Abstract
The current study describes the development of a unique real-time PCR assay for the detection of mutations conferring drug resistance in Mycobacterium tuberculosis. The rifampicin resistance determinant region (RRDR) of rpoB and specific regions of katG and the inhA promoter were targeted for the detection of rifampin (RIF) and isoniazid (INH) resistance, respectively. Additionally, this assay was multiplexed to discriminate Mycobacterium tuberculosis complex (MTC) strains from nontuberculous Mycobacteria (NTM) strains by targeting the IS6110 insertion element. High-resolution melting (HRM) analysis following real-time PCR was used to identify M. tuberculosis strains containing mutations at the targeted loci, and locked nucleic acid (LNA) probes were used to enhance the detection of strains containing specific single-nucleotide polymorphism (SNP) transversion mutations. This method was used to screen 252 M. tuberculosis clinical isolates, including 154 RIF-resistant strains and 174 INH-resistant strains based on the agar proportion method of drug susceptibility testing (DST). Of the 154 RIF-resistant strains, 148 were also resistant to INH and therefore classified as multidrug resistant (MDR). The assay demonstrated sensitivity and specificity of 91% and 98%, respectively, for the detection of RIF resistance and 87% and 100% for the detection of INH resistance. Overall, this assay showed a sensitivity of 85% and a specificity of 98% for the detection of MDR strains. This method provides a rapid, robust, and inexpensive way to detect the dominant mutations known to confer MDR in M. tuberculosis strains and offers several advantages over current molecular and culture-based techniques.
The World Health Organization (WHO) estimates that approximately one-third of the world's population is infected with Mycobacterium tuberculosis, with an estimated 9.27 million new cases reported in 2007 (20). In that year alone, an estimated 1.77 million people died from this treatable disease. Despite this significant burden, only a limited number of tests have been developed and implemented for the rapid diagnosis of tuberculosis (TB). Further, since the majority of TB disease burden occurs in underdeveloped and resource-limited settings, the need for a cost-efficient method is paramount.
The emergence of drug-resistant strains of M. tuberculosis is one of the most critical issues facing TB researchers and clinicians today. Multidrug-resistant (MDR) M. tuberculosis is defined as being resistant to the two best first-line drugs used to treat TB: rifampin (RIF) and isoniazid (INH). Extensively drug-resistant (XDR) M. tuberculosis is defined as having additional resistance to a fluoroquinolone (ciprofloxacin, moxifloxicin, etc.) and an injectable drug (kanamycin, capreomycin, or amakacin), the two best classes of second-line drugs. The WHO estimates that 5% of new TB cases are MDR, with approximately 10% of those actually being XDR (20). Compounding this problem is the fact that no new drugs have been developed and approved for the treatment of TB in the past 30 years (16). The limited number of antibiotics available to treat TB necessitates rapid diagnosis not only to reduce the spread of drug-resistant strains but also to monitor and limit the emergence of newly resistant strains.
While RIF and INH are very effective in the treatment of susceptible strains of M. tuberculosis, drug resistance can emerge quickly, in part due to patient nonadherence to the multidrug regimen or noncontinuous treatment. The molecular basis of resistance to these drugs is well documented. The target of RIF is the β-subunit of bacterial DNA-dependent RNA polymerase, which is encoded by the rpoB gene. At the genetic level, the majority of RIF resistance is due to the accumulation of mutations within an 81-bp region of rpoB, termed the rifampicin resistance determinant region (RRDR). Mutations within this region account for up to 98% of the RIF resistance observed (15). The strong correlation between genotypic changes in this region resulting in phenotypic resistance makes the RRDR an optimal target for the design of rapid molecular diagnostics.
There are two described mechanisms that account for the majority of INH resistance. The most common mechanism involves mutations within the katG gene, which encodes a catalase peroxidase whose activity is required for the activation of INH (9). Nucleotide changes resulting in amino acid substitutions at codon 315 of katG account for up to 50% of the clinical resistance to INH (15). Another less common mutation occurs in the promoter region of the inhA gene, which encodes enoyl-ACP reductase, which is required for mycolic acid biosynthesis (18). Mutations at this locus account for up to 34% of the clinical INH resistance observed and are typically found in combination with additional mutations in katG (15).
The vast majority of mutations that occur within rpoB, katG, and the inhA promoter regions are due to accumulation of single-nucleotide polymorphisms (SNPs), of which there are four classes (8). Class I SNPs, also called transitions, are changes in which a purine is exchanged for a purine (A/G→G/A) or a pyrimidine is exchanged for a pyrimidine (C/T→T/C) (8). Class II, III, and IV SNP changes are collectively referred to as transversions, and all involve the change of a purine to a pyrimidine, or vice versa (17). Class II changes result in A/C→C/A or T/G→G/T transversions, class III changes result in C/G→G/C transversions, and class IV changes result in A/T→T/A changes (8). These genetic mutations often result in phenotypic changes, such as RIF and INH resistance observed in M. tuberculosis, and are excellent targets for rapid molecular diagnostics.
