Abstract
We prospectively tested 95 nasal swabs or nasopharyngeal aspirates taken from 56 adults and 39 children visiting the Reims University Medical Centre (northern France) for influenza-like illnesses (ILI) during the early stage of the French influenza A/H1N1v pandemic (October 2009). Respiratory samples were tested using a combination of two commercially available reverse transcription-PCR (RT-PCR) DNA microarray systems allowing rapid detection of influenza A virus strains, including the new A/H1N1v strain as well as 20 other common or newly discovered respiratory viruses. Concomitantly, a generic and classical real-time RT-PCR assay was performed to detect all circulating influenza A virus strains in the same samples. Of the 95 respiratory samples tested, 30 (31%) were positive for the detection of influenza A/H1N1v virus infection by both RT-PCR DNA microarray and classical real-time RT-PCR detection assays. Among the infections, 25 (83%) were monoinfections, whereas 5 (17%) were multiple infections associating influenza A/H1N1v virus with coronavirus (CoV), human bocavirus (HBoV), respiratory syncytial virus (RSV), or human rhinoviruses (HRVs). Of the 95 respiratory samples tested, 35 (37%) were positive for respiratory viruses other than influenza A/H1N1v virus. Among these infections, we observed 30 monoinfections (HRVs [63%], parainfluenza viruses [PIVs] [20%]), influenza A/H3N2 virus [6%], coronavirus [4%], and HBoV [4%]) and 5 multiple infections, in which HRVs and PIVs were the most frequently detected viruses. No specific single or mixed viral infections appeared to be associated significantly with secondary hospitalization in infectious disease or intensive care departments during the study period (P > 0.5). The use of RT-PCR DNA microarray systems in clinical virology practice allows the rapid and accurate detection of conventional and newly discovered viral respiratory pathogens in patients suffering from ILI and therefore could be of major interest for development of new epidemiological survey systems for respiratory viral infections.
Influenza-like illnesses (ILI) can be attributed to a wide range of respiratory viruses, including seasonal A and B influenza viruses, the new pandemic A/H1N1v influenza virus strain, adenoviruses, respiratory syncytial virus (RSV), enteroviruses (EVs), human rhinoviruses (HRVs), human metapneumovirus (HMPV), human bocaviruses (HBoVs), coronaviruses (CoVs), and parainfluenza viruses (PIVs) (19, 22, 27, 28, 29, 34, 37, 40, 43). Patients infected by these diverse viral pathogens develop widely overlapping symptoms which render clinical diagnosis unreliable and severely limit etio-epidemiological studies (22). The predominant pathogens of ILI are typically the influenza viruses, which cause annual recurrent epidemics affecting an estimated 5 to 15% of the population presenting with upper respiratory tract infections worldwide (29, 40). Influenza can be clinically indistinguishable from diseases caused by other respiratory viruses, such as the common cold. Therefore, rapid and reliable screening of a large panel of respiratory viruses responsible for ILI is of major epidemiological and clinical interest before, during, and after an influenza pandemic wave.
For diagnosis of viral respiratory tract infections responsible for ILI, clinical virology laboratories historically have used traditional methods, such as direct fluorescent-antibody assay (DFA) and culture, for the detection of six or seven conventional respiratory viruses (1, 21, 26). Over the past 10 years, nucleic acid amplification tests, including PCR and nucleic acid sequence-based amplification, have shown greater sensitivity than DFA and culture for the detection of respiratory viruses (5). Multiplex PCR assays have been used to detect the presence of one or more respiratory virus infections in respiratory tract specimens (2, 6, 7, 8, 9, 10, 11). Moreover, the emergence of five new respiratory viruses since 2000, including HMPV, severe acute respiratory syndrome coronavirus (SARS-CoV), CoV strains NL63 and HKU1, avian influenza virus H5N1, and HBoV, has presented challenges for virology laboratories (13, 15, 36, 43). The absence of commercially available tests often leaves laboratories without the ability to diagnose infections with these important emerging viruses (24, 25). There is therefore a need for new and reliable diagnostic tests to diagnose both traditional and emerging respiratory viral infections that are potentially related to ILI (24, 25, 26).
In the present study, we prospectively tested 95 nasal swabs or nasopharyngeal aspirates taken from adults or children visiting the emergency room of the Reims University Medical Centre (Champagne-Ardenne, France) for ILI during October 2009. Respiratory samples were tested using a combination of two commercially available reverse transcription-PCR (RT-PCR) DNA microarray detection systems: one for the specific detection of influenza A virus strains, including A/H1N1v strains, and the second for the detection of 20 common or newly discovered respiratory viruses (influenza A [seasonal A/H1N1 and A/H3N2 strains], B, and C viruses, RSV A and B, PIV-1, PIV-2, PIV-3, PIV-4, PIV-4 A and B, coronavirus E-229, HRVs, HMPV A and B, HBoV, adenoviruses, and enterovirus species B). The results obtained by the association of these two commercially available kits were compared to those of a reference generic real-time RT-PCR assay used daily for the rapid detection of all known influenza A virus strains. The potential application of these commercially available RT-PCR DNA microarray detection systems in virological routine diagnosis of ILI was assessed.
