval was obtained for this protocol, consent was obtained from the parents of all subjects, and assent was obtained from subjects who were at least 9 years old.
(ii) Sampling and clinical data collection.
For the healthy subjects dental plaque was sampled from the (i) healthy enamel. For the subjects with dental caries, plaque was collected separately from the surfaces of each of three types of sites: (ii) intact enamel, (iii) white spot lesions, and (iv) cavitated lesions. In addition, (v) carious dentin was harvested from one tooth. Thus, one sample was collected from each healthy subject and four samples were collected from each subject with caries. Dental plaque was collected by swiping the tooth or lesion surface with a dental explorer and wiping the plaque onto a coarse endodontic paper point. Each plaque sample was obtained by pooling from multiple teeth. Dentin samples were harvested with a spoon excavator. Samples were placed in a sterile 1.5-ml microcentrifuge tube and frozen for storage.
A brief written survey regarding antibiotic and medication history, fluoride status, and exposure to cigarette smoke was completed by the parent of each child. An open-ended account of dietary practices was obtained for each subject. For subjects with caries, existing restorations and all carious surfaces were scored.
Laboratory methods. (i) Sample preparation.
Samples were placed in 300 μl of Tris-EDTA buffer and then beaten with 0.25 g of 0.1-mm glass beads for 60 s at 4,800 rpm in a Biospec Products bead beater. The bacterial DNA was purified using the Qiagen QIAamp DNA minikit according to the manufacturer's instructions and frozen until analysis.
(ii) PCR amplification for clonal analysis.
The 16S rRNA genes were amplified from the purified bacterial DNA using universal primers A17 (5′-GTT TGA TCC TGG CTC AG-3′) and 317 (5′-AAG GAG GTG ATC CAG GC-3′) (Biosynthesis, Lewisville, TX). PCR conditions were as previously described (25). The products of PCR amplification were examined by electrophoresis in 1% agarose and purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA).
(iii) Cloning and sequencing.
Cloning and sequencing of the 16S amplicons generated by PCR were as previously described (25). Amplicons included the 16S hypervariable regions V5 to V9.
(iv) Bacterial 16S rRNA gene sequence identification.
The sequence from each clone was identified by comparing it to a local, curated oral microbiome database (hosted at http://microbiome.osu.edu) using BLAST (3). This database contained 16S sequences from all known oral taxa. Taxonomic assignments were made based on maximum likelihood trees and distance matrices. The majority of sequences could be attributed to named species, and unamed taxa were assigned names according to identifiers from their GenBank entries. Species that could not be distinguished by 16S variability were given combination names. Clinical sequences were required to be ≥98.0% similar to species-level taxonomic units in the database for identification. Sequences with similarity scores of <98% were further analyzed by BLAST against GenBank, the results were manually examined, and chimeric sequences were removed. Sequences that were observed only once and failed to match sequences in GenBank were not considered. Remaining novel sequences were added to the database.
Data management and statistical analyses.
Levels of each species were calculated as the percentage of total bacteria for each sample. Mean bacterial levels and 95% confidence intervals were determined for the most prevalent species by using the JMP program (version 7.0; SAS Institute Inc., Cary, NC). Repeated-measures analysis was performed using PROC MIXED in SAS (SAS 9.1; SAS Institute Inc., Cary, NC) and the default various components structure. The type of site from which each sample was collected was expressed as a level of severity and used in the PROC MIXED analysis. Using this scale, healthy control samples were assigned a value of 1, intact enamel samples were assigned a level of 2, white spot lesions were assigned a level of 3, cavitated lesions were assigned a level of 4, and dentin samples were assigned a level of 5. The PROC MIXED analysis used a linear model to calculate an estimate of the percent change for each species as caries progressed from severity levels 1 through 5 and to determine the significance of the estimate at α = 0.05. The false discovery rate correction for multiple comparisons was determined using SAS. The Shannon diversity index was computed using the formula H′ = −Σpi ln(pi), where pi equals the relative abundance of each of the ith species (30) and was analyzed by PROC MIXED. Post hoc paired and unpaired t tests were used. Pearson correlation was used for exploratory analyses of the contribution of caries severity, dietary patterns, and antibiotic history to bacterial community composition, and t tests were used to analyze fluoride exposure and environmental smoke exposure data.
RESULTS
Twenty-one subjects with caries and 18 healthy controls were recruited for this study. An average of 52 clones (minimum of 47, maximum of 60) were identified per sample, and the total number of clones for all samples was 5,299. The shortest sequence length was 600 bp, and the average sequence length among all clones was 1,061 bp. Chi-square analysis indicated no significant differences between the groups for gender, race, or ethnicity. Ages ranged from 7 to 16 years (mean age was 13), and the difference in means between the caries and healthy groups was not significant by t test. Significantly more children in the healthy control group drank fluoridated water than in the caries group.
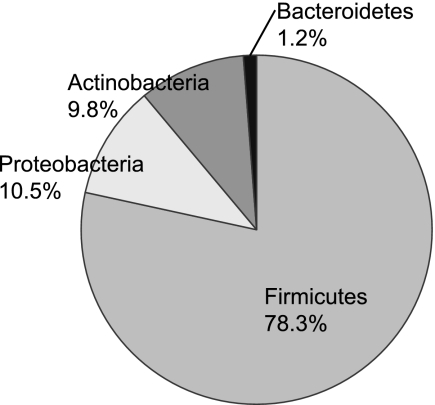
Table 1 lists the 144 species identified in this study. Twelve Lactobacillus species were observed, some at low numbers, but these species showed highly similar distributions by caries severity and were combined into one “total Lactobacillus” group for analysis. The 144 identified species could be assigned to seven bacterial phyla, as shown in Fig. 1. Spirochaetes, Deinococcus-Thermus, and Fusobacteria were found at such low levels that they were not included in the figure. Overall, only 7.6% of total clones were uncultivated.
