Abstract
Prolyl-tRNA synthetase (ProRS) is a class IIa synthetase that, according to sequence analysis, occurs in different organisms with one of two quite distinct structural architectures: prokaryote-like and eukaryote/archaeon-like. The primary sequence of ProRS from the hypothermophilic eubacterium Thermus thermophilus (ProRSTT) shows that this enzyme is surprisingly eukaryote/archaeon-like. We describe its crystal structure at 2.43 Å resolution, which reveals a feature that is unique among class II synthetases. This is an additional zinc-containing domain after the expected class IIa anticodon-binding domain and whose C-terminal extremity, which ends in an absolutely conserved tyrosine, folds back into the active site. We also present an improved structure of ProRSTT complexed with tRNAPro(CGG) at 2.85 Å resolution. This structure represents an initial docking state of the tRNA in which the anticodon stem–loop is engaged, particularly via the tRNAPro-specific bases G35 and G36, but the 3′ end does not enter the active site. Considerable structural changes in tRNA and/or synthetase, which are probably induced by small substrates, are required to achieve the conformation active for aminoacylation.
Keywords: anticodon-binding domain/class IIa synthetase/prolyl-tRNA synthetase/tRNA identity/zinc-binding domain
Introduction
Aminoacyl-tRNA synthetases have been divided into two evolutionarily distinct families, each characterized by a class-defining catalytic domain containing short conserved sequence motifs (Eriani et al., 1990; Cusack et al., 1991; Cusack, 1995; Arnez and Moras, 1997). Within each class, subclasses can be identified containing more closely homologous synthetases that often share a common anticodon-binding module (Cusack, 1995). In class II, the largest subclass is class IIa, which contains the Ser-, Thr-, Pro-, Gly- and His-tRNA synthetases (RSs) (Cusack et al., 1991; Cusack, 1995). All these enzymes, except SerRS, contain a conserved C-terminal anticodon-binding domain whose fold was first observed in the crystal structures of Thermus thermophilus GlyRS (Logan et al., 1995) and subsequently in Escherichia coli and T.thermophilus HisRS (Arnez et al., 1995; Åberg et al., 1997) and E.coli ThrRS (Sankaranarayanan et al., 1999).
With the recent rapid accumulation of primary sequences of aminoacyl-tRNA synthetases, it has become apparent that there are often distinct structural differences, detectable in the primary sequence, between the same synthetase from organisms in the three different taxonomic domains, Archaea, Eubacteria and Eukarya. In a survey of primary sequences of eight different synthetases from a variety of organisms from the three taxa (Shiba et al., 1997), it was found that, in general, Archaea and Eukarya possess ‘eukaryote-like’ synthetases (the prototype being the yeast or human cytoplasmic enzymes) whereas eubacteria and mitochondria possess more or less distinct ‘prokaryote-like’ synthetases (the prototype being the E.coli enzyme). However, there are several exceptions to this rule, concerning for instance the Gly-, Asp-, Ile- and LysRSs, indicative of the complex processes governing the evolution and phylogenetic distribution of synthetases (Doolittle and Handy, 1998). GlyRS is of an unusual α2β2 subunit architecture in the ‘prokaryote-like’ enzyme (e.g. E.coli) but of a more standard α2 class IIa architecture in eukaryotes, archaea and T.thermophilus (Logan et al., 1995). It has been shown that T.thermophilus contains two AspRSs, a ‘prokaryote-like’ version exclusively charging tRNAAsp with aspartic acid, and an ‘archaeon-like’ enzyme which also mischarges tRNAAsn with aspartic acid (Becker and Kern, 1998). Differences in the ‘prokaryote-like’ and ‘eukaryote-like’ IleRSs account for the fact that pseudomonic acid is a potent inhibitor of the former but not the latter. However, it has recently been shown that both the eubacterial Mycobacterium tuberculosis IleRS and certain Staphylococcus aureus strains acquired, probably by horizontal gene tranfer, a ‘eukaryote-like’ IleRS that is resistant to pseudomonic acid (Brown et al., 1998). This example illustrates how recognizing the species differences between human and pathogen synthetases is potentially of great importance in the design of new antibiotics that target these enzymes (Schimmel et al., 1998). Finally, an extreme example is the recent discovery of a completely novel class I LysRS replacing the usual class II enzyme in certain organisms such as the archaebacterium Methanococcus janasschi and the spirochaete Borrelia burgdorferi (Ibba et al., 1997). Systematic differences in synthetases between species are often associated with systematic changes in tRNA identity elements, indicating that synthetases and tRNAs have, to a large extent, co-evolved [e.g. GlyRS (Mazauric et al., 1999), TyrRS (Wakasugi et al., 1998) and ProRS (Stehlin et al., 1998)].
We have cloned and determined the primary sequence of T.thermophilus ProRS (ProRSTT), which shows that it is surprisingly eukaryote/archaeon-like with 45% identity to human ProRSTT, 36% identity to M.janasschi ProRS and much lower identity (20%) to the E.coli enzyme. The enzyme has been crystallized (Yaremchuk et al., 2000a) and the structure determined at 2.43 Å resolution, the first reported structure of a ProRS. This permits visualization of the unique features of the ‘eukaryote/archaeon-like’ ProRS. We have also crystallized the ProRSTT–tRNAPro complex (Yaremchuk et al., 2000b) and preliminary results on its structure have been reported elsewhere (Cusack et al., 1998). Although only determined at 3.5 Å resolution, this structure revealed for the first time the mode of interaction of the class IIa anticodon-binding domain with the tRNA anticodon stem–loop. We report here more detailed data on the structure of tRNAPro and its recognition by ProRSTT resulting from measurements on a superior crystal form of the ProRS–tRNAPro complex that diffracts to 2.8 Å resolution (Yaremchuk et al., 2000b). These crystals have also been soaked with various combinations of small substrates to yield a series of structures representing different steps in the proline activation reaction, which will be presented elsewhere (A.Yaremchuk, M.Tukalo, M.Grøtli and S.Cusack, unpublished results).
Results
Phylogenetic analysis of ProRSs
Using ClustalW (Thompson et al., 1994), together with manual adjustments based on crystal structure, multiple sequence alignment has been done for ProRSs from 21 sources representing the full sequence diversity, including Gram-positive and -negative bacteria, archaea and eukaryotes (cytoplasmic and mitochondrial enzymes). Table I shows an alignment of 10 representative sequences. It is obvious from this analysis and the corresponding phylogenetic tree (Figure 1A) that there are two very distinct types of ProRS. The first group comprises the enzymes from the cytoplasm of eukaryotes, archaea and certain bacteria such as mycoplasmas, spirochaetes and T.thermophilus and which we refer to as eukaryote/archaeon-like. The second, ‘prokaryote-like’, group includes the enzymes from most other bacteria (e.g. E.coli, Bacillus subtilis and Chlamydia trachomatis) and mitochondria of eukaryotes. The sequence difference between the two subclasses of ProRS translates into a striking architectural difference, as shown schematically in Figure 1B. The prokaryote-like ProRSs have typically ∼570 residues/subunit and comprise an N-terminal class II synthetase catalytic domain and a C-terminal class IIa anticodon binding domain. Their characteristic feature is a large insertion domain of ∼180 residues inserted between motifs 2 and 3 in the catalytic domain. The eukaryote/archaeon-like ProRSs typically have ∼480 residues/subunit. Their distinguishing features are the absence of an insertion domain between motifs 2 and 3 and the presence of a unique C-terminal extension of ∼80 residues beyond the class IIa anticodon-binding domain. On a more detailed level, the sequence divergence between the two subclasses is considerable and even extends to residues important in the active site for proline recognition and activation. These systematic differences raise the possibility of targeting prokaryote-like ProRSs of pathogenic bacteria for specific antibiotics that do not inhibit eukaryote ProRSs.