MATERIALS AND METHODS
Patients.
From 1 October to 15 October 2009, 56 adults (mean age = 39.5 years; standard deviation [SD] = 20.4 years) and 39 children (mean age = 5 years; SD = 3.9 years) visiting the emergency room of the Reims University Medical Centre and demonstrating clinical signs compatible with ILI were prospectively included in the present study. A case of ILI was defined as an adult or child demonstrating fever of 38.5°C or higher and one of the following symptoms: cough or sore throat, headache, or myalgia. These symptoms were compatible with a symptomatic infection by seasonal A or B influenza virus or by the new pandemic influenza A/H1N1v virus during October 2009 in France (32, 33). Patients suffering from severe ILI with respiratory distress or febrile confusion, as well as pregnant women and children aged less than 12 months, were hospitalized and potentially treated with oseltamivir or zanamivir (33). The Hospital Ethic's Committee (CHU de Reims, Champagne-Ardenne, France) approved the study, and informed consent was obtained from the patients or from the subjects' families at the time of inclusion. See Table 2 for clinical details of the patients.
Respiratory samples.
For each enrolled child (aged <15 years), nasopharyngeal secretions were collected using sterile physiological saline fluid with a disposable mucus extractor at the time of hospital admission, according to the recent European Respiratory Society guideline (4). For each adult (aged ≥15 years) enrolled in the present study, nasal swabbing using the Virocult system (Corsham, United Kingdom) was performed to collect upper respiratory tract cells. Nasopharyngeal samples were then rapidly transported to the virology laboratory, where they were divided into three sterile tubes and stored at −80°C until used.
RT-PCR DNA microarray detection of human respiratory viruses. (i) Extraction phase.
Total nucleic acid extraction was performed by using a NucliSens easyMAG instrument (bioMérieux, Lyon, France) according to the manufacturer's instructions. Briefly, 200 μl of a nasopharyngeal aspirate sample was treated with 20 μl proteinase K (Merck Novagen, Darmstadt, Germany) for 20 min at 56°C and then added to 2 ml of lysis buffer, and the mixture was incubated for 10 min at room temperature. The treated sample was then transferred to the well of a plastic vessel with 50 μl of silica. This was followed by an automatic magnetic separation phase. Nucleic acids were recovered in 50 μl elution buffer and then stored at −80°C (3, 23).
(ii) Simultaneous detection of 21 human respiratory pathogens by a combination of two RT-PCR DNA microarray detection systems.
Clart PneumoVir and Clart FluAVir kits (Genomica, Madrid, Spain) were used according to the manufacturer's instructions, allowing simultaneous detection and identification of 21 different types and subtypes of human respiratory viruses (influenza A virus [seasonal A/H1N1 and A/H3N2 and new influenza A/H1N1v virus strains], influenza B virus, influenza C virus, parainfluenza virus 1, parainfluenza virus 2, parainfluenza virus 3, parainfluenza virus 4, parainfluenza virus 4 A, parainfluenza virus 4 B, respiratory syncytial virus A, respiratory syncytial virus B, rhinovirus, adenovirus, enterovirus type B, bocavirus, coronavirus E-229, metapneumovirus A, and metapneumovirus B) in 24 serial samples. The diagnostic sensitivity and specificity for each virus were previously determined by a comparative evaluation of these two new nested RT-PCR multiplex systems against classical nested RT-PCR-based molecular biology techniques, as described by Coiras et al. (8, 10). The accuracy of the assay in terms of specificity and sensitivity was strictly controlled during extraction and amplification through use of an internal control and during hybridization, with at least 3 probes per target detected (Genomica, Madrid, Spain). In order to avoid subjective interpretations, the results were processed by a microarray reader using specific software provided by the manufacturer, which allowed automatic detection and interpretation of the results, with a full and specific diagnosis for each analysis in a report. No RT-PCR inhibitors were detected in any of the 95 respiratory samples analyzed in the present study (not shown). In cases of invalid data corresponding to abnormalities occurring during hybridization or incubation with conjugate during the washing phase (n =5) or in the lecture and interpretation phases of the microarray system (n = 6), the samples were fully reanalyzed with new extraction and amplification phases.
(iii) Classical real-time RT-PCR assays for detection and identification of influenza A viruses.
Real-time RT-PCR assays were performed in a final volume of 25 μl containing 5 μl RNA, a 10 μM concentration of each primer, 5 μM probe, and 0.5 U enzyme mix (SuperScriptIII Platinum one-step quantitative RT-PCR system; Invitrogen, Carlsbad, CA). Type A influenza virus RNA was detected by a real-time RT-PCR targeting the conserved matrix gene segment, using GRAM/7Fw (5′-CTTCTAACCGAGGTCGAAACGTA-3′) and GRAM/161Rv (5′-GGTGACAGGATTGGTCTTGTCTTTA-3′) primers and GRAM probe/52/+ probe (5′-6-carboxyfluorescein-TCAGGCCCCTCAAAGCCGAG-BHQ-1-3′) as described previously (39). The quality of the specimens was assessed by a real-time RT-PCR targeting the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as described previously (21). Amplification was performed on a Bio-Rad IQ real-time PCR system (Bio-Rad, Marnes la Coquette, France). Synthetic influenza virus M and GADPH RNA strands obtained previously by in vitro transcription of synthetic plasmids were included in each serial assay and were used as positive controls. Sterile diethyl pyrocarbonate-treated water was used as an RT- and PCR-negative control.
