Abstract
We have recently developed a PCR assay for detection of Mycobacterium spp. at the genus level based on the Cobas Amplicor platform. The sensitivities for smear-positive and smear-negative specimens were found to be 100% and 47.9%, respectively. The specificity was 97.7%, the positive predictive value 84.6%, and the negative predictive value 93.1%. In a follow-up study, we have systematically evaluated the Mycobacterium genus assay in parallel with the Cobas Amplicor Mycobacterium tuberculosis assay on 2,169 clinical specimens, including respiratory and nonrespiratory specimens. Based on the genus assay, nontuberculous mycobacteria were readily detected and identified to the species level by PCR-mediated sequencing. In addition, our data point to a limited specificity of the Cobas Amplicor M. tuberculosis assay.
Nontuberculous mycobacteria (NTM) are frequently associated with human disease. Infections with NTM are increasingly observed in immunocompromised patients (10, 16), although patients without underlying medical conditions are also affected. Diseases caused by NTM include lung disease (e.g., Mycobacterium abscessus, Mycobacterium kansasii), cutaneous ulcers (e.g., Mycobacterium marinum), disseminated infections (e.g., Mycobacterium genavense), lymphadenitis (e.g., Mycobacterium avium), and joint infections (e.g., Mycobacterium haemophilum) (for a review, see references 5 and 16). Rapid and reliable laboratory detection of NTM is crucial for clinical management and proper antibiotic therapy. Molecular genetic assays for the detection of NTM in clinical specimens are only infrequently implemented in routine diagnostics. With a view to developing an assay that is capable of detecting a large number of nontuberculous mycobacteria in clinical specimens, we have recently developed a semiautomated PCR-based assay for NTM on the basis of the Roche Cobas Amplicor platform (8). The sensitivity of the genus assay was 100% for smear-positive specimens and 47.9% for smear-negative specimens. The specificity of the genus assay was 97.7%, the positive predictive value (PPV) 84.6%, and the negative predictive value 93.1% (8). These values are comparable to those published for the Cobas Amplicor Mycobacterium tuberculosis test (11; reviewed in reference 9). We have now extensively evaluated the Mycobacterium genus assay under routine laboratory conditions.
MATERIALS AND METHODS
Decontamination of specimens, microscopy, and culture.
Clinical specimens were decontaminated using the sodium hydroxide method for samples from sterile sites and the N-acetyl-l-cysteine-sodium hydroxide method for respiratory samples (6). Auramine-rhodamine fluorochrome staining was used for microscopic examination; positive microscopy results were confirmed using Ziehl-Neelsen staining (6). For the recovery of mycobacteria from culture, standard media were inoculated (7H11 plates and BBL MGIT [Becton, Dickinson and Company]) and maintained for 7 weeks at 37°C. Mycobacteria were identified by 16S rRNA gene sequence analysis as described previously (7).
Clinical specimens.
Over a period of 12 months (April 2008 to March 2009), the Cobas Amplicor M. tuberculosis and Mycobacterium genus assays were performed in parallel on all specimens for which molecular detection of M. tuberculosis or NTM was requested. These included respiratory as well as nonrespiratory specimens.
DNA extraction.
DNA was extracted from decontaminated samples (0.5 ml) by using the respiratory specimen preparation kit (Roche Diagnostics, Switzerland) according to the instructions for the Cobas Amplicor M. tuberculosis test (Roche Diagnostics, 2007) (12).
Detection of mycobacterial DNA.
The Cobas Amplicor M. tuberculosis test was performed according to the manufacturer's instructions (Roche Diagnostics, Switzerland), and the Mycobacterium genus test was performed as described previously (8).
According to the manufacturer, the Cobas Amplicor M. tuberculosis test is considered valid if the optical density at 660 nm (OD660) of the positive control is >2.0 and the OD660 of the negative control is <0.25. A specimen is scored positive for M. tuberculosis when the OD660 is ≥0.35. A sample is scored negative for M. tuberculosis if the OD660 of the sample is <0.35 and the OD660 of the internal inhibition control is ≥0.35 (12).