A significant obstacle in controlling TB is the amount of time required to reach a diagnosis. Due to the slow growth rate of M. tuberculosis, the initial diagnosis can take up to 6 weeks, with up to an additional 12 weeks to obtain drug susceptibility profiles for clinical isolates, depending on the techniques available to the laboratory. These labor-intensive methods can cause significant delays in identifying MDR or XDR cases, adjusting treatment regimens, and initiating epidemiological investigations. Recently, attention has shifted toward the development of dependable, molecular-based assays that can rapidly detect drug resistance. The development of new methodologies could potentially reduce the time required to diagnose drug resistance so that effective treatment regimens can be established. Direct sequencing of genes known to have a role in antibiotic resistance is one method that is currently used. However, while reliable, it is costly and may not be readily available. Another rapid method, the GenoType MTBDRplus assay (Hain Lifescience GmbH, Nehren, Germany), has made substantial contributions to the area of rapid diagnostics but still requires approximately 8 h to complete the assay and additional training to ensure that results are interpreted correctly (7). High-resolution melt (HRM) analysis is a molecular technique that can be used for detecting subtle genetic changes, such as SNPs conferring drug resistance in M. tuberculosis. By slowly melting the DNA amplicon products of a real-time PCR assay, slight genetic differences can be visualized by changes in dissociation profiles.
The current study describes the use of multiple real-time PCR chemistries and HRM technology to detect RIF, INH, and more importantly, MDR strains of M. tuberculosis. This novel assay design is also capable of distinguishing M. tuberculosis complex bacteria (MTC) from nontuberculous mycobacterium (NTM) strains. This assay provides a rapid, robust, and inexpensive way to identify MDR TB that could result in numerous advantages over current molecular and culture-based techniques.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and drug susceptibility testing.
Clinical M. tuberculosis strains used in this study were obtained from the culture collection at the Mycobacteriology Laboratory Branch, CDC. Information regarding patients linked to the clinical isolates used in this study is protected as described in a protocol approved by the CDC Institutional Review Board. Drug susceptibility testing (DST) was performed using the agar proportion method previously described and in accordance with the guidelines of the Clinical and Laboratory Standards Institute (10). Susceptibility was tested at 1 μg/ml for RIF and 0.2, 1.0, and 5.0 μg/ml for INH.
DNA preparation and sequencing of regions of rpoB, katG, and the inhA promoter.
DNA was prepared from M. tuberculosis cultures grown at 37°C that had reached saturation by using the previously described FastPrep method (13). Amplicons were generated by PCR for regions of rpoB (rpoB-F, 5′-CTTGCACGAGGGTCAGACCA; rpoB-R, 5′-ATCTCGTCGCTAACCACGCC), katG (katG-F, 5′-AACGACGTCGAAACAGCGGC; katG-R, 5′-GCGAACTCGTCGGCCAATTC), and the inhA promoter (inhA-F, 5′-TGCCCAGAAAGGGATCCGTCATG; inhA-R, 5′-ATGAGGAATGCGTCCGCGGA). Amplicons were treated with ExoSAP-IT (Affymetrix, Inc.) according to the manufacturer's instructions and diluted 1:10 for sequencing. The sequencing reaction mixtures for the treated amplicons contained BigDye Terminator v3.1 mix and the BigDye 5× sequencing buffer (Applied Biosystems) with the same primers used for PCR. Sequencing was performed using the ABI 3130xl genetic analyzer. Sequence analysis was performed using the ABI sequence analysis software and aligned using DNASTAR Lasergene 8.0.
Real-time PCR.
Primers and probes for the real-time PCR assays were designed to target four different locations within the M. tuberculosis genome. The primers and probes targeting the rpoB gene and the IS6110 insertion element were combined into a SYBR-based duplex assay. The primers for rpoB were manually designed to selectively amplify a 152-bp amplicon spanning the RRDR (rpoB-F, 5′-GCCGCGATCAAGGAGTTCT; rpoB-R, 5′-ACGTCGCGGACCTCCAG). The primers for the IS6110 insertion element were manually designed to generate a 179-bp amplicon within the MTC-specific region of IS6110 (IS6110-F, 5′-CCACCATACGGATAGGGGA; IS6110-R, 5′-TGGACCGCCAGGGCT). In order to identify SNP transversions within rpoB, two locked nucleic acid (LNA) probes were included in this assay. Beacon Designer software (PREMIER Biosoft) was used to design two LNA probes targeting specific loci within the rpoB amplicon to detect A→T SNP changes in specific strains at Asp516 and His526, respectively (rpoB LNA D516V, 5′-aattcaTggTccAgaAcaa; rpoB LNA H526L, 5′-gtTgaCccTcaAgc; capital letters indicate a locked base). Each LNA probe was designed with a unique 5′ fluorescent label, with Quasar 705 on the LNA D516V probe and CalRd610 on the LNA H526L probe. Both were quenched on the 3′ end with Black hole quencher 2 (BHQ2).