Statistical analyses.
Chi-square tests, Fisher's exact tests (with or without Yates's correction), Student's t tests, and Mann-Whitney tests were carried out when necessary with SAS software, version 8.2 (SAS Institute, Cary, NC). Results were considered statistically significant for two-sided P values of <0.05.
RESULTS
Prevalence of detection of respiratory viral infections and coinfections in patients with ILI by use of two RT-PCR DNA microarray systems.
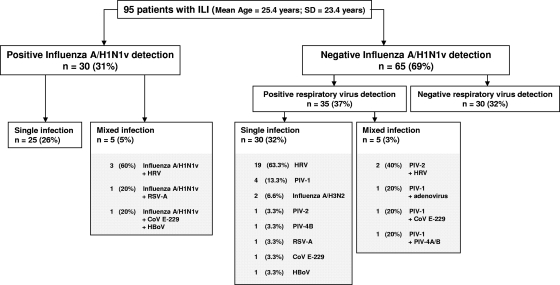
In order to assess the prevalence of upper respiratory tract infections in patients with ILI, we concomitantly used two commercially available RT-PCR DNA microarray systems. Of the 95 respiratory samples tested, 30 (31%) were positive for the detection of influenza A/H1N1v virus infection by both RT-PCR DNA microarray and classical real-time RT-PCR detection assays, whereas 65 samples remained negative for the detection of influenza A/H1N1v virus strains by both techniques (Fig. 1). We concomitantly used a second RT-PCR DNA microarray system allowing the detection of 16 respiratory viruses other than influenza A or B virus, which represented a main advantage compared to the use of only one of the two commercially available systems. Table 1 depicts the prevalences of detection of total, single, and mixed upper respiratory tract viral infections obtained using the combination of these two RT-PCR DNA microarray systems. Interestingly, we identified that among the 30 influenza A/H1N1v virus infection cases, 25 (26%) were monoinfections, whereas 5 were mixed infections (Table 1 and Fig. 1). Among the multiple infections, we identified 3 cases of influenza A/H1N1v virus and human rhinovirus dual infection, 1 case of influenza A/H1N1v virus and RSV A dual infection, and 1 case of influenza A/H1N1v virus, coronavirus E-229, and HBoV triple infection (Fig. 1). In addition, for the 65 samples that were negative for influenza A/H1N1v virus detection, the use of a second RT-PCR and DNA microarray system allowed the identification of 35 viral respiratory infections, with 30 (32%) single infections and 5 (3%) mixed infections. The most frequent viruses responsible for single infections were HRV (63.3%), PIV-1 (13.3%), and influenza A/H3N2 virus (6.6%) (Fig. 1). Among the 5 mixed infections observed, the most frequent viruses detected were HRV and PIV-1, which are well known to be clinically responsible for ILI (Fig. 1) (14, 33). In summary, the combination of the RT-PCR DNA microarray systems allowed us to detect viral respiratory tract infections in 65 (68.4%) of 95 patients with ILI. Of these 65 viral infections, 55 (85%) were single viral infections, whereas 10 (15%) were mixed viral infections (Fig. 1).
FIG. 1.
Prevalence of detection of upper respiratory tract viral infections in patients with ILI by use of a combination of two RT-PCR DNA microarray systems. The distribution (%) of the total number of single (n = 55; 58%) and mixed (n = 10; 10.6%) viral infections among 95 respiratory samples from patients with ILI during the early stage of the French influenza A/H1N1v pandemic (October 2009) was determined using two RT-PCR DNA microarray systems. Only one respiratory sample was tested for each patient. HRV, human rhinovirus; PIV, parainfluenza viruses; CoV E-229, coronavirus E-229; RSV, respiratory syncytial virus; HBoV, human bocavirus.
TABLE 1.