TABLE 1.
Distribution of all species-level taxa
| Species | Rank (%)a |
|---|---|
| Abiotrophia defectiva | 11 (2.15) |
| Achromobacter xylosoxidans | 43 (0.42) |
| Acidaminococcaceae oral taxa135, 145, 148, 155 | 67 (0.21) |
| Actinobaculum sp. oral clone EL030 | 107 (0.04) |
| Actinomyces dentalis | 93 (0.08) |
| Actinomyces georgiae | 118 (0.02) |
| Actinomyces gerencseriae | 58 (0.23) |
| Actinomyces IO077 | 59 (0.23) |
| Actinomyces israelii | 119 (0.02) |
| Actinomyces massiliensis | 120 (0.02) |
| Actinomyces odontolyticus lingnae | 60 (0.23) |
| Actinomyces oral clone IP073 | 108 (0.04) |
| Actinomyces viscosus naeslundii | 14 (1.43) |
| Aggregatibacter segnis | 121 (0.02) |
| Atopobium parvulum | 72 (0.17) |
| Atopobium rimae | 85 (0.11) |
| Bacteroidales oral clone MCE3_262 | 122 (0.02) |
| Bifidobacterium dentium | 74 (0.15) |
| Campylobacter concisus | 69 (0.19) |
| Campylobacter curvus | 123 (0.02) |
| Campylobacter gracilis | 12 (1.92) |
| Campylobacter rectus | 52 (0.28) |
| Campylobacter showae | 75 (0.15) |
| Capnocytophaga gingivalis | 26 (0.87) |
| Capnocytophaga leadbetteri | 94 (0.08) |
| Capnocytophaga ochracea A | 124 (0.02) |
| Capnocytophaga sputigena | 95 (0.08) |
| Cardiobacterium hominis | 76 (0.15) |
| Cardiobacterium valvarum | 101 (0.06) |
| Catonella morbi | 86 (0.11) |
| Catonella sp. AH153 | 125 (0.02) |
| Centipeda periodontii | 70 (0.19) |
| Clostridales oral taxon 07580 (0.13) | |
| Corynebacterium argentoratense | 126 (0.02) |
| Corynebacterium durum | 53 (0.28) |
| Corynebacterium matruchotii | 8 (3.04) |
| Cryptobacterium curtum | 127 (0.02) |
| Dialister clone BS016 | 109 (0.04) |
| Dialister invisus | 42 (0.43) |
| Dialister micraerophilus | 128 (0.02) |
| Dialister pneumosintes | 129 (0.02) |
| Eikenella corrodens | 61 (0.23) |
| Enterococcus casseliflavus | 102 (0.06) |
| Eubacterium BE088 | 62 (0.23) |
| Eubacterium BU014 | 130 (0.02) |
| Eubacterium DO008 | 54 (0.28) |
| Eubacterium IR009 | 31 (0.74) |
| Eubacterium sp. EI074 | 68 (0.21) |
| Eubacterium sulci infirmum | 131 (0.02) |
| Eubacterium yurii subsp. margaretiae yurii | 96 (0.08) |
| Eubacterium yurii subsp. schtitka | 132 (0.02) |
| Fusobacterium nucleatum | 133 (0.02) |
| Gemella haemolysans | 28 (0.83) |
| Gemella morbillorum | 23 (0.91) |
| Gemella sanguinis | 110 (0.04) |
| Granulicatella adiacens | 29 (0.83) |
| Granulicatella elegans | 134 (0.02) |
| Haemophilus parainfluenzae group A | 87 (0.11) |
| Johnsonella sp. CK051 | 135 (0.02) |
| Kingella denitrificans | 44 (0.42) |
| Kingella oralis | 34 (0.53) |
| Lachnospiraceae novel RF01 | 136 (0.02) |
| Lachnospiraceae oral taxon 107 | 63 (0.23) |
| Lachnospiraceae sp. C1 | 55 (0.28) |
| Lactobacillus casei paracasei | 9 (2.81) |
| Lactobacillus crispatus | 49 (0.34) |
| Lactobacillus delbrueckii | 37 (0.51) |
| Lactobacillus fermentum | 35 (0.53) |
| Lactobacillus gasseri johnsonii | 5 (4.62) |
| Lactobacillus oris | 50 (0.32) |
| Lactobacillus parabuchneri kefiri buchneri | 51 (0.32) |
| Lactobacillus plantarum | 41 (0.45) |
| Lactobacillus rhamnosus | 19 (1.09) |
| Lactobacillus salivarius | 27 (0.85) |
| Lactobacillus ultunensis | 45 (0.42) |
| Lactobacillus vaginalis | 13 (1.68) |
| Lautropia FX006 | 97 (0.08) |
| Lautropia mirabilis | 16 (1.19) |
| Megasphaera clone BB166 | 111 (0.04) |
| Megasphaera clone BS073 CS025 | 81 (0.13) |
| Megasphaera micronuciformis | 39 (0.49) |
| Meiothermus timidus | 137 (0.02) |
| Mitsuokella clone EU_669565 | 112 (0.04) |
| Mitsuokella oral taxon 131 | 38 (0.51) |
| Mitsuokella oral taxon 521 | 88 (0.11) |
| Mogibacterium diversum neglectum pumilum vescu | 90 (0.09) |
| Moraxella clone AM_420053 | 138 (0.02) |
| Neisseria AP085 | 113 (0.04) |
| Neisseria bacilliformis | 91 (0.09) |
| Neisseria elongata | 40 (0.47) |
| Neisseria flava mucosa pharyngis sicca | 7 (3.38) |