Table I. Alignment of amino acid sequences of representative ProRSs.
The secondary structure of ProRSTT as defined by DSSP (Kabsch and Sander, 1983) is superposed with α, β and γ representing α-helix, β-strand and 310 helix, respectively. Two-letter codes for the organisms are as in Figure 1A (e.g. PRSBS is prolyl-tRNA synthetase from B. subtilis). The upper seven sequences in each block are eukaryote/archaeon-like ProRS (two ‘anomalous bacteria’, one mycoplasma, two archaeal and two eukaryotic cytoplasmic enzymes). The lower three sequences in each block are prokaryote-like ProRS (one eukaryotic mitochondrial enzyme, one from a Gram-positive bacterium and one from a Gram-negative bacterium). The long insertion in the prokaryote-like ProRS between β9 and β10 is omitted. Residues conserved in all ProRS are boxed with bold type; those conserved differentially in either type of ProRS are boxed in ordinary type. Note the variable conservation of cysteines in the unique C-terminal domain of eukaryote/archaeon-like ProRS (see text). The zince ligands in ProRSTT are marked with an asterisk (*). At the extreme C-terminus of the M.thermoautotrophicum (PRSMTa) sequence, an alternative reading frame restores the conserved terminal tyrosine, strongly implying a frameshift error in the reported protein sequence (gene mt0611). M.janasschi ProRS (PRSMJ) has been reported to have dual proline and cysteine specificity (Stathopoulos et al., 2000).
Fig. 1. (A) Unrooted phylogenetic tree showing the two types of ProRS derived from sequence alignments of representative ProRSs (see Table I). Eukaryotes (cytoplasmic enzyme): Saccharomyces cerevisiae (SC), Drosophila melanogaster (dm), human (hu), Caenorhabditis elegans (ce); archaebacteria: M.jannaschii (mj), Methanobacterium thermoautotrophicum (mta); mycoplasmas: Mycoplasma genitalium (mg), Mycoplasma pneumoniae (mp); ‘anomalous bacteria’ (i.e. those having an eukaryote/archaeon-like rather than a prokaryote-like ProRS): T.thermophilus (tt), B.burgdorferi (bb); eukaryotes (mitochondrial enzyme): S.cerevisiae (scm), Candida albicans (cam); eubacteria: Helicobacter pylori (hp), Mycobacterium tuberculosis (mt), Synechocystis sp. (ss), Haemphilus influenzae (hi), E.coli (ec), Zymomonas mobilis (zm), Aquifex aeolicus (aa), B.subtilis (bs), C.trachomatis (ct). A similar diagram is shown in Stehlin et al. (1998). (B) Schematic diagram of the architecture of prokaryote- and eukaryote/archaeon-like ProRSs with the three motifs of class II synthetases marked. The class IIa anticodon-binding domain is shown in black and the idiosyncratic domains in grey.
Structure determination of T.thermophilus ProRS (ProRSTT)
The crystal structure of substrate-free ProRSTT was initially solved at 2.7 Å resolution using low-resolution phases determined from Bijvoet differences from a single non-isomorphous mercury derivative followed by phase extension using 4-fold non-crystallographic symmetry averaging combined with solvent flattening (see Materials and methods). The structure was then refined using complete data to 2.43 Å resolution to an Rfree of 0.241 (R = 0.216) with tightly restrained stereochemistry. The crystallographic asymmetrical unit contains two ProRSTT dimers, each subunit comprising 477 residues of which zones 1–4 and 78–86 are not visible in the electron density due to disorder.
Overall structure of ProRSTT
The subunit of the ProRSTT dimer is made up of three distinct domains (Figure 2A). The N-terminal class II catalytic domain, containing the three conserved motifs, comprises residues 1–273 and is connected by random coil to the class IIa anticodon-binding domain (residues 290–377). A long α-helix (residues 377–402) then leads to a novel C-terminal zinc-binding domain (residues 402–477). The secondary structure is shown in Table I.
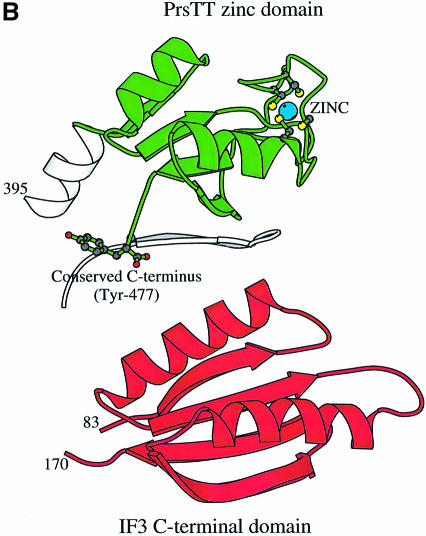
Fig. 2. (A) Ribbon diagram of the ProRSTT monomer. The catalytic domain is grey and motifs 1, 2 and 3 of the class IIa catalytic domain are respectively green, cyan and red. Note the C-terminus folding back to the active site. The class IIa anticodon-binding domains are shown in purple and the novel C-terminal domain in yellow with the zinc atom in cyan. (B) Ribbon diagram of the C-terminal domain of ProRSTT showing the four cysteine ligands of the zinc atom and the conserved C-terminal tyrosine. Also shown in grey is the edge strand (β9, Table I) of the catalytic domain antiparallel β-sheet which makes three main-chain hydrogen bonds with the edge strand (β22) of the C-terminal domain β-sheet. (C) Ribbon diagram of the C-terminal domain of B.stearothermophilus IF3 (Biou et al., 1995; PDB entry 1TIG) orientated to show the identical topology to the C-terminal domain of ProRSTT, although the lengths of the secondary structure elements are clearly different.