Prevalence of detection of single and mixed upper respiratory tract viral infections in 95 patients with ILI by use of two RT-PCR DNA microarray systems
| Virus | Total no. (%) of patients |
||
|---|---|---|---|
| Positive detection by RT-PCR DNA microarray systems | Single virus detection | Mixed virus detection | |
| Influenza A/H1N1v virus | 30 (31.6) | 25 (26.3) | 5 (5.3) |
| HRV | 24 (25.3) | 19 (2) | 5 (5.3) |
| PIV-1 | 7 (7.4) | 4 (4.3) | 3 (3.2) |
| PIV-2 | 3 (3.2) | 1 (1.0) | 2 (2.1) |
| CoV E-229 | 3 (3.2) | 1 (1.0) | 2 (2.1) |
| Influenza A/H3N2 virus | 2 (2.1) | 2 (2.1) | 0 (0) |
| PIV-4 A or B | 2 (2.1) | 1 (1.0) | 1 (1.0) |
| RSV A | 2 (2.1) | 1 (1.0) | 1 (1.0) |
| HBoV | 2 (2.1) | 1 (1.0) | 1 (1.0) |
| Adenovirus | 1 (1.0) | 0 (0) | 1 (1.0) |
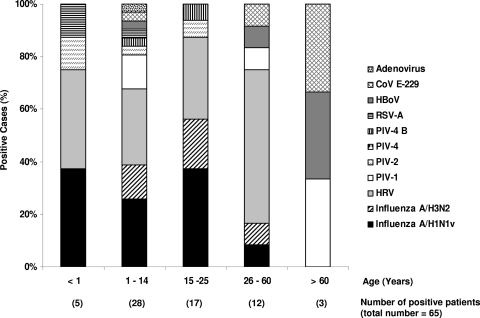
Distribution of positive respiratory viral infection cases by age.
Figure 2 shows the distribution of the 65 positive viral cases by age. Interestingly, we observed a significantly larger number of positive cases for influenza A/H1N1v virus infection in patients aged less than 25 years (P = 0.01). Moreover, the influenza A/H3N2 virus infections appeared to be detected more frequently in patients aged less than 25 years than in patients from the other age classes (P = 0.03). Interestingly, no influenza A/H1N1v or A/H3N2 virus infections were detected in patients aged more than 60 years. During the study period, HRV appeared to be the second most common cause of ILI in patients aged less than 25 years. Moreover PIV, CoVs, and HBoV were identified as causes of ILI in 8 to 34% of patients aged more than 25 years during the study period (Fig. 2).
FIG. 2.
Distribution (%) of positive respiratory viral infections by age class among patients with ILI during the early stage of the influenza A/H1N1v pandemic (northern France, October 2009). HRV, human rhinovirus; PIV, parainfluenza viruses; CoV E-229, coronavirus E-229; RSV, respiratory syncytial virus; HBoV, human bocavirus.
Demographic and clinical features of ILI patient subgroups according to virological findings.
Table 2 depicts the demographic and clinical features of ILI patients according to the detection of a single or mixed infection or the absence of viral infection in the upper respiratory tract. Concerning viral positivity for the upper respiratory tracts of ILI patients, no significant sex ratio was observed (65% of patients were female, and 72% were male; P = 0.33). However, we observed significant proportions of positive viral detection (single and mixed infections) for the male and child subgroups (P < 0.001), whereas this proportion was not significant for the female subgroup (34 versus 18 cases; P = 0.09) (Table 2). Moreover, the time delay between the beginning of clinical signs and the time of respiratory sampling appeared not to be significantly different between the subgroups of patients with and without virus detection in the respiratory tract (P = 0.13). This time delay appeared not to be significantly longer in females with positive viral detection than in those without viral detection in the upper respiratory tract (P = 0.09) (data not shown). During the study period (October 2009), 8 pregnant women were hospitalized and treated with antineuraminidase drugs. Among them, only two were positive for influenza A/H1N1v virus, and none developed any clinical complications such as respiratory bacterial infections or premature delivery (not shown). Moreover, the presence of a mixed viral infection did not appear to have a significant impact on the prescription of antineuraminidase treatment or on the admission of patients into the infectious disease department or intensive care unit (Table 2). No specific viral monoinfections or mixed viral infections appeared to be associated significantly with secondary hospitalization in infectious disease or intensive care departments (P > 0.5). No negative clinical outcomes were observed during the study period (Table 2).
TABLE 2.
Comparison of demographic and clinical features between ILI patient subgroups with single or mixed infections or the absence of viral infection in the upper respiratory tract
| Feature | No. of cases | Mean age (yr) | SD (yr) | No. of single viral infections | No. of mixed viral infections | P valued | No. cases with no viral infection | P valuee |
|---|---|---|---|---|---|---|---|---|
| All patients | 95 | 25.4 | 23.4 | 55 | 10 | <10−3 | 30 | <10−3 |
| Males | 43 | 22.2 | 18.5 | 26 | 5 | <10−3 | 12 | <10−3 |
| Females | 52 | 29.9 | 27.6 | 29 | 5 | <10−3 | 18 | 0.09 |
| Adultsa | 56 | 39.9 | 20.3 | 28 | 4 | <10−3 | 24 | 0.18 |
| Childrenb | 32 | 6.2 | 4.1 | 24 | 4 | <10−3 | 4 | <10−3 |
| Infantsc | 7 | 0.41 | 0.2 | 3 | 2 | 1f | 2 | 0.29f |
| Mean (SD) delay (days) between beginning of clinical signs and time of respiratory sampling | 1.24 (1.41) | 1.77 (1.54) | 0.12g | 1.80 (2.42) | 0.13g | |||
| Oseltamivir or zanamivir treatment | 22 | 36.2 | 29.1 | 7 | 2 | 0.13 | 13 | 0.37 |
| Vaccination against seasonal influenza viruses | 3 | 25.7 | 23.3 | 1 | 0 | 1f | 2 | 1f |
| Hospital admission | 22 | 31.7 | 29.0 | 8 | 0 | <10−2 | 14 | 0.13 |
| Intensive care unit admission | 2 | 66 | 12.7 | 1 | 0 | 1f | 1 | 1f |
| Good clinical outcome | 95 | 25.4 | 23.4 | 41 | 13 | <10−3 | 41 | 0.08 |
Aged ≥15 years.