The Mycobacterium genus test adapted to the Cobas Amplicor platform was performed as described previously by using a genus-specific capture probe (5′-TTTCACGAACAACGCGACAA-3′) coupled to magnetic beads (8). For the Mycobacterium genus assay, a result was considered valid if the OD660 of the positive control was ≥2.0 and the OD660 of the negative control was <0.5. A specimen was considered positive for mycobacteria if the OD660 was ≥0.5 and at least 2-fold higher than the background (negative control). A sample was considered negative if the OD660 of the sample was <0.5 and the OD660 of the internal control was ≥0.35. Specimens were scored positive for NTM if the OD660 was ≥0.50 for the genus assay and the Cobas Amplicor M. tuberculosis result was negative (OD660, <0.35).
Amplification, DNA purification, and sequencing of positive samples and cultures.
Samples that were scored positive by the genus assay or for which the OD660 by the Amplicor M. tuberculosis assay was ≥0.35 and <2.0 were subjected to PCR-mediated 16S rRNA gene sequence analysis. For gene amplification, the Cobas Amplicor pan-Mycobacterium primers KY18 and KY75 or primers 283 and 264 were used in separate PCRs as described previously (8). PCR products were sequenced using primer Mbakt-14 (3) and were analyzed using SmartGene IDNS software and databases (SmartGene, Zug, Switzerland). If unsatisfactory PCR or sequencing results were obtained, reamplification was done with primers KY18 and 259 or with primers 283 and 259. Homology analysis and species identification were carried out as described previously (2, 3, 7).
Sequencing of 16S rRNA genes from cultures was performed using primers 283 and 264 for PCR amplification and primer Mbakt-14 for sequencing (3, 8). In case of M. chelonae complex identification, sequence analysis of rpoB (1) was used for species assignment, e.g., to M. massiliense, M. bolletii, M. abscessus, or M. chelonae. In case of M. kansasii/gastri identification, sequence analysis of hsp65 was used for differentiation (13).
RESULTS
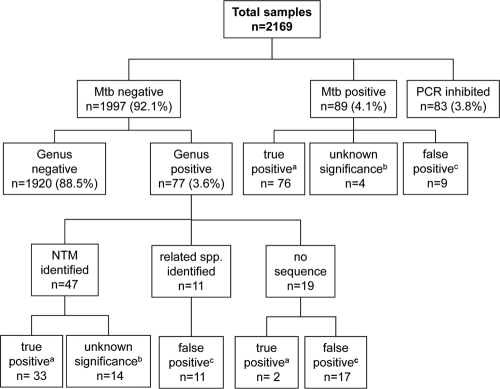
Over a period of 12 months, 2,169 clinical samples were tested for the presence of mycobacterial DNA in a prospective study design (Fig. 1). A negative PCR result in both assays was obtained for 1,920 (88.5%) of the specimens. Inhibition of the PCR, as indicated by a negative internal control as used in the M. tuberculosis assay, occurred for 83 (3.8%) of the specimens. Eighty-nine (4.1%) samples tested positive by the Cobas Amplicor M. tuberculosis assay, indicating the presence of M. tuberculosis. Seventy-seven (3.6%) samples tested positive by the Mycobacterium genus assay and negative by the Cobas Amplicor M. tuberculosis assay, indicating the presence of NTM.
FIG. 1.
Clinical specimens tested by the Cobas Amplicor M. tuberculosis assay and the Mycobacterium genus assay. Mtb, M. tuberculosis. A true-positive result (a) was confirmed by at least two other positive results, e.g., by smear, culture, or another sample from the same patient. A result of unknown significance (b) was obtained by sequence identification, but no other positive result was obtained. A result was considered false positive (c) when no sequence was obtained or when a related species was identified and no other positive result was obtained.