The catalytic domain has the characteristic fold and conserved features of class II synthetases, being based around a six-stranded antiparallel β-sheet with flanking helices. The dimer interface is mediated largely by secondary structural elements of motif 1. As previously described for HisRS (Åberg et al., 1997), the antiparallel symmetry-related interface helices (H2, Table I) are too far apart (15 Å) to permit any side-chain interactions leaving a deep solvent-filled crevice between the two subunits. The only bridging interaction is from Glu48 in helix H2 from one subunit to Asn65 on the strand following H2 on the other subunit. The floor of the crevice is constituted by a hydrophobic core of residues from both subunits. A cross-subunit β-sheet is formed by the strand following H2, which contains the conserved motif 1 Pro69, from one subunit and a β-loop formed by antiparallel strands β1 and β2 (residues 26–40) from the other subunit. An additional inter-subunit four-stranded antiparallel β-sheet is formed by interactions of the hairpin loop formed by residues 90–108 (β5 and β6, Table I) with its symmetry-related counterpart. This loop crosses over towards the active site of the opposite subunit and is particularly extended in eukayote/archaeon-like ProRS (Figure 2A).
Anticodon-binding domain
The anticodon-binding domain is, as expected, structurally homologous to that found in HisRSs (Arnez et al., 1995; Åberg et al., 1997) and GlyRSs (Logan et al., 1995). It has an α/β fold comprising a five-stranded mixed β-sheet surrounded by three α-helices. Details of the interactions of this domain with the anticodon stem–loop of the tRNAPro are presented below. The fold of this domain had hitherto only been found in class IIa aminoacyl-tRNA synthetases. However, it has recently been observed that the accessory subunit of the heterodimeric mitochondrial DNA polymerase γ is homologous to class IIa synthetases, notably GlyRS and ProRS (Carrodeguas et al., 1999; Fan et al., 1999). The anticodon-binding domain of ProRSTT has been used to model part of this subunit from Drosophila DNA polymerase γ (Fan et al., 1999).
C-terminal zinc domain
A long α-helix (H12, residues 377–402), which is bent near its N-terminal end due to the presence of Pro382, connects the anticodon-binding domain to the novel C-terminal domain (residues 402–477) characteristic of eukaryote/archaeon-like ProRSs. This compact domain comprises a four-stranded mixed β-sheet upon which two connecting helices are packed. The β-sheet is contiguous with the large β-sheet of the catalytic domain (Figure 2B). This domain has two notable features. In ProRSTT there are five cysteine residues, all in the C-terminal domain. Four of these, with sequence spacing Cys427X4Cys432X25 Cys458X2Cys461 (where X is any residue), form a tetrahedral binding site for a zinc ion (Figure 2B). The fortuitous replacement of this zinc atom by mercury was essential to the structure determination. The fifth cysteine (Cys446) is free and was also a mercury-binding site. Sequence comparisons show that all four of the zinc-binding cysteines are conserved in most eukaryotic-like ProRSs, although the short spacing between the second pair varies from 1 to 4 (Table I). The exceptions are Methanococcus jannaschii ProRS (which has no cysteines in the C-terminal domain) and Mycoplasma ProRSs (which appear to lack two or more of the required cysteines). These observations, together with the large distance from the active site of the enzyme (26 Å from the α-phosphate position) suggest that the zinc ion plays a non-essential structural role in stabilizing the fold of C-terminal domain. The second interesting feature of the zinc domain is that its C-terminal extremity is folded back towards the active site (Figure 2B). The side-chain of the extreme C-terminal residue, an absolutely conserved tyrosine in eukaryote/archaeon-like ProRSs (Table I), points away from the active site (its hydroxyl group forms hydrogen bonds with Asp220), but the carboxylate group points directly into the active site. Based on known ATP complexes with other class IIa synthetases, the position of this negatively charged group suggests that it may stabilize the position of basic residues interacting with the ATP pyrophosphate. Such a role is consistent with preliminary results on Tyr477Phe and Tyr477Ala mutants of ProRSTT, which show near-wild-type and slightly reduced aminoacylation activity, respectively, suggesting that an aromatic ring is favoured as the terminal residue but is not essential.
A database search using DALI (Holm and Sander, 1993) shows that the topology of the zinc domain is identical to that of the C-terminal domain of Bacillus stearothermophilus initiation factor 3 (IF3; Biou et al., 1995). The strength of the structural similarity in standard deviations above that expected (Z-score) is 3.2 and the root mean square deviation (r.m.s.d.) = 3.5 Å for the 53 superposed Cα atoms with one residue identity. The similarity in fold extends to the α-helix connecting the N-terminal and C-terminal domains of IF3, corresponding to that connecting the anticodon-binding and zinc domains of ProRSTT. However, in IF3, all the secondary structural elements (i.e. four β-strands and two α-helices) are longer than in the ProRSTT domain (Figure 2C), and the elaboration of the loop between the third and fourth β-strands into the zinc-binding site is absent. It remains to be seen whether this structural similarity reflects any functional similarity in, for instance, RNA binding.
Comparison with other class IIa synthetases
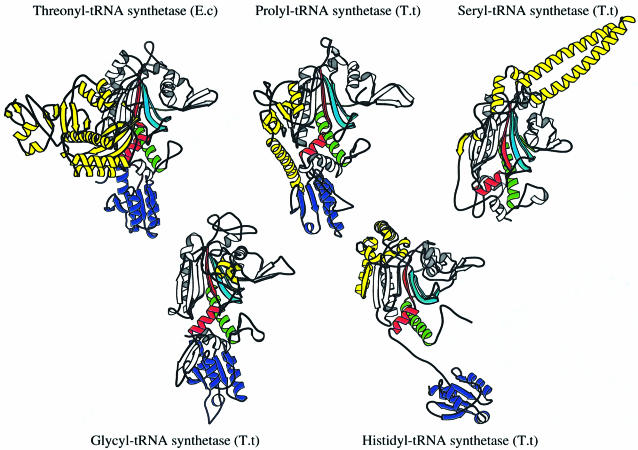
Figure 3 compares the subunit architecture of ProRSTT with the four other class IIa synthetases, SerRSTT (Fujinaga et al., 1993; Cusack et al., 1996a), HisRSTT (Åberg et al., 1997), GlyRSTT (Logan et al., 1995) and E.coli ThrRS (Sankaranarayanan et al., 1999). Superimposition of the catalytic domain with a cut-off of 3.8 Å between corresponding Cα atoms reveals the following structural similarities to ProRSTT: SerRSTT (matched residues, 211; identical residues, 29; r.m.s.d. of Cα atoms, 2.05 Å), GlyRSTT (209, 32, 2.12 Å), HisRSTT (164, 25, 1.82 Å), E.coli ThrRS (159, 23, 2.26 Å). The most extensive structural similarity of ProRSTT is thus with SerRSTT and GlyRSTT.
Fig. 3. Ribbon diagrams of single subunits of representatives of each of the five class IIa tRNA synthetases. All have the class II catalytic domain aligned in the same orientation with motifs 1, 2 and 3 shown in green, cyan and red, respectively. All except SerRS (top right) have the class IIa anticodon-binding domain (purple). System-specific domains are shown in yellow.