| Neisseria flavescens | 32 (0.74) |
| Olsenella clone C1 | 64 (0.23) |
| Olsenella profusa | 73 (0.17) |
| Oribacterium sinus | 98 (0.08) |
| Oribacterium sp. AO068 | 82 (0.13) |
| Parascardovia denticolens | 89 (0.11) |
| Parvimonas micra | 33 (0.55) |
| Peptococcus MCE10_265 | 114 (0.04) |
| Peptostreptococcus stomatis | 92 (0.09) |
| Porphyromonas catoniae | 103 (0.06) |
| Prevotella pallens | 104 (0.06) |
| Propionibacterium BN085 | 105 (0.06) |
| Propionibacterium FMA5 | 17 (1.13) |
| Propionibacterium propionicus A | 65 (0.23) |
| Pseudoramibacter alactolyticus | 140 (0.02) |
| Rothia aeria | 77 (0.15) |
| Rothia dentocariosa | 56 (0.28) |
| Rothia mucilaginosa | 115 (0.04) |
| Scardovia clone C1 | 141 (0.02) |
| Scardovia inopinata | 15 (1.30) |
| Selenomonas AA024 | 21 (1.00) |
| Selenomonas AJ036 | 116 (0.04) |
| Selenomonas CS024 | 47 (0.40) |
| Selenomonas dianae | 46 (0.42) |
| Selenomonas DY027 | 117 (0.04) |
| Selenomonas EW051a | 106 (0.06) |
| Selenomonas EY047 | 83 (0.13) |
| Selenomonas flueggei | 78 (0.15) |
| Selenomonas FT050 | 48 (0.38) |
| Selenomonas infelix | 20 (1.09) |
| Selenomonas noxia | 10 (2.51) |
| Selenomonas sputigena | 18 (1.11) |
| Shuttleworthia satelles | 99 (0.08) |
| Streptococcus anginosus A | 66 (0.23) |
| Streptococcus anginosus B | 36 (0.53) |
| Streptococcus australis | 79 (0.15) |
| Streptococcus cristatus | 24 (0.91) |
| Streptococcus downei | 142 (0.02) |
| Streptococcus gordonii | 6 (4.02) |
| Streptococcus intermedius constellatus | 57 (0.25) |
| Streptococcus massiliensis | 143 (0.02) |
| Streptococcus mitis-S. pneumoniae-S. infantis | 2 (11.40) |
| Streptococcus mutans | 3 (8.10) |
| Streptococcus oligofermentans sinensis | 100 (0.08) |
| Streptococcus parasanguinis | 30 (0.83) |
| Streptococcus sanguinis | 4 (5.17) |
| Streptococcus sobrinus | 25 (0.91) |
| Streptococcus vestibularis salivarius | 22 (0.92) |
| Tannerella oral clone BU063 | 144 (0.02) |
| Treponema socranskii subsp. buccale paredis socranskii | 84 (0.13) |
| Uncultured Capnocytophaga sp. clone SK117 | 139 (0.02) |
| Uncultured Selenomonas sp. clone 03_5_H05 | 71 (0.19) |
| Veillonella atypica dispar parvula | 1 (12.79) |
Rank was assigned in order of decreasing overall prevalence. Values in parentheses show the percentage of total clones represented by the species.
FIG. 1.
Distribution of species by phylum for all samples. Spirochaetes, Deinococcus-Thermus, and Fusobacteria were detected but are not shown because each represented less than 1% of total clones.
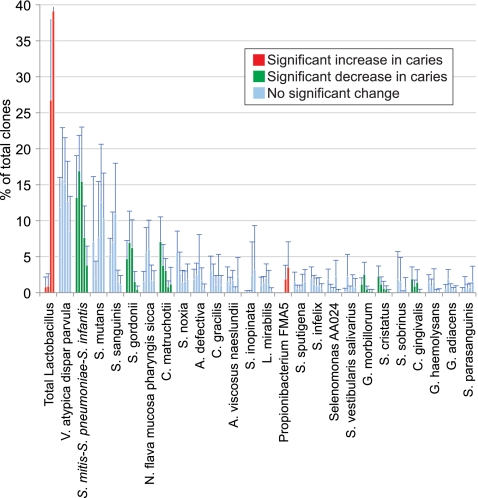
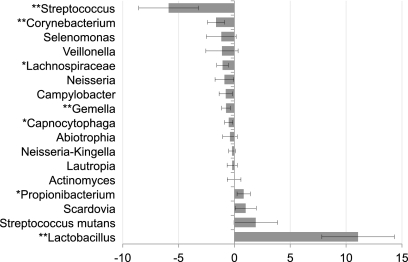
Mean bacterial levels by caries severity are plotted in Fig. 2 for common species (greater than 0.75% of total clones). Figure 3 shows the relationship of bacterial levels to caries severity as determined by PROC MIXED for the most abundant genera (greater than 1% of the total clones) and for S. mutans. Thirteen of these genera showed negative trends by PROC MIXED analysis (levels decreased as caries severity increased), and five of those were significant. Of four genera that had positive estimates by PROC MIXED analysis (levels increased as caries severity increased), only Lactobacillus and Propionibacterium were significant. Levels of S. mutans were not significantly related to caries severity in this study.
FIG. 2.
Mean levels of the most common species as caries severity increased. From left to right within a species, bars indicate levels for healthy control, intact enamel, white spot, cavitated, and dentin samples. Species found at greater than 0.75% of total clones are shown, with 95% confidence intervals. The upper limits for the 95% confidence interval bars for the cavitated and dentin samples of the total Lactobacillus group, which are not shown, were 42.2% and 58.2%, respectively.
FIG. 3.