The 77 samples positive by the Mycobacterium genus assay and negative by the M. tuberculosis assay included 60 respiratory specimens, 8 bronchoalveolar lavage samples, 3 biopsy specimens, 2 wound specimens, 1 cerebrospinal fluid sample, and 3 urine samples; 22 of the 77 genus assay-positive samples were smear positive. For 47 of the 77 samples (61%), 16S rRNA gene sequence analysis of the amplicon resulted in the identification of an NTM species (Table 1). NTM species detected by the genus assay included M. chelonae complex (n = 12), M. gordonae (n = 4), M. fortuitum (n = 2), M. intracellulare (n = 7), M. avium (n = 7), M. kansasii/gastri (n = 4), M. celatum (n = 1), M. mucogenicum/phocaicum (n = 1), M. aubagnense (n = 1), M. frederiksbergense (n = 2), and M. terrae (n = 1). For 5 samples, the 16S rRNA gene sequence obtained had no match of significant homology in the database to allow for species assignment, resulting in assignment to genus level; these sequences were reported as Mycobacterium spp. For 26 of the 47 specimens with assignment of the PCR amplicon to the NTM species level, an NTM was recovered by culture, and the 16S rRNA gene sequence of the NTM was identical to the DNA sequence obtained by molecular analysis of the clinical specimen (Table 1).
TABLE 1.
Analysis of specimens positive by the Mycobacterium genus PCR, where sequence analysis of the PCR product resulted in the identification of NTM (n = 47)
| Specimena | OD660 by: |
Identification by 16S rRNA gene sequenceb | Result by: |
Interpretation | ||
|---|---|---|---|---|---|---|
| Genus assay | M. tuberculosis assay | Smear | Culturec | |||
| Respiratory | >4.00 | <0.35 | M. chelonae complex | Positive | M. abscessus | True positive |
| Respiratory | >4.00 | <0.35 | M. chelonae complex | Positive | M. chelonae complex | True positive |
| Respiratory | >4.00 | <0.35 | M. gordonae | Positive | M. gordonae | True positive |
| Respiratory | >4.00 | <0.35 | M. chelonae complex | Positive | M. abscessus | True positive |
| Respiratory | >4.00 | <0.35 | M. chelonae complex | Positive | M. abscessus | True positive |
| Respiratory | >4.00 | <0.35 | M. fortuitum | Positive | M. fortuitum | True positive |
| BAL | >4.00 | <0.35 | M. intracellulare | Positive | M. intracellulare | True positive |
| Respiratory | >4.00 | <0.35 | M. intracellulare | Positive | M. intracellulare | True positive |
| Respiratory | 3.95 | <0.35 | M. chelonae complex | Positive | M. abscessus | True positive |
| Respiratory | 3.78 | <0.35 | M. chelonae complex | Positive | M. abscessus | True positive |
| Wound | 3.76 | <0.35 | M. intracellulare | Positive | M. intracellulare | True positive |
| Respiratory | 3.64 | <0.35 | M. gordonae | Positive | M. gordonae | True positive |
| Wound | 3.64 | <0.35 | M. intracellulare | Positive | M. intracellulare | True positive |
| Respiratory | 3.47 | <0.35 | M. avium | Positive | M. avium | True positive |
| Respiratory | 3.46 | <0.35 | M. kansasii/gastri | Positive | M. kansasii | True positive |
| Respiratory | 3.25 | <0.35 | M. avium | Positive | M. avium | True positive |
| Respiratory | 3.01 | <0.35 | M. avium | Positive | M. avium | True positive |
| Respiratory | 2.95 | <0.35 | M. intracellulare | Positive | M. intracellulare | True positive |
| BAL | 2.27 | <0.35 | M. kansasii/gastri | Positive | M. kansasii | True positive |
| Respiratory | >4.00 | <0.35 | M. intracellulare | Negative | M. intracellulare | True positive |