In the four enzymes with the class IIa anticodon-binding domain, the position and orientation of this domain relative to the catalytic domain can vary significantly, although in each case it occupies roughly the same spatial position as the N-terminal anticodon-binding domain in class IIb synthetases (Cusack, 1995). In ProRSTT, as in E.coli ThrRS (Sankaranarayanan et al., 1999), the anticodon-binding domain exclusively packs against the catalytic domain of the same subunit (but see discussion below on conformational flexibility). In HisRS the anticodon-binding domain is connected to the catalytic domain by an extended peptide and exclusively packs against the catalytic domain of the other subunit in the dimer (Arnez et al., 1995; Åberg et al., 1997), whereas in GlyRSTT it packs against both subunits (Logan et al., 1995). The system-specific domains (yellow in Figure 3) are quite different between these enzymes. In ProRSTT it is a C-terminal domain, which, however, is similarly positioned relative to the catalytic domain to the domains inserted between motifs 2 and 3 found in HisRS and all three class IIb synthetases (Berthet-Colominas et al., 1998). GlyRS, unusually, has an insertion domain of ∼85 residues between motifs 1 and 2 rather than between motifs 2 and 3, which is only partially ordered in the crystal structure and which has been proposed to interact with the minor groove of the tRNA acceptor stem (Logan et al., 1995). In the co-crystal structure of the ThrRS–tRNAThr complex, the large N-terminal extra domain has been shown to interact with the minor groove of the tRNA acceptor stem (Sankaranarayanan et al., 1999), whereas in seryl-tRNA synthetase, the N-terminal coiled-coil recognizes the cognate tRNA long variable arm (Biou et al., 1994).
Structure of the ProRSTT–tRNAPro(CGG) complex
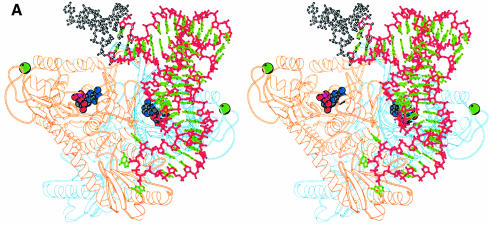
The structure of a ProRSTT–tRNAPro complex has been previously described at 3.5 Å resolution (Cusack et al., 1998). A closely related crystal form of the same complex was later grown with superior diffraction quality, permitting data to be collected up to 2.85 Å resolution (Yaremchuk et al., 2000b). Here we describe in more detail the structure of the ProRSTT–tRNAPro complex. Data have been obtained on the binary complex ProRSTT–tRNAPro(CGG) at 3 Å resolution and on a ternary complex comprising ProRSTT–tRNAPro(CGG) and a sulfamoyl analogue of prolyl-adenylate (ProAMS) at 2.85 Å resolution. With regard to the features that will be presented here, namely the structure of tRNAPro and the protein–tRNA interactions, the two structures are equivalent and so we consider only the structure at the higher resolution. The structure has been refined to an R factor (Rfree) of 21.0% (24.2%). More details on the data quality and refinement statistics are given in Tables II and III. The overall structure of the complex is shown in Figure 4A. Only one tRNA is bound to the synthetase although there is no reason to suppose that two tRNAs could not bind simultaneously to the dimeric enzyme. Furthermore, the complex in the crystal is clearly non-catalytically active for aminoacylation, since the acceptor end does not enter the active site and the 3′ and 5′ ends of the tRNA are disordered. Possible reasons for this are discussed below.
Table II. Data collection statistics.
| Crystal content |
|||
|---|---|---|---|
| ProRSTT native | ProRSTT soaked withmercury aniline | ProRSTT + tRNAPro(CGG)soaked with ProAMS | |
| Beamline | BM14 | BM14 | ID2 |
| Detector | 300 mm MarResearch Image Plate (Mar IP) | 150 mm Mar IP | 345 mm Mar IP |
| Wavelength (Å) | 0.826 | 1.00 (LIII edge) | 0.99 |
| Exposure (s)/image (°) | 170/0.7 | 70/1 | 20/1 |
| Space group | P21212 | P21212 | P43212 |
| Cell dimensions (Å) | a = 132.3, b = 191.1, c = 125.1 | a = 132.6, b = 193.5, c = 125.2 | a = b = 140.8, c = 237.0 |
| Number of crystals | 1 | 1 | 1 |
| Resolution (Å) | 15–2.43 | 18–4.1 | 20–2.85 |
| Total reflections | 448 321 | 93 054 | 221 758 |
| Unique reflections (anomalous pairs) | 117 904 | 25 110 (20 406) | 55 013 |
| Average redundancy | 3.8 | 3.7 | 4.0 (2.8) |
| Completeness | 98.6 | 97.6 | 98.2 (96.1) |
| Rmerge (highest bin) | 0.053 (0.244) | 0.031 (0.036) | 0.084 (0.298) |
Table III. Refinement statistics.
| ProRSTT native | ProRSTT + tRNAPro(CGG)soaked with Pro-AMS | |
|---|---|---|
| Resolution (Å) | 12–2.43 | 20–2.85 |
| Solvent content (%) | 64.8 | 71.7 |
| Work reflections | 111 925 | 52 192 |
| Test reflections | 5941 (5%) | 2797 (5%) |
| Rfree | 0.240 | 0.242 |
| Rwork | 0.211 | 0.210 |
| Number of protein atoms | 14 980 (two dimers) | 7502 (one dimer) |
| Number of substrate atoms | – | 60 (two Pro-AMS) |
| Number of solvent molecules | 738 water | no water, four sulphate ions |
| Number of metal atoms | four zinc | two zinc |
| <B> (Å2) | ||
| protein | 35.35 | 48.8 |
| solvent | 37.12 | – |
| substrate | – | 40.4 (Pro-AMS) |
| 56.2 (tRNA) | ||
| r.m.s. bonds | 0.005 | 0.007 |
| r.m.s. angles | 1.250 | 1.383 |
| Ramachandran plot | ||
| favourable (%) | 91.7 | 88.4 |
| additional (%) | 7.7 | 10.2 |
| generous (%) | 0.6 | 1.1 |
| disallowed (%) | 0.0 | 0.2a |
<B>, mean atomic temperature factor over all atoms.
aIn the ProRSTT–tRNAPro (CGG)–ProAMS ternary complex, Phe150 in both subunits, which is in good electron density, moves into the disallowed region of the Ramachandran plot due to the binding of ProAMS in the nearby active site.
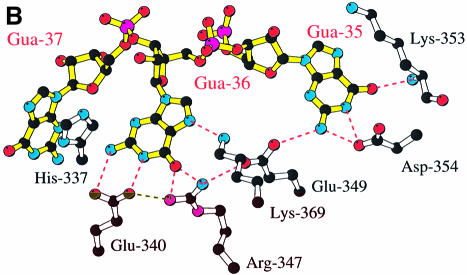
Fig. 4. (A) Stereo diagram showing the overall structure of the ProRSTT– tRNAPro(CGG)–ProAMS complex in which one tRNA molecule binds to the tRNA synthetase dimer. The tRNA backbone is shown in red and the bases in green, with the disordered acceptor extremity of the tRNA shown in white. In each active site there is a molecule of ProAMS. (B) Diagram showing specific recognition of the tRNAPro-specific anticodon bases G35 and G36 by a network of hydrogen bonds to charged side-chains in the anticodon-binding domain. These residues are highly conserved in eukaryote/archaeon-like, but not prokaryote-like ProRSs (Table I). (C) Stereo diagram showing details of the cross-contacts from one subunit to the anticodon stem of the tRNA largely bound on the other subunit. This feature is also observed in tRNA complexes of other class IIa synthetases, such as SerRS (Cusack et al., 1996a) and ThrRS (Sankaranarayanan et al., 1999). The arrow represents β-strand β12.