The katG and inhA promoter mutations were targeted independently in single-plex assays. The primers were designed to span a region within each gene known to contain mutations conferring INH resistance. The primers for the katG gene were generated using Beacon Designer software (PREMIER Biosoft) and produced a 123-bp amplicon (katG-F, 5′-TCGTATGGCACCGGAACC; katG-R, 5′-CAGCTCCCACTCGTAGCC). The software was also used to design an LNA probe to bind within the amplicon and detect the Ser315Thr mutation, a G→C SNP change found in resistant strains (katG LNA S315T, 5′-atcaCcaCcgGcaTcg; capital letters indicate a locked base). The LNA S315T probe was labeled on the 5′ end with Q670 and quenched with BHQ3 on the 3′ end. The primers for the inhA promoter region were designed manually and generated a 132-bp amplicon (inhA-F, 5′-CGTTACGCTCGTGGACATAC; inhA-R, 5′-GTTTCCTCCGGTAACCAGG).
The SYBR reaction mixture was prepared using the Universal SYBR GreenER qPCR kit (Invitrogen), containing the following components per reaction mixture in all three assays: 12.5 μl of 2× master mix, 5 μl of the template, and nuclease-free water (Promega), for a total reaction volume of 25 μl. Primers and probes were added to the appropriate master mix in the following concentrations: 100 nM rpoB, IS6110, katG, and inhA primers; 250 nM katG LNA probe; 500 nM rpoB LNA516 probe; 1 μM rpoB LNA526 probe. Amplification was performed on the Rotor-Gene 6000 system (Qiagen) under the following conditions: 1 cycle of 95°C for 2 min, followed by 45 cycles of 95°C for 15 s and 63°C for 30 s, with data acquired on the 63°C step in the green, orange, red, and crimson channels. Following amplification, HRM was performed between 80°C and 89°C at a rate of 0.02°C per step. All samples were tested in duplicate. All control and unknown DNA samples were diluted such that threshold cycle values fell between cycles 19 and 29.
Melt curve and HRM data analyses.
Melt curves were generated by taking the derivative of the raw fluorescence level during each HRM step. The resulting dF/dT plot contains specific peaks at the melting temperature of the double-stranded products.
The HRM curves were analyzed by selecting two normalization regions, one occurring prior to the melting of the amplicon and one following complete separation of the two strands. The normalization regions utilized for each assay were as follows: rpoB/IS6110 assay, ∼85°C to 85.5°C (region one) and ∼88°C to 88.5°C (region two); katG assay, ∼80°C to 80.5°C (region one) and ∼83.5°C to 84°C (region two); inhA assay, ∼81.5°C to 82°C (region one) and ∼85°C to 85.5°C (region two). HRM curves were viewed in “replicate mode,” a user option whereby a single melt curve is derived by averaging all of the replicates for that sample. The “difference graph” was generated by normalizing the HRM profile of the wild-type (WT) control strain to zero and highlighting any deviations (i.e., resistant isolates) as distinct curves.
RESULTS
The rpoB/IS6110 and katG/inhA assays were used to screen a panel of DNA from 252 M. tuberculosis clinical isolates obtained from the Mycobacterial Laboratory Branch. The panel included 29 different types or combinations of mutations within rpoB (Table 1), 5 unique types or combinations of mutations within katG, and 2 distinct types or combinations of mutations within the promoter region of inhA (Table 2). The results from the assay were compared to the phenotypic DST and sequencing results for the targeted loci.
TABLE 1.