| Respiratory | >4.00 | <0.35 | M. kansasii/gastri | Negative | M. kansasii | True positive |
| Respiratory | 3.95 | <0.35 | M. avium | Negative | M. avium | True positive |
| Respiratory | 3.95 | <0.35 | M. avium | Negative | M. avium | True positive |
| Respiratory | 3.78 | <0.35 | M. chelonae complex | Negative | M. abscessus | True positive |
| Respiratory | 3.07 | <0.35 | M. gordonae | Negative | M. gordonae | True positive |
| Respiratory | 0.53 | <0.35 | M. chelonae complex | Negative | M. bolletii | True positive |
| Respiratory | >4.00 | <0.35 | M. chelonae complex | Positive | Negative | True positive |
| Respiratory | >4.00 | <0.35 | M. chelonae complex | Positive | Negative | True positive |
| Respiratory | 3.47 | <0.35 | M. chelonae complex | Negative | Negative | True positived |
| BAL | 2.76 | <0.35 | M. celatum | Negative | Negative | True positived |
| Respiratory | 2.62 | <0.35 | M. gordonae | Negative | Negative | True positived |
| Respiratory | 2.35 | <0.35 | M. intracellulare | Not done | Negative | True positived |
| Respiratory | 1.65 | <0.35 | M. avium | Not done | Negative | True positived |
| Respiratory | 3.65 | <0.35 | M. aubagnense | Negative | Negative | Unknown significance |
| Respiratory | 3.34 | <0.35 | M. avium | Negative | Negative | Unknown significance |
| Respiratory | 3.21 | <0.35 | Mycobacterium sp. | Negative | Negative | Unknown significance |
| Respiratory | 2.82 | <0.35 | M. frederiksbergense | Negative | Negative | Unknown significance |
| Respiratory | 2.73 | <0.35 | Mycobacterium sp. | Negative | Negative | Unknown significance |
| Respiratory | 2.17 | <0.35 | M. kansasii/gastri | Negative | Negative | Unknown significance |
| Respiratory | 0.89 | <0.35 | M. mucogenicum | Negative | Negative | Unknown significance |
| Respiratory | 0.77 | <0.35 | M. fortuitum | Negative | Negative | Unknown significance |
| Respiratory | 0.68 | <0.35 | Mycobacterium sp. | Negative | Negative | Unknown significance |
| Respiratory | 0.54 | <0.35 | Mycobacterium sp. | Negative | Negative | Unknown significance |
| Respiratory | 0.52 | <0.35 | M. chelonae complex | Negative | Negative | Unknown significance |
| Respiratory | 0.52 | <0.35 | M. frederiksbergense | Negative | Negative | Unknown significance |
| BAL | 0.51 | <0.35 | M. terrae | Negative | Negative | Unknown significance |
| CSF | 0.50 | <0.35 | Mycobacterium sp. | Not done | Not done | Unknown significance |
BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid.
Sequencing was performed on PCR products obtained by amplification of nucleic acids extracted from the clinical sample.
Identification by 16S rRNA sequence analysis, followed when necessary by sequencing and homology analysis of rpoB (in the case of M. chelonae complex) or hsp65 (in the case of M. kansasii/gastri).
A corresponding NTM species was recovered by culture in another patient sample or samples.
For 30 samples that were positive by the Mycobacterium genus assay, sequence analysis of the PCR product did not result in NTM species assignment (Table 2); for 2 of these samples, NTM were recovered by culture. For 11/28 samples, sequencing of the PCR product revealed species of closely related genera, e.g., Corynebacterium (n = 10) and Gordonia (n = 1) (Table 2). For 17/28 samples, no readable 16S rRNA gene sequence was obtained, even though two different PCR protocols were used.
TABLE 2.