The structure of tRNAPro
As in most prokaryotes, tRNAPro(CGG) contains 77 nt with an insertion of a nucleotide in the D-loop (C17a) and a 5 nt variable loop. The three-dimensional structure of the tRNA is rather similar to that of yeast tRNAPhe (Holbrook et al., 1978) apart from the anticodon loop distortion induced by binding to the synthetase (see below) and a protruding bulge on the D-loop side of the molecule formed by the stacking of C17 with the extra base C17a. In particular, the nucleotides and hence tertiary interactions in the core of the tRNA are nearly identical to yeast tRNAPhe, the only sequence difference being that the G22–C13 base-pair in tRNAPro is an A23–U13 base-pair in tRNAPhe. In tRNAPro, A25 and G44 form a highly buckled non-Watson–Crick base-pair and the triplets C25–G10:G45, C23–G12:A9 and G22–C13:G46 are formed. The A14–U8 base-pair interacts with A21 via a hydrogen bond between U8 2′-OH to A21 N1. There is no evidence for a hydrogen bond interaction between the 2′-OH of U8 and the amino group of G46 (the distance between the U8 2′-OH and G46 N2 is 3.8 Å) as proposed for E.coli tRNAPro by Yap and Musier-Forsyth (1995). This point will be discussed further below in relation to possible conformational changes in the tRNA required to form a functional complex. A further difference between tRNAPro and tRNAPhe concerns nt 47 in the variable loop. In tRNAPhe, base U47 is flipped out into the solvent, whereas in tRNAPro the base of G47 folds the other way into the broadened tRNA major groove stacking against the edges of A21 and G46 and making hydrogen bonds to the phosphates of A21 on one side and G46 on the other side.
Anticodon stem–loop recognition
The only observed synthetase–tRNA contacts are to the anticodon stem–loop. The major interactions are between the anticodon bases and the anticodon-binding domain of subunit B (Figure 4B) but there are additional cross-interactions between the upper part of the anticodon stem and two loops from synthetase subunit A (Figure 4C). Based on the original 3.5 Å structure of the complex, a preliminary description of these interactions has been given (Cusack et al., 1998). The reader is referred to Figures 3 and 4 in that article for a diagram of the distorted anticodon stem–loop interacting with the anticodon- binding domain and an annotated sequence alignment of the anticodon-binding domain from representative class IIa synthetases. Here we show details of the specific recognition of the two anticodon bases G35 and G36, which are sufficient to identify any tRNAPro isoacceptor uniquely, by an extensive network of hydrogen bonds involving charged residues (Figure 4B). Hydrogen bonds are made to all Watson–Crick positions of both bases as well as to the N7 of G36. Intimately involved in this network are residues Arg347B and Glu349B, which are highly conserved in eukaryote/archaeon-like ProRSs (including human) but usually replaced by small hydrophobic residues in prokaryote-like ProRSs (Table I). The implied systematic difference in the strength of interactions with G35 and G36 is consistent with these two bases being less important as identity elements in, for instance, the E. coli system than in the human system (Liu et al., 1995; Stehlin et al., 1998; see also discussion in Cusack et al., 1998). Bases G35 and G36 sit upon a hydrophobic patch formed by residues Ile295, Pro332 and Phe336. In particular, Phe336 (which, by sequence alignment, is a conserved aromatic or other large hydrophobic residue in all class IIa anticodon-binding domains) forms an edge on interaction with the base of G36, as frequently observed in other protein–RNA complexes. Base G37 stacks against His337 (Figure 4B) but is not otherwise specifically recognized. The higher-resolution structure here shows that there is a putative hydrogen bond between the side-chain of Asp298B and the N4 position of the wobble base C34, although this was not apparent at lower resolution. Interestingly, when the tRNA-bound and -unbound structures are compared, the loop 296B–306B shows the greatest mobility in the anticodon-binding domain, presumably allowing adaptation to the presence of any base in the wobble position as occurs in different tRNAPro isoacceptors. Other segments that adapt via small concerted movements to anticodon binding are residues 331B–334B and 353B–356B. The former region contains Gly333, which is absolutely conserved in all ProRSs (Table I) and indeed in most class IIa synthetases, and which cannot be a replaced by a larger amino acid as this would cause steric hindrance with the tRNA backbone at nt G36.
The cross-subunit interactions involve residues in loops 126A–128A and 245A–249A contacting the tRNA backbone of nts 28 and 29 (Figure 4C). For instance, Trp127A, which is sandwiched between Arg128A and Arg247A, makes a hydrogen bond to the phosphate of nt 29, via the Nε atom, and Ser126A hydrogen bonds to the phosphate of nt 28. This is in very good agreement with phosphate protection studies (S.P.Egorova and M.A.Tukalo, unpublished results). The side-chain of Gln245A further cross-links the tRNA to the protein by making hydrogen bonds to the N2 of base G43 via its Oε1 and to the main-chain carbonyl oxygen of Leu249A via its Nε2 group.
Discussion
Conformational flexibility of ProRSTT
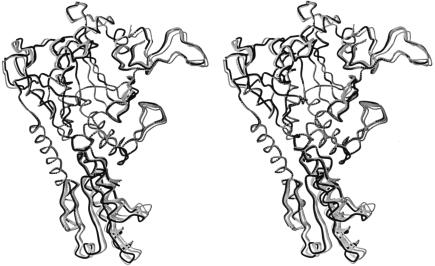
The asymmetrical unit of the native ProRSTT crystals contains two dimers and several regions of each monomer had to be released from the tight non-crystallographic restraints due to significant deviations, often due to crystal contacts. Superimposition of the catalytic domain of the four crystallographically independent ligand-free subunits allows visualization of the conformational flexibility of the molecule (Figure 5). Significant differences are observed in the orientation of the anticodon-binding domain and also in the conformation of various active site loops as well as the motif 1 loop, which forms part of the dimer interface. On the other hand, the extended β-sheet, which encompasses the catalytic and C-terminal zinc domains, is rigid. Depending on the exact position of the anticodon-binding domain, there is a variable degree of contact between Gln329 in this domain and Arg128 of the catalytic domain of the opposite subunit, ranging from no contact to van der Waals contact of side-chain extremities to a more intimate contact involving two cross-subunit side-chain to main-chain hydrogen bonds. The observed orientation of the tRNA-bound, anticodon-binding domain in the binary tRNA complex is at one extreme of the conformations observed in the native structure (Figure 5). In this case, there is interaction of the side-chains Gln329B and Arg128A, the latter residue also interacting with the tRNA (Figure 4C). It has been shown elsewhere that a small change in the observed orientation of the anticodon-binding domain of the non-tRNA-bound subunit is partly responsible for the existence of two different crystal forms of the ProRSTT–tRNAPro complex (Yaremchuk et al., 2000b). The possible importance of the hinge motion of the anticodon-binding domain is discussed below. The functionally important local conformational changes associated with proline, ATP and prolyl-adenylate binding in the active site will be described in more detail elsewhere (A.Yaremchuk, M.Tukalo, M.Grøtli and S.Cusack, unpublished results).