Aged >12 months and <15 years.
Aged ≤12 months.
Single versus mixed viral infection. P values of <0.05 were considered significant.
Viral infection (single and mixed) versus absence of viral infection. P values of <0.05 were considered significant.
Fisher's test.
Student's t test.
DISCUSSION
ILI can be related to seasonal A and B influenza viruses, to the new pandemic influenza A/H1N1v virus strain, and also to a large number of common or newly discovered respiratory viruses (13, 15, 16, 36, 43). Therefore, rapid and reliable screening of a large panel of respiratory viruses responsible for ILI is of major epidemiological and clinical interest for monitoring an influenza pandemic wave (33). In the present study, we prospectively tested the presence of respiratory viruses in the upper respiratory tracts of 56 adults and 39 children visiting the emergency room of the Reims University Medical Centre (northern France) for ILI in October 2009, at the beginning of the epidemic circulation of influenza A/H1N1v virus in France (32). Interestingly, the presence of respiratory viruses in the upper respiratory tracts of ILI patients was tested using a combination of two commercially available RT-PCR DNA microarray detection systems allowing, concomitantly, the specific detection, typing, and subtyping of influenza A virus strains, including A/H1N1v, and the detection of 20 other common or newly discovered respiratory viruses. Moreover, the results obtained by the association of these two commercially available kits were compared to those of a reference generic real-time RT-PCR assay used for the rapid detection of all known influenza A virus strains (39). Finally, our data allowed us to assess the potential application of two commercially available RT-PCR DNA microarray detection systems for routine virological diagnosis or weekly epidemiological survey of respiratory viruses responsible for ILI.
In the present study, the combination of the RT-PCR DNA microarray systems allowed us to detect viral respiratory tract infections in 65 (68.4%) of the 95 patients with ILI (Fig. 1). A Peruvian study reported a 2-year prospective investigation, from 2006 to 2008, and demonstrated positive respiratory viral infections in 43% of 6,835 ILI patients by use of a combination of real-time PCR and classical cell culture techniques (22). Moreover, Wallace et al. and Hasman et al. reported positive viral infections by use of RT-PCR multiplex techniques for 60% of 240 and 68% of 154 ILI patients, respectively (19, 37). Our levels of positive detection appeared to be in accordance with those obtained in these two studies. However, our incidence data appeared to be higher than those obtained in the Peruvian study (68.4% versus 43%; P < 0.001) (22); this could be explained by the larger number of common and newly discovered respiratory pathogens potentially detectable by the RT-PCR DNA microarray detection systems as well as by the high sensitivity of this detection technology, allowing limits of detection ranging from 10 to 100 copies of viral genomes per amplification tube. Interestingly, in the present study, influenza A/H1N1v virus was detected in only 30 (31%) of the 95 tested patients, and influenza A/H3N2 virus was detected in only 2 (2.1%) of the 95 tested patients (Fig. 1). Numerous previous studies detected the presence of influenza viruses in the respiratory tracts of ILI patients by use of real-time RT-PCR systems during 2007 to 2008 (18, 30, 41, 42). These studies reported levels of positive influenza A virus seasonal strain detection ranging from 25% in Peru to 40% in Greece (18, 22). Our level of influenza A virus detection (33.1%) appears to be in agreement with previous published worldwide studies (17, 22, 30, 41, 42; L. Finelli, U.S. Centers for Disease Control and Prevention, personal communication). Moreover, the use of a reference generic influenza A virus real-time RT-PCR system confirmed all of the positive and negative results obtained by the tested influenza A virus RT-PCR DNA microarray systems (data not shown). Taken together, these data suggest that the use of new commercially available RT-PCR DNA microarray systems is valuable for the detection of influenza A virus strains in clinical virology practice. However, further prospective and large multicenter clinical studies would be useful to assess the specificity and sensitivity of the detection of influenza A virus strains in a larger number of clinical samples by the use of new commercially available RT-PCR DNA microarray systems.