Changes in distributions of the most common genus groups as caries severity increased. S. mutans was considered separately because of its importance in caries. Estimates of the percent change per one level of severity were determined by PROC MIXED analysis. Genus groups found present at greater than 1% of total clones are shown, with 95% confidence intervals. P values were adjusted using a false discovery rate correction. *, corrected P < 0.05; **, corrected P < 0.01.
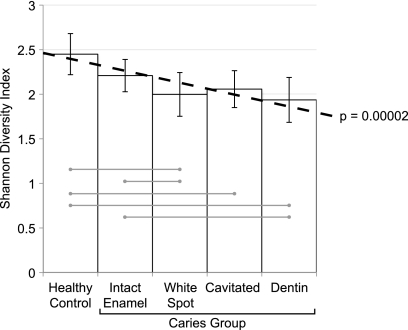
The relationship between caries severity and bacterial community diversity is shown in Fig. 4. Overall, bacterial diversity decreased significantly (P = 0.00002) as caries progressed from healthy to cavitated and deep dentinal lesions.
FIG. 4.
Decreasing species diversity as caries severity increased. The decrease in severity was modeled using PROC MIXED analysis and is shown as a dotted line (estimate, −0.13; P < 0.00002). Mean Shannon diversity indices at the species level are shown with 95% confidence intervals for each level of caries severity. Samples that were statistically different according to post hoc t tests and paired t tests are connected by light gray lines.
Subjects were divided into four groups based on the presence of common caries pathogens in their cavitated samples, including an S. mutans-dominated group, a Lactobacillus-dominated group, a group with both S. mutans and Lactobacillus, and a group with neither Lactobacillus nor S. mutans. Exploratory analyses showed no relationship between bacterial profile and caries severity measures, water fluoride status, dietary history, antibiotic use, smoke exposure, or demographic variables (data not shown).
DISCUSSION
This study, which utilized open-ended 16S rRNA gene cloning and sequencing, confirms the presence of S. mutans and Lactobacillus in caries but provides additional insight into the heterogeneous and complex nature of supragingival oral biofilms in severe caries of the young permanent dentition.
The ecologic plaque hypothesis and bacterial community diversity.
Dental caries results from a substrate-driven disruption of the bacterial ecosystem, conceptualized by Marsh (32) and termed the “ecologic plaque hypothesis.” Caries results when the fermentation of readily available carbohydrates to lactate by acid-producing species first lowers pH and then leads to suppression of acid-sensitive species and overgrowth of acid-tolerant species. The net result is a reduction in bacterial community diversity as caries progresses, and a shrinking number of species can survive the harsh conditions. Evidence to support this ecologic disruption has been reported by Li et al., who determined that the mean species richness and Shannon diversity index were greater in caries-free children than in those with severe early childhood caries (27). The current study provides additional support for the ecologic disruption model of caries pathobiology, confirming a decline in diversity between healthy sites and all stages of caries (Fig. 4) and an increase in the fraction of the microbiota accounted for by cariogenic species (Fig. 5). As an example, advanced caries samples from two subjects consisted entirely of lactobacilli, indicating a complete loss of pH-modulating species and providing an example of a classic catastrophic ecological disruption.
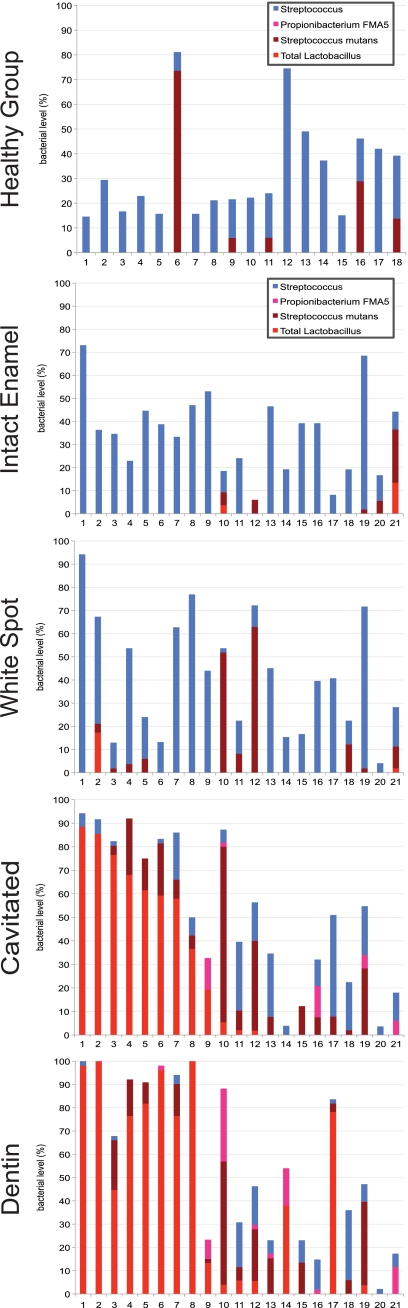
FIG. 5.
Levels of Streptococcus spp., Propionibacterium FMA5, Streptococcus mutans, and Lactobacillus spp. at each level of caries experience. The Streptococcus group includes levels of health-associated streptococci S. mitis-S. pneumoniae-S. infantis, S. oralis, S. cristatus, S. gordonii, and S. sanguinis. At each level in the caries group, subjects are presented in the same order, according to level of Lactobacillus in cavitated samples.
Uncultivated bacteria.
In contrast to previous investigations of the oral microbiome that reported at least 50% uncultivated species (2, 24, 25, 40, 42), less than 8% of the sequences generated for this study were attributed to uncultivated species. To some extent this difference can be explained by differences between supragingival and subgingival bacterial communities. In large part, however, it can be attributed to an increase in information about these communities and the analytic techniques used to study them. Importantly, as data accumulate on 16S rRNA gene sequences for named species, many sequences previously labeled as novel, uncultivated phylotypes can now be identified as named species. With the large amount of sequence data available locally and in public databases, it has also become possible to screen out more sequencing artifacts, such as chimeric sequences.