Summary of strains containing mutations within the RRDR of rpoBa
| Amino acid change(s) | SNP(s) | No. of strains |
|---|---|---|
| Asp516Ala | GAC→GCC | 1 |
| Asp516Tyr | GAC→TAC | 2 |
| Asp516Tyr, Ser531Leu | GAC→TAC, TCG→TTG | 1 |
| Asp516Val | GAC→GTC | 9 |
| Phe586Val, Asp516Val | TTC→GTC, GAC→GTC | 1 |
| Insertion at nt 1295 | GCC insertion | 2 |
| Gln490Arg | CAG→CGG | 1 |
| Gln513His, Leu533Pro | CAA→CAC, CTG→CCG | 3 |
| Gln513Leu | CAA→CTA | 1 |
| His526Arg | CAC→CGC | 3 |
| His526Arg, Arg529Gln | CAC→CGC, CGA→CAA | 1 |
| His526Asn, Ser531Leu | CAC→AAC, TCG→TTG | 1 |
| His526Asp | CAC→GAC | 4 |
| His526Gln, Leu533Pro | CAC→CAG, CTG→CCG | 1 |
| His526Gly | CAC→GGC | 1 |
| His526Leu | CAC→CTC | 2 |
| His526Tyr | CAC→TAC | 14 |
| Ile572Leu, Asp516Gly | ATC→CTC, GAC→GGC | 1 |
| Leu459Arg, Asp516Tyr | CTG→CGG, GAC→TAC | 1 |
| Leu459Arg, His526Tyr | CTG→CGG, CAC→TAC | 1 |
| Leu511Pro | CTG→CCG | 1 |
| Leu533Pro | CTG→CCG | 3 |
| Phe514Phe, Ser531Leu | TTC→TTT, TCG→TTG | 1 |
| Ser522Gln | TCG→CAG | 2 |
| Ser531Leu | TCG→TTG | 87 |
| Ser531Leu, His526Tyr | TCG→TTG, CAC→TAC | 1 |
| Ser531Leu, Ile480Val | TCG→TTG, ATC→GTC | 1 |
| Ser531Phe | TCG→TTC | 1 |
| Ser531Trp | TCG→TGG | 2 |
| Thr481Ala, Ser531Leu | ACC→GCC, TCG→TTG | 1 |
| WT | WT | 101 |
Of the 252 strains analyzed, 151 contained mutations within the RRDR of rpoB.
TABLE 2.
Summary of strains containing mutations in the inhA promoter or katGa
| Gene or promoter | Amino acid change(s) | SNP(s) | No. of strains |
|---|---|---|---|
| inhA promoter | C(−15)→T | C→T | 26 |
| inhA promoter | T(−8)→C | T→C | 3 |
| katG | Gly273Ser | GGT→AGT | 1 |
| katG | Ser315Asn | AGC→AAC | 1 |
| katG | Ser315Ile | AGC→ATC | 1 |
| katG | Ser315Thr | AGC→ACC | 139 |
| katG | Ser315Thr, Ile335Val | AGC→ACC, ATC→GTC | 2 |
| katG | WT | WT | 97 |
Of the 252 strains analyzed, 173 contained mutations in the inhA promoter, katG, or both.
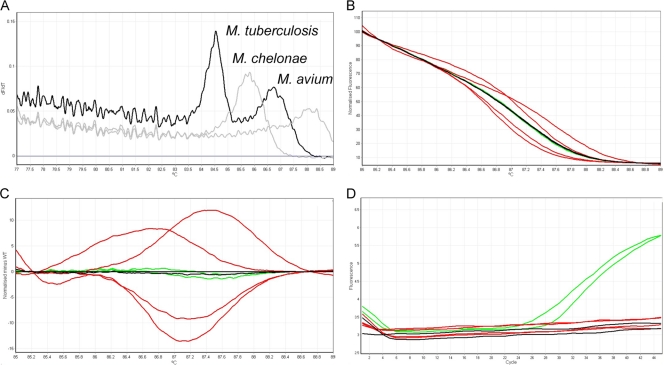
The work flow algorithm (Fig. 1) describes the assay setup and data analysis used to determine if a strain was MTC and RIF/INH resistant. The foundation of the assay is SYBR-based real-time PCR followed by HRM analysis. As both transition and transversion SNPs within rpoB and katG confer drug resistance, HRM analysis alone cannot adequately detect all possible mutations, especially transversions. For this reason we supplemented the assay with LNA probes designed to target the most common transversion SNPs, which significantly improved assay performance. The first tube was a multiplexed PCR mixture containing primers specific for internal fragments of IS6110 and the RRDR of rpoB from MTC. The IS6110 marker was analyzed using melt curve analysis (MCA) of the PCR amplicons to confirm that samples were MTC strains. MCA profiles for MTC strains contained two peaks, one corresponding to rpoB and another to IS6110 (Fig. 2A). The primers also amplified rpoB from other NTM bacteria, including Mycobacterium avium subsp. avium, Mycobacterium chelonae, and Mycobacterium abscessus (Fig. 2A), but not Mycobacterium fortuitum, Mycobacterium gordonae, or Mycobacterium intracellulare (data not shown). Typically, if amplification did occur, only one peak was observed due to the absence of IS6110 from NTM strains. Occasionally, no IS6110 peak was observed in strains previously confirmed as MTC. This observation is likely due to the fact that the IS6110 insertion element is present in varying amounts between strains of M. tuberculosis. However, it is important to note that the assay as a whole is only compatible with MTC. When subjecting NTM strains to the assay, the rpoB amplicons had distinct melting curves compared to MTC, and the katG and inhA portions of the assay were not reactive to NTM strains. Due to the composition of our sample set, all 252 strains used in this study were confirmed to be MTC, as expected.