Interestingly, the combination of two RT-PCR DNA microarray systems allowed us to detect not only influenza A virus strains but also 17 other common or newly discovered respiratory viruses in the same sample. This approach demonstrated that HRV and PIV were important causes of ILI, with 25 and 10.5% prevalence, respectively (Fig. 1). These levels of HRV and PIV detection appeared significantly higher than those previously described in Peru (PIV prevalence of 3.2% and HRV prevalence of 2.6%; P < 0.001), indicating that our RT-PCR DNA microarray system might be able to detect a larger number of genotypes of PIV and HRV, with a higher sensitivity, than the classical real-time PCR systems (22). This potential capacity to detect a large number of genotypes among these two groups of pathogens might be of major epidemiological and clinical interest before, during, and after an influenza pandemic wave. Moreover, the use of RT-PCR DNA microarray systems allowed us to demonstrate the presence of 10 (10.5%) mixed infections in the upper respiratory tracts of ILI patients. HRVs were identified in 50% of these 10 mixed infection cases, whereas PIV and coronaviruses were identified in 5 and 2 of the 10 mixed infections, respectively (Fig. 1). Interestingly, no mixed infections with two different influenza viruses were identified during our study period, suggesting that such events are rare, as described previously for classical influenza A epidemic seasons (14). Previous studies using real-time PCR techniques reported viral coinfection rates in the 5 to 10% range (18, 19, 22, 37). During these studies, influenza A virus, RSV, PIV, and HMPV were the most commonly implicated viruses in mixed infections (18, 19, 22, 37). In concomitant viral respiratory infections, virus detection by RT-PCR techniques could be linked to past, ongoing, or beginning viral respiratory infections in the upper respiratory tract (31, 38). Because our RT-PCR DNA microarray systems are only qualitative, not quantitative, detection techniques, it was not possible to determine which virus was predominant and therefore could be considered the etiological cause of ILI. However, in cases of mixed infection, it has been hypothesized that picornaviruses could serve as a clinical illness promotion factor, functioning additively or synergistically in the pathogenesis of lower respiratory syndromes such as bronchiolitis (27). Therefore, preexisting asymptomatic HRV airway infections may enhance the risk of ILI. Confirmation of a cofactor role of HRV infection will require longitudinal studies over several seasons, comparing disease severity with frequency of coinfection for cohorts of ILI patients.
In the present study, we compared demographic and clinical features between ILI patient subgroups with single or mixed infections or the absence of viral infection in the upper respiratory tract by using a combination of two RT-PCR DNA microarray systems. Interestingly, the distribution of positive respiratory influenza A/H1N1v and A/H3N2 viral infection cases by age class demonstrated a significant larger proportion of positive viral detection cases among patients aged less than 25 years (P = 0.01), whereas the patients aged more than 60 years appeared to be exempted from influenza virus infection. These data are in agreement with 2007-2008 data published by European and Indian studies demonstrating that the proportion of laboratory-confirmed influenza cases was higher among participants aged 6 to 20 years (18, 35, 42). Moreover, an Australian study from April to September 2009 reported the same significantly higher proportion of positive influenza A/H1N1v virus infections among patients aged 0 to 24 years, with a median age of 15 years (17). In the present study, the median age of patients with notifiable cases was 12.8 years (range, 6 months to 31 years); this was in contrast to the median age of patients during the early stages of the pandemic in the United States (20 years) and Spain (22 years) (12, 17). However, the median age of pandemic patients in the United States had dropped from 20 to 13 years by the time approximately 10,000 cases had been reported (Finelli, personal communication). These findings and ours might reflect amplification of the pandemic in school-aged children and intense follow-up and testing of this age group (17). Moreover, the absence of influenza A virus infection in patients aged more than 60 years was probably due to previous exposure to antigenically similar H1N1 viruses, therefore possibly explaining the presence of peripheral blood anti-influenza A/H1N1 virus neutralizing antibodies capable of inducing protective cross-reactivity against the new pandemic strain (20). Moreover, we observed a significant proportion of positive viral detection in males (single and mixed infections, adults and children [P < 0.01]), whereas this was not significant for the female subgroup (Table 2). These data were not explained by the time delay between the beginning of clinical signs and the time of respiratory sampling but could possibly be explained by intense follow-up and Health Ministry-recommended testing of females, and specifically pregnant women, for whom compulsory hospital visits were observed during the early stages of the French influenza A/H1N1v pandemic (data not shown). Finally, during the study period, no negative clinical outcomes were observed for patients with notified ILI cases who were infected or not by influenza A/H1N1v virus (Table 2). However, only larger national studies of ILI might be able to compare the clinical profiles and mortality rates between the 2009 influenza A/H1N1v pandemic and previous influenza seasons.
In conclusion, the use of RT-PCR DNA microarray systems in clinical virology practice allows for rapid and accurate detection of conventional and newly discovered viral respiratory pathogens in children and adults suffering from ILI. These molecular assays could be of major interest for developing new epidemiological survey systems for respiratory viral infections in hospitalized subjects and outpatients. Moreover, the use of such rapid and reliable molecular tools is needed to provide not only epidemiological and virological data but also the opportunity to understand the emergence of novel influenza virus strains.
Acknowledgments
We are indebted to the physicians who prospectively included the children in the present study.
This work was supported in part by a grant for clinical and virological research (IFR53/EA-4303) from the Medical University and School of Medicine of Reims, France. Fanny Renois was supported by an official grant from the French Army department (Bourse DGA [Délégation Générale de l'Armement], Ministère de la Défense).