Lactate-producing species.
The distribution among subjects of the primary acid-producing species was complex and is shown by caries stage in Fig. 5. Although it did not significantly increase in this study, Streptococcus mutans was included because of its established relationship with caries. As caries progressed to cavitated and dentin lesions, some subjects had high levels of Lactobacillus species or S. mutans, others had high levels of both, and some had neither Lactobacillus nor S. mutans.
S. mutans and Lactobacillus.
The percent S. mutans was not linearly related to caries progression from healthy to deep caries (estimate by PROC MIXED; not significant). Although initially surprising, examination of levels of S. mutans as caries progressed (Fig. 2) was helpful in understanding the relationship of S. mutans to the natural history of caries. S. mutans levels were elevated in many healthy subjects, and this is consistent with other studies that have demonstrated a low specificity for S. mutans as a predictive test for caries. The standard deviation was very high in this set of healthy samples, and the elevated mean was influenced by a single sample with 74% S. mutans. The percent S. mutans was high in the biofilms over white spots and even higher over cavitated lesions, consistent with a recognized, key role in caries. However, levels of S. mutans were lower in dentin lesions than in cavitated lesions. Because the PROC MIXED estimates were testing a linear relationship, an overall significant increase was not found by this analysis.
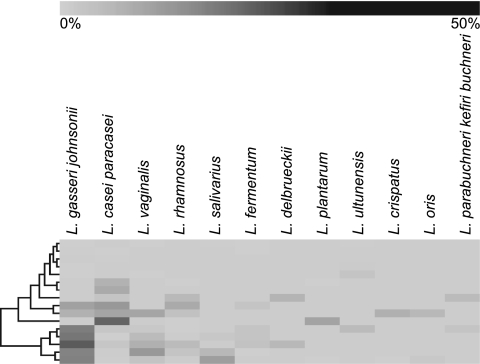
Lactobacillus species represented nearly 14% of the total clones. Twelve Lactobacillus species were identified (Table 1). The most common species were L. gasseri-L. johnsonii (combined because they cannot be distinguished by 16S sequence analysis) and L. casei-L. paracasei (similarly combined). A cluster diagram (Fig. 6) illustrates how the distribution of Lactobacillus species varied among subjects with caries. This distribution of species was similar to that observed in a previous study of carious dentin (8). High species diversity within subjects and high heterogeneity between individuals were observed, consistent with previous findings (9).
FIG. 6.
Hierarchical cluster analysis of Lactobacillus species present in all caries samples.
Levels of all Lactobacillus species increased as caries progressed. It has been previously observed that Lactobacillus species are functionally similar (36), so they were combined for analysis to capture the significance of the group as a whole. Lactobacillus species are acidogenic and aciduric and were the first organisms to be identified as potential caries pathogens (51). They have been found to cause caries when inoculated into gnotobiotic rats (17, 44) but are normally considered to be associated with the progression of caries (48, 51). Lactobacilli have been previously detected in carious dentin (8, 10, 29). It is interesting that in the present study lactobacilli were found only in subjects with caries and rarely in healthy sites in subjects with caries, suggesting that microbiologic screening tests that relied on this group would have a high specificity for diagnosis of established caries.
The lactobacilli became dominant late in the caries process, corroborating previous reports associating Lactobacillus species with progressive caries, but not initiation (51). It can be seen in Fig. 2 that white spot lesions had high levels of S. mutans and virtually no lactobacilli. In cavitated lesions, both S. mutans and Lactobacillus increased, and in dentin, for most subjects S. mutans levels declined and Lactobacillus levels rose steeply. It has been suggested that because they are weakly adherent, lactobacilli depend on S. mutans and other acid producers to initiate the caries lesions that are their ideal niches. It has also been shown that lactobacilli have antimicrobial properties and can inhibit the growth of S. mutans (45, 46). It is unclear what determines whether S. mutans or lactobacilli are the dominant bacteria in late-stage caries, since exploratory analyses comparing subjects with community profiles dominated by S. mutans and Lactobacillus showed no differences in caries severity, fluoride exposure, dietary habits, antibiotic history, smoke exposure, gender, or race.
Other acid producers.
Although Lactobacillus or S. mutans was the numerically dominant acid-producing species in over half of the subjects, they were at low levels or not present in many samples. Other groups of bacteria have been shown to make lactic acid at low pH, and some potential acid producers were observed at high levels in the present study. These included strains of Selenomonas, Neisseria, and Streptococcus mitis. None of these species was significantly higher in caries compared to healthy samples in the current study, so it is likely that only specific strains have cariogenic potential. As discussed below, some strains of S. mitis may produce bases and Neisseria may metabolize lactic acid under some conditions, making it difficult to sort out the role of these species in caries. Metagenomic studies and metabolic profiling are needed to determine the role(s) of these bacteria in caries progression.
Utilization of lactate and other organic acids and caries pathobiology.
The production of lactate in caries-associated ecosystems is clearly critical, but another metabolic activity coupled to the production of lactate is its utilization as a substrate. Caries-associated biofilms often contain high levels of bacteria that utilize organic acids as an energy source. The contribution of lactate-utilizing species to caries pathobiology is not clear. It has been suggested that the net contribution might be an increase in pH due to acid removal. Clinical and laboratory data, however, suggest that the removal of lactate can be part of a detoxification consortium that allows continuing production of high levels of lactate and that ultimately contributes to lowered pH (6). In the present study acid-utilizing bacteria accounted for large fractions of the bacteria at caries sites, including Veillonella, Propionibacterium, and Campylobacter species. Some groups may be capable of both acid production and utilization and thus have a potentially more complex role. These include Neisseria and Selenomonas.