FIG. 1.
MDR TB real-time PCR and HRM assay algorithm.
FIG. 2.
Representative analysis and interpretation of the IS6110/rpoB portion of the assay. (A) The representative melting curve analysis for IS6110 is shown. MTC strains (black) contain two peaks, while NTM strains (gray) contain only one peak. (B) HRM graph displayed in normalized mode, depicting the HRM profiles of WT strains (black), A→T transversion SNPs (green), and all other classes of SNPs (red). (C) Genotype difference graph with WT strains (black) normalized to zero. Differences in strains with A→T transversion SNPs (green) and all other classes of SNPs (red) are shown as deviations from the WT melting pattern. (D) Positive amplification signals in the crimson channel, indicating the presence of an A→T transversion SNP for Asp516Val (green).
RIF resistance was predicted based on a combination of real-time PCR, HRM analysis, and LNA probes. HRM analysis of the rpoB marker was performed to determine RIF susceptibility by observing differences in the melting profiles. The HRM profile of each isolate was compared to control strains included in each run, including WT, a strain containing a C→G SNP transversion mutation, and two strains that each contained an A→T SNP transversion mutation at distinct loci. Separation of melting profiles between WT strains and strains containing a nontransversion SNP began at ∼85°C and ended at ∼89°C. The unique HRM profiles of WT strains and strains containing SNP transition and transversion mutations are shown in Fig. 2B in the normalized graph mode. Figure 2C displays the same HRM profiles viewed as a difference graph. Both views show unique melting profiles for strains containing SNP transitions (red) compared to WT (black). Those strains with melt profiles that deviated from the WT control were classified as RIF resistant by this assay, and no further analysis was necessary (125 of 252 strains). However, SNP transversions (green) were less distinct, requiring LNA probes to detect specific SNP A→T changes at Asp516Val and His526Leu of the RRDR. A positive amplification signal in either one of the LNA probe channels (crimson channel for Asp516Val or orange channel for His526Leu) indicated RIF resistance (12 of 252 strains), whereas no amplification in either channel indicated a RIF-susceptible strain (Fig. 2D).
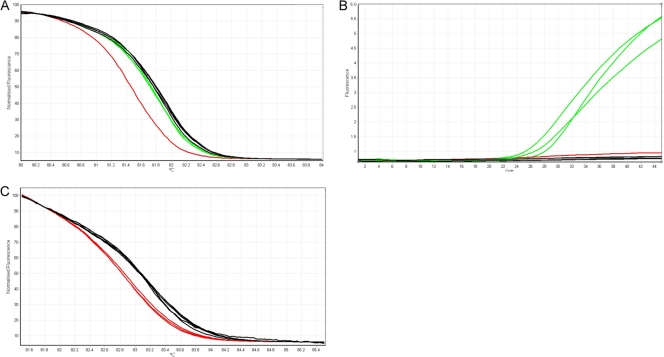
Following the classification of a strain as MTC and the determination of RIF resistance, the real-time PCR and HRM analysis for katG/inhA were performed. The two markers were analyzed independently of each other, as depicted in the algorithm shown in Fig. 1. INH resistance identified by the katG marker was detected using HRM analysis and positive amplification using an LNA probe to detect the Ser315Thr mutation. HRM analysis was done by comparing the melting pattern of each sample to those of WT, SNP transition mutation (G→A), and LNA probe SNP transversion mutation (G→C) control strains included in each run. Separation of melting profiles between WT strains (black) and strains containing transition SNP mutations (red) occurred between ∼80°C and ∼85°C (Fig. 3A). Samples with melt profiles that deviated from WT were classified as resistant (2 of 252 stains). Only two strains harboring either a Ser315Asn or Ser315Ile mutation were detected by melt analysis alone. An additional strain harbored a Gly273Ser mutation that was outside the target amplicon. Following HRM analysis, any samples with a melting profile indistinguishable from WT strains (green and black, respectively) were further analyzed for positive amplification in the LNA probe channel. Samples with a positive signal in the LNA probe channel (red channel for Ser315Thr) were identified as INH resistant (141 of 252 strains), and those with a negative LNA probe signal were analyzed further for mutations in the inhA promoter region (Fig. 3B).
FIG. 3.