Fig. 5. Stereo diagram of five independent ProRSTT subunits, all with empty active sites, superimposed by means of the catalytic domain. The four grey structures correspond to the four crystallographically independent subunits in the native crystal. The black structure corresponds to the subunit in the ProRSTT–tRNAPro(CGG) binary complex whose anticodon-binding domain binds the single tRNAPro molecule. The superimposition shows the hinge movement of the anticodon-binding domain, as well as a looseness of several loops near the active site, which disappears when small substrates are bound.
Comparison of the ProRSTT–tRNAPro complex with the E.coli ThrRS–tRNAThr complex
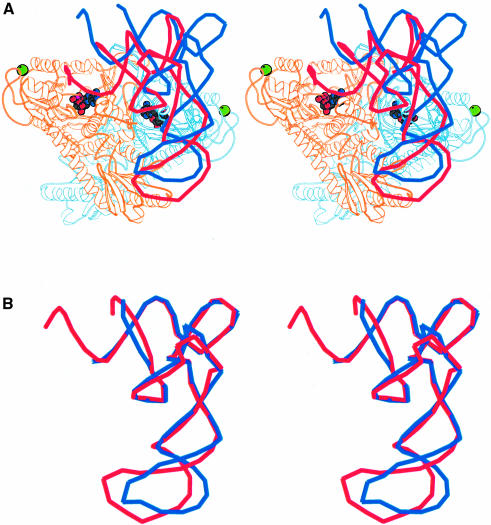
Recently the structure of the E.coli ThrRS–tRNAThr complex in a conformation with the 3′ end of the tRNA ordered in the active site has been published (Sankaranarayanan et al., 1999). Since this is a closely related class IIa system, it can be used to model the active ProRSTT–tRNApro complex. Figure 6A shows the position of the tRNAThr relative to the ProRSTT–tRNApro complex after superimposition of the conserved active site domain of each enzyme. This model suggests that the position of the 3′ end of the tRNA could be stabilized by interactions with the C-terminal zinc domain. A dramatic reorientation of the tRNA is clearly required for it to enter the active site and this could happen either through distortion of the tRNA, or by movement of the anticodon-binding domain carrying with it the tRNA, or both. Figure 6B shows that the core regions of tRNAThr and tRNAPro are remarkably similar in conformation. The only significant difference is in the distortion of the anticodon stem–loop, which may be due to the fact that base 38 is flipped out in tRNAThr but is base-paired to base 32 in tRNAPro (Sankaranarayanan et al., 1999). In view of the network of specific interactions between ProRSTT and the tRNAPro anticodon stem–loop (Figure 4B and C), it seems unlikely that a major rearrangement of this region would occur. On the other hand, a small change in the orientation of the anticodon-binding domain (and the discussion above shows that this is possible) could have a large effect on the position of the acceptor stem of the tRNA due to a lever arm effect. Another possibility is that there is a distortion within the core of the tRNA causing it to bend into the active site. This could then explain the dependence of aminoacylation on an abnormal interaction of the 2′-OH of U8 (Yap and Musier-Forsyth, 1995), which is otherwise not explained by our structure. A crystal structure of a catalytically functional ProRSTT–tRNAPro complex is clearly required to resolve this issue.
Fig. 6. (A) Stereo diagram of tRNAThr superimposed on the ProRSTT–tRNAPro(CGG)–ProAMS ternary complex. The tRNAThr is coloured red and the tRNAPro is coloured blue. The position of the tRNAThr is that obtained after superimposition of the catalytic domains of ProRSTT and E.coli ThrRS (Sankaranarayanan et al., 1999; PDB entry 1QF6) in their respective tRNA complexes. The red tRNA molecule is thus a good model of tRNAPro functionally bound in the active site of ProRSTT although it does not simultaneously correctly engage with the anticodon-binding domain as the latter is differently orientated with respect to the catalytic domain in E.coli ThrRS. The figure shows that a considerable change in orientation of the tRNAPro from the observed position is required to reach the active conformation (see text). It also suggests that the C-terminal zinc domain could contact the 3′ strand of the acceptor stem. (B) Superimposition of the backbone structures of tRNAPro (blue) and tRNAThr (red) in their respective inactive and active complexes. This shows that, apart from the different distortion of the anticodon loop, the two tRNAs have a remarkably similar conformation. The difference in the anticodon loop is largely due to the fact that base 38 is flipped out in tRNAThr but is base-paired to base 32 in tRNAPro (Sankaranarayanan et al., 1999).
Why is the current ProRSTT–tRNAPro complex not in an aminoacylation-competent state? In all our ProRSTT and ProRSTT–tRNAPro structures, the peptide 77–88 is disordered except when prolyl-adenylate or its sulfamoyl analogue, ProAMS, have been bound in the active site by soaking experiments, proline alone not being sufficient (data not shown). This peptide carries an aromatic residue, Phe87, which is highly conserved in homologous class IIa synthetases (e.g. Tyr208 in SerRSTT and Tyr313 in E.coli ThrRS). In the structure of the E.coli ThrRS–tRNAThr complex, the base of A76 is observed to stack between this aromatic residue and the class II conserved motif 2 arginine, an arrangement that is likely to be general. Only in the prolyl-adenylate- or ProAMS-bound structures of ProRSTT is Phe87 in the correct position to fulfil this function. We therefore think that correct binding of the 3′ end of the tRNA in the active site requires the conformational changes induced by the presence of the prolyl-adenylate, this being an additional means of ensuring proline-specific aminoacylation. Verification of this hypothesis would require a structure of a functional ProRSTT–tRNApro complex presumably co-crystallized in the presence of proline-adenylate or a suitable analogue. In this context we note that all ProRSTT–tRNApro complex crystals available to date have been grown in the absence of substrates. Soaking in substrates afterwards (e.g. ProAMS) affects the active site, but has no effect on tRNA conformation, probably because extensive crystal contacts holding the tRNAPro in its inactive conformation (e.g. the G19–C56 tertiary base-pair of one tRNA is stacked on G36 of another tRNA) prevent a rearrangement. Three other T.thermophilus tRNA synthetase–tRNA binary complexes have been crystallized in which the tRNA is partially bound to the synthetase, but not functionally engaging the active site [SerRSTT (Biou et al., 1994), LysRSTT (Cusack et al., 1996b) and AspRSTT (D.Moras, personal communication)]. It could be that a contributory factor to the trapping of these intermediately-docked states in crystals, other than the absence of the small substrates, is that thermophilic enzymes have insufficient mobility at low temperatures to allow the necessary conformational changes to occur for full binding. However, the structure of a non-functional binary complex of yeast ArgRS–tRNAArg has recently been determined whereas the ternary complex with arginine does show the 3′ end binding correctly in the active site (B.Delagoutte, D.Moras and J.Cavarelli, personal communication). This would seem to confirm the importance, at least in some cases, of prior binding of small substrates as a prerequisite for ordered and functional binding of the tRNA 3′ end. This phenomenon may be more important in systems that lack strong identity elements in the acceptor stem or discriminator base.