None of the authors have a commercial or other association that might pose a conflict of interest.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Andréoletti, L., M. Lesay, A. Deschildre, V. Lambert, A. Dewilde, and P. Wattré. 2000. Differential detection of rhinoviruses and enteroviruses RNA sequences associated with classical immunofluorescence assay detection of respiratory virus antigens in nasopharyngeal swabs from infants with bronchiolitis. J. Med. Virol. 3:341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellau-Pujol, S., A. Vabret, L. Legrand, J. Dina, S. Gouarin, J. Petitjean-Lecharbonnier, B. Pozzetto, C. Ginevra, and F. Freymuth. 2005. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 126:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. Van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 3:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouscambert, M., B. Lina, A. Trompette, J. Motte, and L. Andréoletti. 2005. Detection of human metapneumovirus RNA sequences in nasopharyngeal aspirates of young French children with acute bronchiolitis by real-time reverse transcriptase PCR and phylogenetic analysis. J. Clin. Microbiol. 43:1411-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briese, T., G. Palacios, M. Kokoris, O. Jabado, Z. Liu, N. Renwick, V. Kapoor, I. Casas, F. Pozo, R. Limberger, P. Perez-Brena, J. Ju, and W. I. Lipkin. 2005. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg. Infect. Dis. 11:310-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownie, J., S. Shawcross, J. Theaker, D. Whitcombe, R. Ferrie, C. Newton, and S. Little. 1997. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 25:3235-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coiras, M. T., J. C. Aguilar, M. L. Garcia, I. Casas, and P. Perez-Brena. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 72:484-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coiras, M. T., M. R. Lopez-Huertas, G. Lopez-Campos, J. C. Aguilar, and P. Perez-Brena. 2005. Oligonucleotide array for simultaneous detection of respiratory viruses using a reverse-line blot hybridization assay. J. Med. Virol. 76:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coiras, M. T., P. Perez-Brena, M. L. Garcia, and I. Casas. 2003. Simultaneous detection of influenza A, B and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested-PCR assay. J. Med. Virol. 69:132-144. [DOI] [PubMed] [Google Scholar]
- 11.Conejero-Goldberg, C., E. Wang, C. Yi, T. E. Goldberg, L. Jones-Brando, F. M. Marincola, M. J. Webster, and E. F. Torrey. 2005. Infectious pathogen detection arrays: viral detection in cell lines and postmortem brain tissue. Biotechniques 39:741-751. [DOI] [PubMed] [Google Scholar]
- 12.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 25:2605-2615. [DOI] [PubMed] [Google Scholar]
- 13.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguière, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Müller, V. Rickerts, M. Stürmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1978. [DOI] [PubMed] [Google Scholar]
- 14.Falchi, A., C. Arena, L. Andréoletti, J. Jacques, N. Lévêque, T. Blanchon, B. Lina, C. Turbelin, Y. Dorléans, A. Flahault, J. P. Amoros, G. Spadoni, F. Agostini, and L. Varesi. 2008. Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. J. Clin. Virol. 2:148-151. [DOI] [PubMed] [Google Scholar]
- 15.Falsey, A. R., E. E. Walsh, and F. G. Hayden. 2002. Rhinovirus and coronavirus infection associated hospitalisations among older adults. J. Infect. Dis. 185:1338-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 17.Fielding, J., N. Higgins, J. Gregory, K. Grant, M. Catton, I. Bergeri, R. Lester, and H. Kelly. 2009. Pandemic H1N1 influenza surveillance in Victoria, Australia, April-September, 2009. Euro. Surveill. 42:19368. [DOI] [PubMed] [Google Scholar]
- 18.Gioula, G., D. Chatzidimitriou, A. Melidou, M. Exindari, and V. Kyriazopoulou-Dalaina. 2010. Contribution of human metapneumovirus to influenza-like infections in North Greece, 2005-2008. Euro. Surveill. 9:19499. [DOI] [PubMed] [Google Scholar]
- 19.Hasman, H., C. T. Pachucki, A. Unal, D. Nguyen, T. Devlin, M. E. Peeples, and S. A. Kwilas. 2009. Aetiology of influenza-like illness in adults includes parainfluenzavirus type 4. J. Med. Microbiol. 58:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, K. Takahashi, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 7258:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacques, J., H. Moret, F. Renois, N. Lévêque, J. Motte, and L. Andréoletti. 2008. Human bocavirus quantitative DNA detection in French children hospitalized for acute bronchiolitis. J. Clin. Virol. 43:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laguna-Torres, V. A., J. Gómez, V. Ocaña, P. Aguilar, T. Saldarriaga, E. Chavez, J. Perez, H. Zamalloa, B. Forshey, I. Paz, E. Gomez, R. Ore, G. Chauca, E. Ortiz, M. Villaran, S. Vilcarromero, C. Rocha, O. Chincha, G. Jiménez, M. Villanueva, E. Pozo, J. Aspajo, and T. Kochel. 