Representative analysis and interpretation of the katG and inhA portion of the assay. (A) HRM plot for the katG marker, shown in normalized graph mode, illustrating the HRM profile separation between WT strains (black) and strains containing mutations (red and green). (B) A positive amplification signal in the red channel indicates the presence of a G→C transversion SNP (green). (C) HRM plot for the inhA marker, displayed in normalized graph mode, comparing the HRM profiles of WT strains (black) and strains with mutations (red).
INH resistance as identified by the inhA marker was detected using HRM analysis alone. The melting profile of each sample was compared to those of WT and SNP transition (C→T or T→C) control strains included in each run. Separation of melting profiles between WT strains and strains containing mutations began at ∼80°C and ended at ∼86°C. Figure 3C illustrates a normalized graph of the inhA HRM profiles between WT strains (black) and strains containing a mutation (red). Samples with melt profiles that deviated from WT were classified as resistant (29 of 252 strains). We were able to detect 10 INH-resistant strains harboring mutations in only the inhA promoter region that were not associated with katG mutations. Isolates identified as containing a mutation in either katG, the inhA promoter region, or both were considered INH resistant.
Each of the 252 isolates was tested using this assay, and results were compared to direct sequencing of targeted loci as well as to phenotypes determined by DST. The rpoB/IS6110 portion of the assay correctly identified 140 of the 154 RIF-resistant isolates and 96 of the 98 RIF-susceptible isolates, resulting in a sensitivity of 91% and a specificity of 98%. The katG/inhA portion of the assay correctly identified 151 of the 174 INH-resistant strains and 78 of the 78 INH-susceptible strains, resulting in a sensitivity of 87% and a specificity of 100%. Combined, the assay correctly identified 126 of the 148 MDR strains and 102 of the 104 non-MDR strains, resulting in an overall sensitivity of 85% and a specificity of 98%.
DISCUSSION
We have described the development of a novel real-time PCR assay that combines HRM analysis and multiple fluorescent chemistries to detect SNP mutations conferring drug resistance in M. tuberculosis. This assay rapidly detects the dominant mutations responsible for conferring RIF and INH resistance and can subsequently identify MDR strains of M. tuberculosis. After screening 252 strains containing a comprehensive assortment of mutation types and combinations, sensitivity and specificity data were similar to those obtained with direct sequencing of the targeted loci. The assay was able to reliably detect the most common mutations associated with clinical resistance observed in the population (Table 3). For example, the His526Tyr mutation within the RRDR of the rpoB gene has been reported to account for up to 30% of RIF-resistant strains in the United States (6). In our sample set, the Asp516Val, His526Tyr, and Ser531Leu mutations accounted for 72% of the strains evaluated that were RIF resistant by DST. The assay correctly identified 100% of these strains as RIF resistant. The majority of isolates with rare mutations, and even samples known to contain mixed populations, were also detected using this assay (data not shown).
TABLE 3.
Summary of assay performance for detection of the most common mutations
| Gene or promoter | Amino acid change | No. of strains with mutation | No. of strains identified correctly |
|---|---|---|---|
| rpoB | Asp516Val | 9 | 9 |
| rpoB | His526Tyr | 14 | 14 |
| rpoB | Ser531Leu | 87 | 87 |
| inhA promoter | C(−15)→T | 26 | 26 |
| katG | Ser315Thr | 139 | 138 |
Occasionally, rare mutations within rpoB were not detected. For instance, SNP transversions at locations not targeted by either LNA probe or strains with two SNPs that compensated for the deviation from the WT melting pattern were not detected (Table 4). Because class III and IV SNP changes involve bases exchanges that do not change the number of hydrogen bonds between the bases, HRM alone was not sufficient to detect these SNPs, and this remains a limitation of this technique for use in diagnostics. However, we were able to circumvent this potential shortcoming by including LNA probes designed to target the most common SNP transversions that would have otherwise been missed. As expected, the assay was not able to predict resistance in cases where mutations conferring resistance were outside the RRDR or when strains contained no mutation in rpoB but were determined to be RIF resistant by DST (6 of 252 strains) (Table 4). Similarly, INH resistance occurred in strains containing no mutations in the targeted loci but that were resistant by DST (22 of 252 strains). These strains are likely to be resistant by other mechanisms or by unknown mechanisms of INH resistance, and this possibility has been previously discussed (15). In each portion of the assay, false positives occurred in strains possessing a mutation but susceptible by DST.
TABLE 4.