Very recently it was reported that the M.janasschi ProRS has a dual synthetase activity and can, remarkably, synthesize both Pro-tRNAPro and Cys-tRNACys with complete specificity (Stathopoulos et al., 2000). This observation is even more remarkable in that M.janasschi ProRS is 36% identical to T.thermophilus ProRS, whose structure is described here, and the sequence alignment (Table I) shows no obviously special features that might explain the extra ability of the M.janasschi enzyme.
Materials and methods
Crystals and data collection
Wild-type ProRS was purified from T.thermophilus strain HB-8 and single crystals of ligand-free ProRSTT were grown by the hanging drop method in 32% ethylene glycol as described (Yaremchuk et al., 2000a). The crystals are of space-group P21212 with cell dimensions at 100 K of a = 132 Å, b = 192 Å, c = 125Å and diffract to ∼2.4 Å resolution. There are two homodimers in the asymmetrical unit (Vm = 3.63 Å3/Da; solvent content = 65%). Diffraction data were collected on crystals cryoprotected with 35% ethylene glycol at 110 K on beamline BM14 at the ESRF (Table III). The data were integrated using MOSFLM (Leslie, 1992) and all subsequent processing was done with the CCP4 package (1994). Crystals soaked with various heavy metal compounds were screened by low resolution (4.5 Å) data collections using BM14 with the wavelength set to the corresponding absorption edge to maximize the anomalous scattering contribution. The only useful derivative was obtained by soaking a crystal with 1 mM p-acetoxymercuril aniline for 26 h which was measured just above the mercury LIII edge (12.284 keV).
Crystallization of the ProRSTT–tRNAPro complex is described elsewhere (Yaremchuk et al., 2000b). Crystal were cryoprotected with 27% glycerol. Data collection parameters and statistics for the ternary complex with ProAMS are given in Table II.
Structure determination
The mercury-aniline-soaked crystal of apo-ProRSTT gave a strong anomalous signal to 5 Å resolution. The anomalous difference Patterson (resolution 4.8–12 Å) contained many strong peaks and was interpreted using RSPS to give five initial mercury sites, whereas the isomorphous difference Patterson was essentially flat presumably due to non-isomorphism. Using MLPHARE, the five initial sites were refined using anomalous differences. Derived phases were used to calculate an anomalous difference Fourier map, from which a further nine weaker mercury sites were identified. These were refined to give an overall mean figure of merit of 0.20. Careful examination of the 14 sites permitted a non-crystallographic 2-fold axis to be identified, which was used to average the 4.8 Å native map using the program DM. The improved map could be correctly interpreted using characteristic features of class II synthetases, notably the antiparallel helices at the dimer interface. The two non-crystallographic dimer axes were thus identified and it became apparent that the initial 2-fold axis, found from the heavy atom sites, was in fact a pseudo 2-fold axis relating the two dimers in the asymmetrical unit. Using a mask derived from the known structure of GlyRS (Logan et al., 1995), 4-fold averaging with phase extension to 2.8 Å and automatic refinement of transformation matrices was then applied using DM. Rfree improved from 0.60 to 0.25 in 250 cycles. The resultant map was excellent and permitted an essentially complete model to be built.
Structure determination of the complex with tRNAPro was by molecular replacement as described (Cusack et al., 1998).
Refinement
Refinement of the apo-structure was done with XPLOR version 3.1 (Brünger, 1992) using standard protocols and including solvent correction and overall anisotropic temperature factor. Progress was monitored with Rfree. The large number of independent reflections (∼120 000) at 2.43 Å resolution allowed tight non-crystallographic symmetry restraints between the four equivalent subunits to be removed at later stages of refinement over 189 of the 464 residues in the model to take account of conformational variability. A very large number of ordered solvent molecules is also visible (≥700 in the current model). Refinement statistics and structure quality parameters are given in Table III.
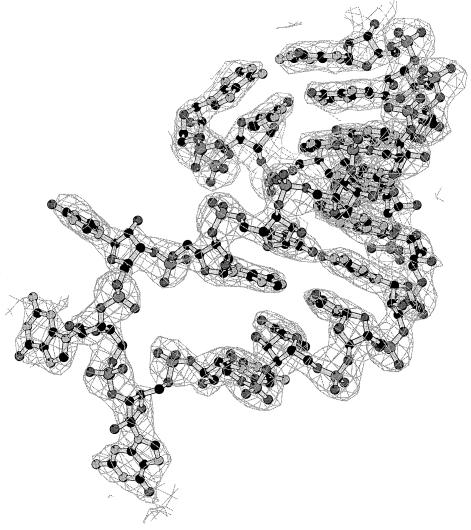
Refinement of the ProRSTT–tRNApro ternary complex with ProAMS was performed with CNS (Brünger et al., 1998). A segment of the map is shown in Figure 7 and the refinement statistics given in Table III. The asymmetrical unit contains one ProRSTT dimer and one tRNAPro(CGG). Due to the asymmetry resulting from tRNA binding to only one subunit as well as to crystal contacts, only 260/472 residues were restrained by the non-crystallographic symmetry. Ribose puckers were restrained to C3′ endo except for nts 7, 9, 16, 18–20, 34–37, 48, 58 and 60, which were restrained to C2′ endo.
Fig. 7. Final 2Fo – Fc map of the anticodon loop of tRNAPro(CGG) contoured at 1.5σ.
Acknowledgments
Acknowledgements
We thank members of the EMBL–ESRF Joint Structural Biology Group, especially Andrew Thompson, Bjarne Rasmussen and Jonathan Grimes, for assistance on the ESRF synchrotron beamlines. We thank Ivan Krikliviy for help in purification of T.thermophilus tRNAPro and Morton Grøtli for the synthesis of ProAMS. The research of M.T. was supported in part by an International Research Scholar’s award from the Howard Hughes Medical Institute. Illustrations of structures were prepared with MOLSCRIPT (Kraulis, 1991) or BOBSCRIPT (Esnouf, 1997).