Neisseria.
In the current study Neisseria species were found at relatively high levels in the cavitated samples of two subjects that had no Lactobacillus and only small amounts of S. mutans. Neisseria species have the ability both to metabolize glucose, producing organic acids, and to utilize lactic acid. Type of activity may depend on growth conditions, as strains of N. meningitidis and N. gonorrhoeae have been shown to utilize glucose to produce acetate and lactate, depending on culture conditions (4, 35). N. meningitidis can also utilize lactate (16), and Neisseria strains isolated from dental plaque have been shown to degrade lactate to acetate (21). Thus, the role of Neisseria species in the oral cavity may be complex.
Selenomonas.
In the present study levels of Selenomonas spp. were not significantly associated with disease, but they were found at relatively high levels in three subjects whose cavitated samples did not contain Lactobacillus or S. mutans, suggesting a role in caries. In previous studies uncultivated Selenomonas species were associated with root caries in elderly patients (42) and coronal caries in young children (12). The oral species S. sputigena has been shown to grow on lactate and to produce acetate, propionate, and succinate (43). Selenomonas species have been shown to both ferment glucose and utilize lactate in studies of rumen bacteria (18, 41), suggesting a potentially complex role for Selenomonas in caries progression.
Streptococcus mitis.
In several cavitated samples, S. mitis was observed at higher levels than either Lactobacillus or S. mutans. Overall, increased levels of S. mitis were associated with health (discussed below). However, the contribution of S. mitis to the progression of caries may also be complex. S. mitis can produce ammonia and raise pH, as discussed below. However, it is also known that specific strains of S. mitis are capable of producing acid rapidly, even at low pH (13), and these strains may be important in caries.
Veillonella atypica dispar parvula.
In the current study, Veillonella species were detected in most samples, and levels showed no significant relationship to caries status. Veillonella relies solely on lactate and other organic acids as an energy source, producing lower-pKa acids and potentially raising pH. This might suggest a beneficial effect, but in previous molecular studies Veillonella species were found to be significantly associated with caries in children (1, 5). It has also been shown in vitro that combinations of S. mutans and V. alcalescens have higher acid production than colonies of either species alone (38). Thus, the contribution of Veillonella to caries remains incompletely understood, and its presence may be important to permit the survival of acidogenic species under low-pH conditions.
Propionibacterium.
Propionibacterium FMA5 was significantly associated with caries in the current study. It was observed only in advanced lesions, with mean levels less than 5%. Propionibacterium species have been previously linked to caries in primary and permanent teeth (1, 10) and with root caries in elderly patients (42). Propionibacterium species can metabolize sugars, but it has been reported that when lactate is present, it is the preferred substrate (49). These findings, taken together, suggest that like Veillonella, Propionibacterium may enhance acid production by other species.
Campylobacter.
In the current study, although Campylobacter did not increase overall as caries progressed, it may have been important in certain subjects as part of a functional consortium with Selenomonas species. Campylobacter was the dominant genus in one subject who had no measurable Lactobacillus or S. mutans at caries sites, and this subject had high levels of Selenomonas. Another sample containing Selemononas as the apparently dominant acid producer also contained Campylobacter. Campylobacter species utilize organic acids as their energy source and like, Veillonella and Propionibacterium, may contribute to caries by serving as an acid sink. Other epidemiologic evidence is unclear: Campylobacter showae was recently associated with health and Campylobacter gracilis with caries in permanent teeth (1).
Health-associated species.
Levels of most species decreased as caries progressed, and for nine species these changes were statistically significant, including Streptococcus mitis-S. pneumoniae-S. infantis (combined because they cannot be distinguished by 16S sequence analysis), Corynebacterium matruchotii, Streptococcus gordonii, Streptococcus cristatus, Capnocytophaga gingivalis, Eubacterium IR009, Campylobacter rectus, and Lachnospiraceae sp. C1. Recent clinical studies have found strains of S. mitis and S. oralis (1, 11), S. cristatus (1, 11, 42), and C. gingivalis and C. rectus (1) to be associated with oral health. C. matruchotii is associated with calculus formation (34), which occurs at high pH. S. gordonii has been associated with sound surfaces in a study of nursing bottle caries (31) and has been found to reduce the effects of many S. mutans virulence factors (26, 52). The roles of Eubacterium IR009 and Lachnospiraceae sp. C1 are unknown. Surprisingly, S. sanguinis was not identified as health associated, although it has been in many previous studies (5, 11, 39).
Production of base.
The pH shifts that produce dental caries have primarily been attributed to the production of acid, but health-associated species play an active role by producing basic substances to lower pH (7). They produce ammonia either by catabolizing salivary urea via urease (37) or by catabolizing arginine in the arginine deiminase system. S. mitis, S. pneumoniae, S. gordonii, and S. cristatus express arginine deiminase (14, 15, 20, 28), the first enzyme in the arginine deiminase system, which converts arginine to ammonia, ornithine, and carbon dioxide. Loss of these species may have led to lower pH and greater demineralization.
Catabolism of acid by acid-sensitive species.
As discussed above, some acid-utilizing species can survive at low pH and may ultimately contribute to the survival of cariogenic communities. But acid-utilizing species associated with health may not survive highly acid conditions. The health-associated species C. matruchotii has been shown to utilize carbohydrates and lactate and acetate (22), and Campylobacter species in general utilize organic acids, especially formate and fumarate (47) and lactate (33), which may have implications for the ability of oral Campylobacter species to maintain pH homeostasis in a healthy oral biofilm.
Limitations and future studies.