Summary of discordant results for the RIF portion of the assay
| Discordant result | Amino acid change(s) | SNP(s) | No. of strains | HRMa | DSTa |
|---|---|---|---|---|---|
| False negative | Gln513Leu | CAA→CTA | 1 | S (1 of 1) | R |
| False negative | His526Arg, Arg529Gln | CAC→CGC, CGA→CAA | 1 | S (1 of 1) | R |
| False negative | His526Asp | CAC→GAC | 4 | S (3 of 4) | R |
| False negative | Ser522Gln | TCG→CAG | 2 | S (1 of 2) | R |
| False negative | Ser531Trp | TCG→TGG | 2 | S (2 of 2) | R |
| False negative | WT | WT | 6 | S (6 of 6) | R |
| False positive | Leu511Pro | CTG→CCG | 1 | R (1 of 1) | S |
| False positive | Leu533Pro | CTG→CCG | 3 | R (3 of 3) | S |
S, strain predicted to be susceptible by the indicated method; R, strain predicted to be resistant by the indicated method.
Compared to other HRM assays that have been developed for the rapid diagnosis of drug resistance in M. tuberculosis (5, 12), this assay is more robust and informative, providing additional critical details about strains. First, unlike other assays, this assay is able to discern MTC from NTM strains. This is useful in a clinical setting where patients, especially those who are immunocompromised, could possibly be infected with NTM. Additionally, other HRM assays rely solely on the identification of SNPs associated with RIF resistance as the predictor for MDR. While up to 97% of RIF strains have in fact been determined to be MDR, mono-RIF resistance is possible, albeit rare (19). The lack of information provided by other assays regarding INH susceptibility remains a shortcoming and could incorrectly suggest that INH would not be effective if included in a drug treatment regimen. By using RIF resistance as the sole predictor for MDR, it potentially removes INH from drug regimens where it would be more effective and have fewer side effects than other drugs. The assay described here has been designed to detect resistance to both RIF and INH simultaneously, which offers a significant improvement over current comparable diagnostic methods.
The advantages of this assay over current molecular or culture-based techniques are numerous. This comprehensive assay rapidly provides a significant amount of information at a much lower cost than culture-based or sequencing methods. Within approximately 5 h of obtaining DNA, the assay is able to confirm the sample as MTC and determine the RIF and INH resistance patterns, and therefore provide a preliminary MDR diagnosis. The quick turnaround time is due, in part, to the fact that the methods have been optimized so that the same PCR conditions are used for all portions. Comparable assays require that PCR conditions be modified for each target loci (11) or sometimes require the use of multiple platforms (2). This assay is compatible with several methods of DNA preparation and is valid over a 3-log range of DNA concentrations (data not shown), eliminating the need for time-consuming standardized DNA extractions and quantification. The assay detects SNP transversion mutations with LNA probes rather than resorting to the commonly used, yet cumbersome, approach of spiking the sample with secondary control DNA (5). As a result, sample tubes remain closed between real-time PCR and HRM steps, minimizing the chance for contamination. The use of LNA probes to detect certain SNP transversions greatly reduces user subjectivity and increases confidence in the results. The use of several real-time PCR chemistries in one tube is made possible by the multichannel format of the Qiagen Rotor-Gene 6000 platform. Although other multiplex assays have been developed, to our knowledge this assay is the first of its kind to combine multiple fluorescent chemistries together in one reaction to achieve maximum discrimination.
An additional appealing feature of this assay is the flexibility it allows the user. Components can be adjusted to perform the assay with or without specific markers, providing countless options as to how to meet the needs of the laboratory. Because of the molecular basis of the techniques, real-time PCR and HRM analysis cannot detect drug resistance conferred by unknown mutations or by unknown mechanisms, and this limitation is common among all sequence-dependent detection methods. However, as new mutations are identified or become more prevalent in a population, additional probes could easily be designed and added to the assay, being limited only by the number of channels available with a specific platform, making it more robust and comprehensive. This adaptability is an important feature, because patterns of drug resistance can vary widely based on the drugs used in a particular geographic area (14).
The most significant application of this assay is the potential for improving TB treatment. Currently, in the United States, the standard empirical treatment for adults who have not previously received TB therapy consists of four drugs, RIF, INH, ethambutol (EMB), and pyrazinamide (PZA), until DST results are available (1). It is worth noting, however, that in 2008, 8.2% of previously untreated TB patients were infected with strains that were INH resistant (3). Because of this relatively high frequency of INH resistance, EMB is included in the treatment regimen in order to prevent the emergence of RIF resistance (1). However, because of the added cost and potential side effects of EMB (4), such as optic neuritis, it is preferable to omit it from the treatment regimen when its use is not necessary. In this case, the assay is useful not only to predict drug resistance but also to predict susceptibility, thereby identifying potentially more desirable and effective drugs that could be added to enhance treatment while eliminating those with harsher side effects that are not as clinically effective. Additionally, by using this assay as a preliminary indicator for drug resistance, those strains that give positive results can be fast tracked for second-line DST, as opposed to waiting several weeks to obtain first-line DST results. Taken together, these applications illustrate the clinical value of the assay.