2009. Influenza-like illness sentinel surveillance in Peru. PLoS One 4:e6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loens, K., K. Bergs, D. Ursi, H. Goossens, and M. Ieven. 2007. Evaluation of NucliSens easyMAG for automated nucleic acid extraction from various clinical specimens. J. Clin. Microbiol. 45:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahony, J. B., S. Chong, F. Merante, S. Yaghoubian, T. Sinba, C. Lisle, and R. Janeczko. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahony, J. B. 2008. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 21:707-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahony, J. B., G. Blackhouse, J. Babwah, M. Smieja, S. Buracond, S. Chong, W. Ciccotelli, T. O'Shea, D. Alnakhli, M. Griffiths-Turner, and R. Goeree. 2009. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J. Clin. Microbiol. 47:2812-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMillan, J. A., L. B. Weiner, A. M. Higgins, and V. J. Lamparella. 1993. Pharyngitis associated with herpes simplex virus in college students. Pediatr. Infect. Dis. J. 4:280-284. [DOI] [PubMed] [Google Scholar]
- 28.McMillan, J. A., L. M. Leonard, A. M. Weiner, K. Higgins, and M. T. Macknight. 1993. Rhinovirus infection associated with serious illness among pediatric patients. Pediatr. Infect. Dis. 12:321-325. [DOI] [PubMed] [Google Scholar]
- 29.Monto, A. S. 2002. Epidemiology of viral respiratory infections. Am. J. Med. 112(Suppl. 6A):4S-12S. [DOI] [PubMed] [Google Scholar]
- 30.Njouom, R., S. A. Mba, D. N. Noah, V. Gregory, P. Collins, P. Cappy, A. Hay, and D. Rousset. 2010. Circulation of human influenza viruses and emergence of oseltamivir-resistant A(H1N1) viruses in Cameroon, Central Africa. BMC Infect. Dis. 10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nokso-Koivisto, J., T. J. Kinnari, P. Lindalh, T. Hovi, and A. Pitkäranta. 2002. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J. Med. Virol. 66:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nougairede, A., L. Ninove, C. Zandotti, S. D. Thiberville, C. Gazin, B. La Scola, R. N. Charrel, and X. de Lamballerie. 2010. Interim report on the A/H1N1 influenza virus pandemic in Marseille, France, April-November 2009. Clin. Microbiol. Infect. 16:322-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nougairede, A., L. Ninove, C. Zandotti, X. de Lamballerie, C. Gazin, M. Drancourt, B. La Scola, D. Raoult, and R. N. Charrel. 2010. Point of care strategy for rapid diagnosis of novel A/H1N1 influenza virus. PLoS One 17:e9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portes, S. A., E. E. Da Silva, M. M. Siqueira, A. M. De Filippis, and M. M. Krawczuk. 1998. Enteroviruses isolated from patients with acute respiratory infections during seven years in Rio de Janeiro (1985-1991). Rev. Inst. Med. Trop. Sao Paulo 40:337-342. [DOI] [PubMed] [Google Scholar]
- 35.Surveillance Group for New Influenza A(H1N1) Virus Investigation and Control in Spain. 2009. New influenza A(H1N1) virus infections in Spain, April-May 2009. Euro. Surveill. 19:19209. http://www.eurosurveillance.org. [DOI] [PubMed] [Google Scholar]
- 36.Van den Hoogen, B., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered pneumovirus isolated from young children with respiratory disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace, L. A., T. C. Collins, J. D. Douglas, S. McIntyre, J. Millar, and W. F. Carman. 2004. Virological surveillance of influenza-like illness in the community using PCR and serology. J. Clin. Virol. 31:40-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waner, J. L. 1994. Mixed viral infections: detection and management. Clin. Microbiol. Rev. 7:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whiley, D. M., and T. P. Sloots. 2005. A 5′-nuclease real-time reverse transcriptase-polymerase chain reaction assay for the detection of a broad range of influenza A subtypes, including H5N1. Diagn. Microbiol. Infect. Dis. 53:335-337. [DOI] [PubMed] [Google Scholar]
- 40.Williams, B. G., E. Gouws, C. Boschi-Pinto, J. Bryce, and C. Dye. 2002. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect. Dis. 1:25-32. [DOI] [PubMed] [Google Scholar]
- 41.Yang, P., W. Duan, M. Lv, W. Shi, X. Peng, X. Wang, Y. Lu, H. Liang, H. Seale, X. Pang, and Q. Wang. 2009. Review of an influenza surveillance system, Beijing, People's Republic of China. Emerg. Infect. Dis. 10:1603-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaman, R. U., A. S. Alamgir, M. Rahman, E. Azziz-Baumgartner, E. S. Gurley, M. A. Sharker, W. A. Brooks, T. Azim, A. M. Fry, S. Lindstrom, L. V. Gubareva, X. Xu, R. J. Garten, M. J. Hossain, S. U. Khan, L. I. Faruque, S. S. Ameer, A. I. Klimov, M. Rahman, and S. P. Luby. 2009. Influenza in outpatient ILI case-patients in national hospital-based surveillance, Bangladesh, 2007-2008. PLoS One 12:e8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zambon, M. C., J. Stockton, J. Clewley, and D. F. Fleming. 2001. Contribution of influenza and respiratory syncytial virus to community cases of influenza-like illness: an observational study. Lancet 358:1410-1416. [DOI] [PubMed] [Google Scholar]