With sequences for just 50 clones per sample, species present at low levels have a low probability of detection, and the accuracy of levels for more common species is limited. The next generation of sequencing technologies are currently being used for deep sequencing of bacterial samples and offer promise for caries studies. However, the length of reads on the 454 pyrosequencing platform does not yet provide adequate resolution for the streptococci. As technological advances allow longer reads, deep sequencing will provide a powerful tool for studying the microbiology of caries. Primer bias is also an issue with 16S-based studies, and multiple, degenerate primer sets could potentially provide a more representational amplification. Finally, the resolving power of even full-length 16S genes is limited, and metagenomic studies that provide sequence information on whole genomes to allow functional profiling may be necessary to elucidate the microbial etiology of caries. Limitations of the clinical study design include cross-sectional sampling and the pooling of samples from multiple sites, and future studies that examine individual sites over time as caries progresses are needed.
Summary and conclusions.
The bacterial community composition in health and severe caries of the young permanent dentition was compared using Sanger sequencing of the ribosomal 16S rRNA genes. It appears that caries has a heterogeneous etiology, with multiple profiles of acid-producing species observed. Lactobacillus species were the dominant bacteria in this rampant caries cohort, and levels rose significantly as caries progressed from initial to deep lesions. Lactobacilli were not detected in healthy subjects. S. mutans was often observed at high levels in the early stages of caries, but also in some healthy subjects, and it was not statistically significantly associated with caries progression in the overall model. Propionibacterium FMA5 was significantly associated with caries progression but was not found at high levels. Additional candidates include strains of S. mitis, Selenomonas, and Neisseria. An overall loss of community diversity occurred as caries progressed, and a number of species that significantly decreased were identified, including Streptococcus mitis-S. pneumoniae-S. infantis, Corynebacterium matruchotii, Streptococcus gordonii, Streptococcus cristatus, Capnocytophaga gingivalis, Eubacterium IR009, Campylobacter rectus, and Lachnospiraceae sp. C1. The relationship of acid-base metabolism to 16S-based species assignments appears to be complex, and in future studies metagenomic approaches that allow functional profiling of entire genomes will be helpful in elucidating the microbial pathogenesis of caries.
Acknowledgments
This work was supported by grants DE16125, T32DE14320, and F30DE019339 from the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Footnotes
Published ahead of print on 8 September 2010.
REFERENCES
- 1.Aas, J. A., A. L. Griffen, S. R. Dardis, A. M. Lee, I. Olsen, F. E. Dewhirst, E. J. Leys, and B. J. Paster. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Baruque-Ramos, J., H. Hiss, A. Converti, V. M. Goncalves, and I. Raw. 2006. Accumulation of organic acids in cultivations of Neisseria meningitidis C. J. Ind. Microbiol. Biotechnol. 33:869-877. [DOI] [PubMed] [Google Scholar]
- 5.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw, D. J., and P. D. Marsh. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 32:456-462. [DOI] [PubMed] [Google Scholar]
- 7.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Byun, R., M. A. Nadkarni, K. L. Chhour, F. E. Martin, N. A. Jacques, and N. Hunter. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42:3128-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caufield, P. W., Y. Li, A. Dasanayake, and D. Saxena. 2007. Diversity of lactobacilli in the oral cavities of young women with dental caries. Caries Res. 41:2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhour, K. L., M. A. Nadkarni, R. Byun, F. E. Martin, N. A. Jacques, and N. Hunter. 2005. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 43:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corby, P. M., W. A. Bretz, T. C. Hart, N. J. Schork, J. Wessel, J. Lyons-Weiler, and B. J. Paster. 2007. Heritability of oral microbial species in caries-active and caries-free twins. Twin Res. Hum. Genet. 10:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corby, P. M., J. Lyons-Weiler, W. A. Bretz, T. C. Hart, J. A. Aas, T. Boumenna, J. Goss, A. L. Corby, H. M. Junior, R. J. Weyant, and B. J. Paster. 2005. Microbial risk indicators of early childhood caries. J. Clin. Microbiol. 43:5753-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Soet, J. J., B. Nyvad, and M. Kilian. 2000. Strain-related acid production by oral streptococci. Caries Res. 34:486-490. [DOI] [PubMed] [Google Scholar]
- 14.Dong, Y., Y. Y. Chen, J. A. Snyder, and R. A. Burne. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Encheva, V., S. E. Gharbia, R. Wait, S. Begum, and H. N. Shah. 2006. Comparison of extraction procedures for proteome analysis of Streptococcus pneumoniae and a basic reference map. Proteomics 6:3306-3317. [DOI] [PubMed] [Google Scholar]
- 16.Erwin, A. L., and E. C. Gotschlich. 1993. Oxidation of D-lactate and L-lactate by Neisseria meningitidis: purification and cloning of meningococcal D-lactate dehydrogenase. J. Bacteriol. 175:6382-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzgerald, R. J., H. V. Jordan, and H. O. Archard. 1966. Dental caries in gnotobiotic rats infected with a variety of Lactobacillus acidophilus. Arch. Oral Biol. 11:473-476. [DOI] [PubMed] [Google Scholar]
- 18.Hespell, R. B., B. J. Paster, and F. E. Dewhirst. 2006. The genus Selenomonas, p. 982-990. In M. Dworkin and S. Falkow (ed.), The prokaryotes: a handbook on the biology of bacteria, 3rd ed., vol. 4. Springer, New York, NY. [Google Scholar]
- 19.Hicks, M. J., and C. M. Flaitz. 1993. Epidemiology of dental caries in the pediatric and adolescent population: a review of past and current trends. J. Clin. Pediatr. Dent. 18:43-49. [PubMed] [Google Scholar]
- 20.Hiraoka, B. Y., M. Mogi, K. Fukasawa, and M. Harada. 1986. Coordinate repression of arginine aminopeptidase and three enzymes of the arginine deiminase pathway in Streptococcus mitis. Biochem. Int. 12:881-887. [PubMed] [Google Scholar]
- 21.Hoshino, E., and A. Araya. 1980. Lactate degradation by polysaccharide-producing Neisseria isolated from human dental plaque. Arch. Oral Biol. 25:211-212. [DOI] [PubMed] [Google Scholar]
- 22.Howell, A., Jr., and L. Pine. 1961. The classification of organisms termed Leptotrichia (Leptothrix) buccalis. IV. Physiological and biochemical characteristics of Bacterionema matruchotii. Bacteriol. Rev. 25:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanasi, E., I. Johansson, S. C. Lu, N. R. Kressin, M. E. Nunn, R. Kent, Jr., and A. C. Tanner. 2010. Microbial risk markers for childhood caries in pediatricians' offices. J. Dent. Res. 89:378-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. U. S. A. 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuramitsu, H. K., and B. Y. Wang. 2006. Virulence properties of cariogenic bacteria. BMC Oral Health 6(Suppl. 1):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y., Y. Ge, D. Saxena, and P. W. Caufield. 2007. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J. Clin. Microbiol. 45:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, X., R. J. Lamont, J. Wu, and H. Xie. 2008. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J. Bacteriol. 190:4367-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loesche, W. J., and S. A. Syed. 1973. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 7:201-216. [DOI] [PubMed] [Google Scholar]
- 30.Magurran, A. E. 2004. Measuring biological diversity. Wiley-Blackwell, Hoboken, NJ.