Analysis of specimens positive by the Mycobacterium genus PCR, where sequence analysis of the PCR product did not result in NTM species assignment (n = 30)
| Specimena | OD660 by: |
Identification by 16S rRNA gene sequenceb | Result by: |
Interpretation | ||
|---|---|---|---|---|---|---|
| Genus assay | Cobas Amplicor M. tuberculosis test | Smear | Culture | |||
| Biopsy | 2.63 | <0.35 | No sequence | Negative | M. intracellulare | True positive |
| Respiratory | 0.99 | <0.35 | No sequence | Positive | M. avium | True positive |
| Respiratory | 3.64 | <0.35 | Corynebacterium sp. | Negative | Negative | False positive |
| Urine | 3.14 | <0.35 | C. genitalium | Not done | Negative | False positive |
| Respiratory | 2.67 | <0.35 | Corynebacterium sp. | Negative | Negative | False positive |
| Urine | 2.53 | <0.35 | C. amycolatum | Not done | Negative | False positive |
| BAL | 1.56 | <0.35 | C. pseudogenitalium | Negative | Negative | False positive |
| Biopsy | 1.47 | <0.35 | C. afermentans | Negative | Negative | False positive |
| Respiratory | 0.88 | <0.35 | C. propinquum | Negative | Negative | False positive |
| BAL | 0.63 | <0.35 | Gordonia sp. | Negative | Negative | False positive |
| Respiratory | 0.58 | <0.35 | C. propinquum | Negative | Negative | False positive |
| Respiratory | 0.51 | <0.35 | C.argentoratense | Negative | Negative | False positive |
| Respiratory | 0.50 | <0.35 | C. propinquum | Negative | Negative | False positive |
| Respiratory | 3.95 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 2.04 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 1.64 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 1.29 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 1.09 | <0.35 | No sequence | Negative | Not done | False positive |
| Respiratory | 0.91 | <0.35 | No sequence | Negative | Not done | False positive |
| Respiratory | 0.89 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 0.89 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 0.74 | <0.35 | No sequence | Negative | Negative | False positive |
| BAL | 0.68 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 0.61 | <0.35 | No sequence | Negative | Negative | False positive |
| BAL | 0.58 | <0.35 | No sequence | Negative | Negative | False positive |
| Biopsy | 0.55 | <0.35 | No sequence | Negative | Negative | False positive |
| Urine | 0.53 | <0.35 | No sequence | Not done | Negative | False positive |
| Respiratory | 0.53 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 0.52 | <0.35 | No sequence | Negative | Negative | False positive |
| Respiratory | 0.50 | <0.35 | No sequence | Negative | Negative | False positive |
BAL, bronchoalveolar lavage.
Sequencing was performed on PCR products obtained by amplification of nucleic acids extracted from the clinical sample. “No sequence” means that no sequence was obtained from the amplification product.
To determine the relevance of a positive Mycobacterium genus PCR result, we scored positive samples as true positives if either (i) the sample was smear positive, (ii) the corresponding NTM was recovered by culture, or (iii) the corresponding NTM was recovered from other samples from the patient. According to these criteria, 35 of the 77 genus assay-positive samples (45%) were regarded as true positive (Fig. 1). For another 14 samples (18%), sequencing of the PCR product revealed NTM, but none of the criteria defined above was fulfilled; we considered these results to be of unknown significance. In 28 samples (36%), we failed to identify an NTM by sequence analysis of the PCR product, and we considered these genus PCR results to be false positive. The majority of true-positive samples showed OD660 values of >2.0 (32/35); in comparison, the majority of samples that were scored as false positive had OD660 values of <2.0 (22/28).
Eighty-nine (4.1%) samples were positive by the Cobas Amplicor M. tuberculosis test. All samples with OD660 values of ≥2.0 (n = 61 [69%]) were either smear positive or positive for M. tuberculosis by culture. For samples with OD660 values of <2.0 (n = 28 [31%]), we analyzed the specificity of the Cobas Amplicor M. tuberculosis assay by sequencing the PCR products obtained with the primers used in the Cobas Amplicor system and the PCR product obtained with more-specific primers for Mycobacterium, in order to prevent preferential amplification of related genera due to larger DNA amounts (Table 3). A sample was scored as true positive if either (i) the sequence analysis of the 16S rRNA gene PCR product obtained directly from the clinical sample revealed M. tuberculosis complex, (ii) the sample was smear positive, or (iii) M. tuberculosis was cultured from the corresponding sample or from other samples of the patient. A sample was scored negative for M. tuberculosis if the sample and at least two additional samples from the patient were negative in all tests. Otherwise, the positive PCR result of the sample was considered to be of unknown significance.
TABLE 3.