References
- Åberg A., Yaremchuk,A., Tukalo,M., Rasmussen,B. and Cusack,S. (1997) Crystal structure analysis of activation of histidine by T. thermophilus histidyl-tRNA synthetase. Biochemistry, 36, 3084–3094. [DOI] [PubMed] [Google Scholar]
- Arnez J.G. and Moras,D. (1997) Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci., 22, 189–232. [DOI] [PubMed] [Google Scholar]
- Arnez J.G., Harris,D.C., Mitschler,A., Rees,B., Francklyn,C.S. and Moras,D. (1995) Crystal structure of histidyl-tRNA synthetase from E.coli complexed with histidyl-adenylate. EMBO J., 14, 4143–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H.D. and Kern,D. (1998) Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl Acad. Sci. USA, 95, 12832–12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet-Colominas C., Seignovert,L., Härtlein,M., Grøtli,M., Cusack,S. and Leberman,R. (1998) The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J., 17, 2947–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biou V., Yaremchuk,A., Tukalo,M. and Cusack,S. (1994) The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science, 263, 1404–1410. [DOI] [PubMed] [Google Scholar]
- Biou V., Shu,F. and Ramakrishnan,V. (1995) X-ray crystallography shows that translational initiation factor IF3 consists of two compact α/β domains linked by an α-helix. EMBO J., 14, 4056–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.R., Zhang,J. and Hodgson,J.E. (1998) A bacterial antibiotic resistance gene with eukaryotic origins. Curr. Biol., 8, R365–R367. [DOI] [PubMed] [Google Scholar]
- Brünger T.A. (1992) X-PLOR Version 3.1. Yale University Press, New Haven, CT. [Google Scholar]
- Brünger A.T. et al. (1998) Crystallographic and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carrodeguas J.A., Kobayashi,R., Lim,S.E., Copeland,W.C. and Bogenhagen,D.F. (1999) The accessory subunit of Xenopus laevis mitochondrial DNA polymerase γ increases processivity of the catalytic subunit of human DNA polymerase γ and is related to class II aminoacyl-tRNA synthetases. Mol. Cell. Biol., 19, 4039–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Rees,B., Ruff,M., Thierry,J.-C. and Moras,D. (1993) Yeast tRNAAsp recognition by its class II aminoacyl-tRNA synthetase. Nature, 362, 181–184. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cusack S. (1995) Eleven down and nine to go. Nature Struct. Biol. 2, 824–831. [DOI] [PubMed] [Google Scholar]
- Cusack S, Härtlein,M and Leberman,R. (1991) Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res., 19, 3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (1996a) The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNASer and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J., 15, 2834–2842. [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (1996b) The crystal structures of T.thermophilus lysyl-tRNA synthetase complexed with E.coli tRNALys and a T.thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J., 15, 6321–6334. [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A, Krikliviy,I. and Tukalo,M. (1998) tRNAPro anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure, 6, 101–108. [DOI] [PubMed] [Google Scholar]
- Doolittle R.F. and Handy,J. (1998) Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr. Opin. Genet. Dev., 8, 630–636. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue,M., Poch,O., Gangloff,J. and Moras,D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequences motif. Nature, 347, 203–206. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MOLSCRIPT that includes greatly enhanced coloring capabilities. J. Mol. Graph., 15, 133–138. [DOI] [PubMed] [Google Scholar]
- Fan L., Sanschagrin,P.C., Kaguni,L.S. and Kuhn,L.A. (1999) The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl Acad. Sci. USA, 96, 9527–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga M., Berthet-Colominas,C., Yaremchuk,A.D., Tukalo,M.A. and Cusack,S. (1993) Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 Å resolution. J. Mol. Biol., 234, 222–233. [DOI] [PubMed] [Google Scholar]
- Holbrook S.R., Sussman,J.L., Warrant,R.W. and Kim,S.H. (1978) Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J. Mol. Biol., 123, 631–660. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Ibba M, Morgan,S, Curnow,A.W., Pridmore,D.R., Vothknecht,U.C., Gardner,W., Lin,W., Woese,C.R. and Söll,D. (1997) An euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science, 278, 1119–1122. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Leslie A.G.W. (1992). Joint CCP4 and ESF–EACBM Newsletter on Protein Crystallography, no. 26. Daresbury Laboratory, Warrington, UK. [Google Scholar]
- Liu H., Peterson,R., Kessler,J. and Musier-Forsyth,K. (1995) Molecular recognition of tRNAPro by E.coli proline tRNA synthetase in vitro. Nucleic Acids Res., 23, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D.T., Mazauric,M.-H., Kern,D. and Moras,D. (1995) Crystal structure of glycyl-tRNA synthetase from T.thermophilus. EMBO J., 14, 4156–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazauric M.H., Roy,H. and Kern,D. (1999) tRNA glycylation system from Thermus thermophilus. tRNAGly identity and functional interrelation with the glycylation systems from other phylae. Biochemistry, 38, 13094–13105. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S., Dock-Bregon,A.-C., Romby,P., Caillet,J., Springer,M., Rees,B., Ehresmann,C., Ehresmann,B. and Moras,D. (1999) The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell, 97, 371–381. [DOI] [PubMed] [Google Scholar]
- Schimmel P., Tao,J. and Hill,J. (1998) Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J., 12, 1599–1609. [PubMed] [Google Scholar]
- Shiba K., Motegi,H. and Schimmel,P. (1997) Maintaining genetic code through adaptations of tRNA synthetases to taxonomic domains. Trends Biochem. Sci., 22, 453–457. [DOI] [PubMed] [Google Scholar]
- Stathopoulos C., Li,T., Longman,R., Vothknecht,U.C., Becker,H.D., Ibba,M. and Soll,D. (2000) One polypeptide with two aminoacyl-tRNA synthetase activities. Science, 287, 479–82. [DOI] [PubMed] [Google Scholar]
- Stehlin C., Heacock,D.H.,ii, Liu,H. and Musier-Forsyth,K. (1997) Chemical modification and site-directed mutagenesis of the single cysteine in motif 3 of class II E. coli prolyl-tRNA synthetase. Biochemistry, 36, 2932–2938. [DOI] [PubMed] [Google Scholar]
- Stehlin C., Burke,B., Yang,F., Liu,H., Shiba,K. and Musier-Forsyth,K. (1998) Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry, 37, 8605–8613. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K., Quinn,C.L., Tao,N. and Schimmel,P. (1998) Genetic code in evolution: switching species-specific aminoacylation with a peptide transplant. EMBO J., 17, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap L-P. and Musier-Forsyth,K. (1995) Transfer RNA aminoacylation: identification of a critical ribose 2′-hydroxyl–base interaction. RNA, 1, 418–424. [PMC free article] [PubMed] [Google Scholar]
- Yaremchuk A., Cusack,S. and Tukalo,M. (2000a) Crystallization and preliminary X-ray diffraction analysis of Thermus thermophilus prolyl-tRNA synthetase. Acta Crystallogr. D, 56, 195–196. [DOI] [PubMed] [Google Scholar]
- Yaremchuk A., Kriklivyi,I., Cusack,S. and Tukalo,M. (2000b) Improved crystals of Thermus thermophilus prolyl-tRNA synthetase complexed with cognate tRNA obtained by crystallisation from precipitate. Acta Crystallogr. D, 56, 197–199. [DOI] [PubMed] [Google Scholar]