We have described a novel assay for the detection of MDR M. tuberculosis that exhibits several advantages over similar rapid diagnostics that are currently available. This assay has been designed to identify strains as MTC and to determine resistance to RIF and INH by combining several real-time PCR chemistries with HRM in a three-tube format. The ability to rapidly and accurately detect drug-resistant strains with the confidence provided by this assay has positive implications for an improvement in clinical diagnosis and treatment of TB.
Acknowledgments
We thank Tracy Dalton and Melisa Wilby for their critical reading of the manuscript and suggestions.
This research was supported in part by an appointment to the Emerging Infectious Diseases (EID) Fellowship Program administered by the Association of Public Health Laboratories (APHL) and funded by the CDC.
Footnotes
Published ahead of print on 1 September 2010.
REFERENCES
- 1.Anonymous. 2003. Treatment of tuberculosis. MMWR Recomm. Rep. 52:1-77. [PubMed] [Google Scholar]
- 2.Bravo, L. T., M. J. Tuohy, C. Ang, R. V. Destura, M. Mendoza, G. W. Procop, S. M. Gordon, G. S. Hall, and N. K. Shrestha. 2009. Pyrosequencing for rapid detection of Mycobacterium tuberculosis resistance to rifampin, isoniazid, and fluoroquinolones. J. Clin. Microbiol. 47:3985-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2009. Reported tuberculosis in the United States, 2008. U.S. Department of Health and Human Services, CDC, Atlanta, GA.
- 4.Chung, H., Y. H. Yoon, J. J. Hwang, K. S. Cho, J. Y. Koh, and J. G. Kim. 2009. Ethambutol-induced toxicity is mediated by zinc and lysosomal membrane permeabilization in cultured retinal cells. Toxicol. Appl. Pharmacol. 235:163-170. [DOI] [PubMed] [Google Scholar]
- 5.Hoek, K. G., N. C. Gey van Pittius, H. Moolman-Smook, K. Carelse-Tofa, A. Jordaan, G. D. van der Spuy, E. Streicher, T. C. Victor, P. D. van Helden, and R. M. Warren. 2008. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 46:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapur, V., L. L. Li, S. Iordanescu, M. R. Hamrick, A. Wanger, B. N. Kreiswirth, and J. M. Musser. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacoma, A., N. Garcia-Sierra, C. Prat, J. Ruiz-Manzano, L. Haba, S. Roses, J. Maldonado, and J. Dominguez. 2008. GenoType MTBDRplus assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis strains and clinical samples. J. Clin. Microbiol. 46:3660-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liew, M., R. Pryor, R. Palais, C. Meadows, M. Erali, E. Lyon, and C. Wittwer. 2004. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 50:1156-1164. [DOI] [PubMed] [Google Scholar]
- 9.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NCCLS. 2003. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic Actinomycetes. Approved standard, NCCLS document M24-A. NCCLS, Wayne, PA. [PubMed]
- 11.Ong, D. C., W. C. Yam, G. K. Siu, and A. S. Lee. 2010. Rapid detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis by high-resolution melting analysis. J. Clin. Microbiol. 48:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietzka, A. T., A. Indra, A. Stoger, J. Zeinzinger, M. Konrad, P. Hasenberger, F. Allerberger, and W. Ruppitsch. 2009. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J. Antimicrob. Chemother. 63:1121-1127. [DOI] [PubMed] [Google Scholar]
- 13.Plikaytis, B. B., R. H. Gelber, and T. M. Shinnick. 1990. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J. Clin. Microbiol. 28:1913-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 15.Riska, P. F., W. R. Jacobs, Jr., and D. Alland. 2000. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuber. Lung Dis. 4:S4-S10. [PubMed] [Google Scholar]
- 16.Sacks, L. V., and R. E. Behrman. 2008. Developing new drugs for the treatment of drug-resistant tuberculosis: a regulatory perspective. Tuberculosis (Edinb.) 88(Suppl. 1):S93-S100. [DOI] [PubMed] [Google Scholar]
- 17.Vignal, A., D. Milan, M. SanCristobal, and A. Eggen. 2002. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 34:275-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilcheze, C., and W. R. Jacobs, Jr. 2007. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu. Rev. Microbiol. 61:35-50. [DOI] [PubMed] [Google Scholar]
- 19.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world: fourth global report. WHO, Geneva, Switzerland.
- 20.World Health Organization. 2009. Global tuberculosis control: epidemiology, strategy, financing. WHO report 2009. WHO, Geneva, Switzerland.