Analysis of specimens positive by the Cobas Amplicor M. tuberculosis test with OD660 values of <2.0 (n = 28)
| Specimena | OD660 by the M. tuberculosis test | Identification by 16S RNA sequencingb | Result by: |
Detection of M. tuberculosis in other patient sample(s)d | Interpretation | |
|---|---|---|---|---|---|---|
| Smear | Culturec | |||||
| Respiratory | 0.44 | M. tuberculosis complex | Positive | M. tuberculosis | ND | True positive |
| Biopsy | 1.65 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| BAL | 1.26 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| Respiratory | 0.37 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| Respiratory | 1.52 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| Biopsy | 1.16 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| Biopsy | 0.39 | No sequence | Negative | M. tuberculosis | ND | True positive |
| Biopsy | 1.01 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| Respiratory | 0.74 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| BAL | 1.36 | M. tuberculosis complex | Negative | M. tuberculosis | ND | True positive |
| Respiratory | 0.59 | No sequence | Negative | M. tuberculosis | ND | True positive |
| Biopsy | 1.84 | M. tuberculosis complex | Negative | Negative | Negative | True positive |
| Respiratory | 1.64 | M. tuberculosis complex | Negative | Negative | Negative | True positive |
| Stool | 0.60 | M. tuberculosis complex | Negative | Negative | Positive | True positive |
| Biopsy | 0.83 | M. tuberculosis complex | Negative | Negative | Positive | True positive |
| Respiratory | 0.74 | C. propinquum | Negative | Negative | No other samples | Unknown significance |
| Respiratory | 0.46 | M. xenopi | Negative | Negative | No other samples | Unknown significance |
| Respiratory | 1.16 | C. pseudodiphtheriticum, C. durum | Negative | Negative | Positive | Unknown significance |
| Respiratory | 0.68 | C. propinquum | Negative | Negative | No other samples | Unknown significance |
| Respiratory | 0.96 | M. abscessus | Positive | M. abscessus | Negative | False positive |
| Respiratory | 0.62 | Dietzia sp. | Negative | Negative | Negative | False positive |
| Respiratory | 0.40 | C. propinquum | Negative | Negative | Negative | False positive |
| Respiratory | 0.76 | C. pseudodiphtheriticum | Negative | Negative | Negative | False positive |
| Respiratory | 1.68 | No sequence | Negative | Negative | Negative | False positive |
| Respiratory | 0.45 | C. durum | Negative | Negative | Negative | False positive |
| BAL | 1.38 | No sequence | Negative | Negative | Negative | False positive |
| Respiratory | 0.46 | No sequence | Negative | Negative | Negative | False positive |
| Respiratory | 0.42 | C. propinquum | Negative | Negative | Negative | False positive |
BAL, bronchoalveolar lavage.
Sequencing was performed from PCR products directly obtained by amplification of nucleic acids extracted from the clinical sample. “No sequence” means that no sequence was obtained from the amplification product.
Identification by 16S rRNA sequence analysis.
ND, not determined.
Investigation of the 28 samples positive by the Cobas Amplicor M. tuberculosis assay with OD660 values of <2.0 (Table 3) showed that 11/28 were culture positive for M. tuberculosis; for 9 of these, sequence analysis of the PCR product obtained by amplification of nucleic acid extracted from the clinical samples revealed a 16S RNA sequence characteristic of M. tuberculosis. Seventeen of 28 samples did not show growth of M. tuberculosis in culture. Sequence analysis of the PCR products obtained from these 17 clinical samples revealed the presence of M. tuberculosis in 4. For 10 specimens, sequence analysis revealed an NTM (n = 2) or a species of a closely related genus, such as Corynebacterium spp. (n = 7) and a Dietzia sp. (n = 1). For 5 samples positive by the M. tuberculosis assay, no readable sequence could be obtained (Table 3). Altogether, 76 of the 89 samples (85.4%) that were positive by the Cobas Amplicor M. tuberculosis assay were considered true positive. For 4 specimens (4.5%), the positive result was considered to be of unknown significance, and for 9 samples (10.1%), the Cobas Amplicor M. tuberculosis result was considered false positive. For each patient with a positive result of unknown significance or a false-positive result, the clinical history was evaluated. No history of Mycobacterium infections was found for any of these patients.