- 31.Marchant, S., S. R. Brailsford, A. C. Twomey, G. J. Roberts, and D. Beighton. 2001. The predominant microflora of nursing caries lesions. Caries Res. 35:397-406. [DOI] [PubMed] [Google Scholar]
- 32.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 33.Mohammed, K. A., R. J. Miles, and M. A. Halablab. 2004. The pattern and kinetics of substrate metabolism of Campylobacter jejuni and Campylobacter coli. Lett. Appl. Microbiol. 39:261-266. [DOI] [PubMed] [Google Scholar]
- 34.Moorer, W. R., J. M. Ten Cate, and J. F. Buijs. 1993. Calcification of a cariogenic Streptococcus and of Corynebacterium (Bacterionema) matruchotii. J. Dent. Res. 72:1021-1026. [DOI] [PubMed] [Google Scholar]
- 35.Morse, S. A., S. Stein, and J. Hines. 1974. Glucose metabolism in Neisseria gonorrhoeae. J. Bacteriol. 120:702-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nascimento, M. M., V. V. Gordan, C. W. Garvan, C. M. Browngardt, and R. A. Burne. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol. Immunol. 24:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noorda, W. D., D. J. Purdell-Lewis, A. M. van Montfort, and A. H. Weerkamp. 1988. Monobacterial and mixed bacterial plaques of Streptococcus mutans and Veillonella alcalescens in an artificial mouth: development, metabolism, and effect on human dental enamel. Caries Res. 22:342-347. [DOI] [PubMed] [Google Scholar]
- 39.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 40.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paynter, M. J., and S. R. Elsden. 1970. Mechanism of propionate formation by Selenomonas ruminantium, a rumen micro-organism. J. Gen. Microbiol. 61:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Preza, D., I. Olsen, J. A. Aas, T. Willumsen, B. Grinde, and B. J. Paster. 2008. Bacterial profiles of root caries in elderly patients. J. Clin. Microbiol. 46:2015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ricke, S. C., and D. M. Schaefer. 1996. Growth and fermentation responses of Selenomonas ruminantium to limiting and non-limiting concentrations of ammonium chloride. Appl. Microbiol. Biotechnol. 46:169-175. [DOI] [PubMed] [Google Scholar]
- 44.Rosen, S., W. S. Lenney, and J. E. O'Malley. 1968. Dental caries in gnotobiotic rats inoculated with Lactobacillus casei. J. Dent. Res. 47:358-363. [DOI] [PubMed] [Google Scholar]
- 45.Simark-Mattsson, C., C. G. Emilson, E. G. Hakansson, C. Jacobsson, K. Roos, and S. Holm. 2007. Lactobacillus-mediated interference of mutans streptococci in caries-free vs. caries-active subjects. Eur. J. Oral Sci. 115:308-314. [DOI] [PubMed] [Google Scholar]
- 46.Strahinic, I., M. Busarcevic, D. Pavlica, J. Milasin, N. Golic, and L. Topisirovic. 2007. Molecular and biochemical characterizations of human oral lactobacilli as putative probiotic candidates. Oral Microbiol. Immunol. 22:111-117. [DOI] [PubMed] [Google Scholar]
- 47.Tanner, A., C. H. Lai, and M. Maiden. 1992. Characteristics of oral gram negative species, p. 299-341. In J. Slots and M. A. Taubman (ed.), Contemporary oral microbiology and immunology, 1st ed. Mosby-Year Book, St. Louis, MO.
- 48.Tanzer, J. M., J. Livingston, and A. M. Thompson. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028-1037. [PubMed] [Google Scholar]
- 49.Tyree, R. W., E. C. Clausen, and J. L. Gaddy. 1990. The production of propionic acid from sugars by fermentation through lactic acid as an intermediate. J. Chem. Technol. Biotechnol. 50:157-166. [Google Scholar]
- 50.U.S. Department of Health and Human Services. 2000. Oral health in america: a report of the Surgeon General. Executive summary. National Institute of Dental and Craniofacial Research, National Institutes of Health, Rockville, MD.
- 51.van Houte, J. 1994. Role of micro-organisms in caries etiology. J. Dent. Res. 73:672-681. [DOI] [PubMed] [Google Scholar]
- 52.Wang, B. Y., and H. K. Kuramitsu. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 71:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]