DISCUSSION
We have recently developed a Mycobacterium genus assay based on the Cobas Amplicor platform (8). Here we report on a 12-month prospective evaluation for detection and identification of Mycobacterium spp. in clinical samples in which we combined the Cobas Amplicor M. tuberculosis test with the Mycobacterium genus assay. All clinical samples submitted for Cobas Amplicor M. tuberculosis testing were subjected to the Mycobacterium genus test in parallel. Direct 16S rRNA gene amplification of nucleic acids extracted from the clinical sample, followed by sequence analysis, facilitated rapid identification of the NTM at the species level for most of the samples positive by the genus assay (Table 1). The majority of NTM identified were found to represent well-established clinical pathogens, while we consider M. frederiksbergense (17), M. aubagnense, M. terrae, M. gordonae, and the unclassified Mycobacterium spp. to be of little clinical relevance, if any. Specimens with OD660 values of ≥2.0 by the Mycobacterium genus assay were frequently confirmed by other criteria for the presence of NTM, in contrast to specimens with OD660 values of <2.0. Such low OD660 values are probably due to cross-reactivity of the genus probe or very low numbers of NTM. Cross-reactivity of the genus probe with other closely related species was indicated by sequence analysis of the amplicon that resulted in the identification of closely related species, such as Corynebacterium and Gordonia spp. (Table 2). In silico homology analysis of the Mycobacterium genus probe shows 3 or 4 mismatches, which are predominantly positioned in the middle of the sequence, with the most closely related genera (Table 4). The temperature conditions of the Cobas Amplicor protocol cannot be changed. To increase the specificity of the Mycobacterium genus assay, the sequence of the PCR product must be analyzed before a positive result is reported. In 47 of the genus assay-positive samples, a nontuberculous mycobacterium was identified by sequence analysis; of these, 33 were true positive as judged by additional criteria. The other 14 samples were considered to be of unknown significance. On the basis of these results, patients were not treated. For each patient, we have evaluated the clinical history and have not found additional positive clinical samples or a history of Mycobacterium infections.
TABLE 4.
Homology analysis of the Mycobacterium genus probe with species of the most closely related genera
| Bacterial species | GenBank accession no.a | DNA sequenceb |
|---|---|---|
| Mycobacterium genusc | 5′-TTTCACGAACAACGCGACAA-3′ | |
| Corynebacterium propinquum | AY244785 | 5′-......AG..G...T.....-3′ |
| Corynebacterium durum | AF537593 | 5′-......AG..G........C-3′ |
| Corynebacterium afermentans | X82054 | 5′-..A...A...G.........-3′ |
| Corynebacterium pseudodiphtheriticum | AJ439343 | 5′-......AG..G...T.....-3′ |
| Corynebacterium pseudotuberculosis | X81916 | 5′-......AG..G.........-3′ |
| Corynebacterium tuberculostearicum | AJ438044 | 5′-......AG..G.........-3′ |
| Corynebacterium amycolatum | FN668737 | 5′-......AG..G.........-3′ |
| Corynebacterium argentoratense | AF537589 | 5′-......AG..G.........-3′ |
| Nocardia pneumoniae | GQ853075 | 5′-......AG..G.........-3′ |
| Gordonia bronchialis | CP001802 | 5′-......AG..G.........-3′ |
| Tsukamurella pulmonis | AY741505 | 5′-......AG..G...T.....-3′ |
| Dietzia maris | X79290 | 5′-......AG..G...T.....-3′ |
| Rhodococcus equi | AY741716 | 5′-......AG..G.........-3′ |
For the corresponding 16S rRNA gene sequence.
Dots represent nucleotides identical to those in the Mycobacterium genus probe sequence.
The Mycobacterium genus probe was published previously (7).
The Cobas Amplicor M. tuberculosis assay is commonly regarded as a reliable assay for the majority of specimens tested (4, 11, 15). However, 9 of 28 samples with OD660 values of <2.0 could not be confirmed as positive for M. tuberculosis by other criteria, such as smear positivity, sequence analysis of the amplicon, recovery from culture, or the presence of M. tuberculosis in additional samples from the patient. This resulted in a PPV of 89.9%, which is in accordance with the results of other studies (9). The detection in these samples of closely related bacterial species of the genera Corynebacterium and Dietzia suggests cross-reactivity of the Cobas Amplicor M. tuberculosis assay, resulting in false-positive test results. It has been noted previously that the Cobas Amplicor pan-Mycobacterium primers KY18 and KY75 are able to amplify the 16S rRNA genes of species closely related to Mycobacterium, such as Corynebacterium, Nocardia, and Rhodococcus species (14). Consequently, the corresponding genus assay values of these 9 false-positive test results did not distinguish true- from false-positive results, probably due to the cross-reactivity of the genus probe, as discussed above.
We conclude that (i) a molecular detection assay for NTM, followed by nucleic acid sequence analysis allowing species assignment, is a valuable tool for the rapid detection of NTM in clinical specimens and (ii) sequence analysis of the amplicon is required when Cobas Amplicor M. tuberculosis results with OD660 values of <2.0 are obtained.
Acknowledgments
This work was supported in part by the University of Zurich and the BAG (Federal Office of Public Health; Switzerland).
We have no conflict of interest to disclose.
Footnotes
Published ahead of print on 8 September 2010.
REFERENCES
- 1.Adékambi, T., P. Colston, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., R. Zbinden, S. Abels, B. Böddinghaus, M. Altwegg, and E. C. Böttger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eing, B. R., A. Becker, A. Sohns, and R. Ringelmann. 1998. Comparison of Roche COBAS Amplicor Mycobacterium tuberculosis assay with in-house PCR and culture for detection of M. tuberculosis. J. Clin. Microbiol. 36:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heifets, L. 2004. Mycobacterial infections caused by nontuberculous mycobacteria. Semin. Respir. Crit. Care Med. 25:283-295. [DOI] [PubMed] [Google Scholar]
- 6.Isenberg, H. D. (ed.). 1992. Clinical microbiology procedures handbook, vol. 1. American Society for Microbiology, Washington, DC.
- 7.Kirschner, P., J. Rosenau, B. Springer, K. Teschner, K. Feldmann, and E. C. Böttger. 1996. Diagnosis of mycobacterial infections by nucleic acid amplification: 18-month prospective study. J. Clin. Microbiol. 34:304-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peter-Getzlaff, S., J. Lüthy, B. Böddinghaus, E. C. Böttger, and B. Springer. 2008. Development and evaluation of a molecular assay for detection of nontuberculous mycobacteria by use of the COBAS Amplicor platform. J. Clin. Microbiol. 46:4023-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piersimoni, C., and C. Scarparo. 2003. Relevance of commercial amplification methods for direct detection of Mycobacterium tuberculosis complex in clinical samples. J. Clin. Microbiol. 41:5355-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reischl, U., N. Lehn, H. Wolf, and L. Naumann. 1998. Clinical evaluation of the automated COBAS Amplicor MTB assay for testing respiratory and nonrespiratory specimens. J. Clin. Microbiol. 36:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roche Diagnostics. 2007. Cobas Amplicor Mycobacterium tuberculosis test: instruction manual. Roche Diagnostics, Mannheim, Germany.
- 13.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tevere, V. J., P. L. Hewitt, A. Dare, P. Hocknell, A. Keen, J. P. Spadoro, and K. K. Young. 1996. Detection of Mycobacterium tuberculosis by PCR amplification with pan-Mycobacterium primers and hybridization to an M. tuberculosis-specific probe. J. Clin. Microbiol. 34:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tortoli, E., M. Tronci, C. P. Tosi, C. Galli, F. Lavinia, S. Natili, and A. Goglio. 1999. Multicenter evaluation of two commercial amplification kits (Amplicor, Roche and LCx, Abbott) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. Diagn. Microbiol. Infect. Dis. 33:173-179. [DOI] [PubMed] [Google Scholar]
- 16.Tortoli, E. 2009. Clinical manifestations of nontuberculous mycobacteria infections. Clin. Microbiol. Infect. 15:906-910. [DOI] [PubMed] [Google Scholar]
- 17.Willumsen, P., U. Karlson, E. Stackebrandt, and R. M. Kroppenstedt. 2001. Mycobacterium frederiksbergense sp. nov., a novel polycyclic aromatic hydrocarbon-degrading Mycobacterium species. Int. J. Syst. Evol. Microbiol. 51:1715-1722. [DOI] [PubMed] [Google Scholar]