Abstract
Infections caused by the Mycobacterium avium complex (MAC) are on the rise in both human and veterinary medicine. A means of effectively discriminating among closely related yet pathogenetically diverse members of the MAC would enable better diagnosis and treatment as well as further our understanding of the epidemiology of these pathogens. In this study, a five-target multiplex PCR designed to discriminate MAC organisms isolated from liquid culture media was developed. This MAC multiplex was designed to amplify a 16S rRNA gene target common to all Mycobacterium species, a chromosomal target called DT1 that is unique to M. avium subsp. avium serotypes 2 and 3, to M. avium subsp. silvaticum, and to M. intracellulare, and three insertion sequences, IS900, IS901, and IS1311. The pattern of amplification results allowed determination of whether isolates were mycobacteria, whether they were members of the MAC, and whether they belonged to one of three major MAC subspecies, M. avium subsp. paratuberculosis, M. avium subsp. avium, and M. avium subsp. hominissuis. Analytical sensitivity was 10 fg of M. avium subsp. paratuberculosis genomic DNA, 5 to 10 fg of M. avium subsp. avium genomic DNA, and 2 to 5 fg of DNA from other mycobacterial species. Identification accuracy of the MAC multiplex was evaluated by testing 53 bacterial reference strains consisting of 28 different mycobacterial species and 12 nonmycobacterial species. Identification accuracy in a clinical setting was evaluated for 223 clinical MAC isolates independently identified by other methods. Isolate identification agreement between the MAC multiplex and these comparison assays was 100%. The novel MAC multiplex is a rapid, reliable, and simple assay for discrimination of MAC species and subspecies in liquid culture media.
Since the early 1980s, there has been an increase in disease caused by organisms broadly categorized as nontuberculous mycobacteria (NTM), a generic term for mycobacteria not in the Mycobacterium tuberculosis complex and other than M. leprae (32). Of these NTM, Mycobacterium avium complex (MAC) species are the most common cause of human and animal disease globally (6, 14, 16, 24). The clinical relevance of the MAC in humans has been amplified in recent decades with the increasing population of immunocompromised individuals resulting from longer life expectancy, immunosuppressive chemotherapy, and the AIDS pandemic (27). The MAC is divided into two main species: M. avium and M. intracellulare. M. avium is further subdivided (per Turenne et al.) into four subspecies: M. avium subsp. avium, M. avium subsp. hominissuis, M. avium subsp. paratuberculosis, and M. avium subsp. silvaticum (39).
Members of the family Mycobacteriaceae, comprising the MAC, differ in virulence and ecology. Those designated M. avium subsp. hominissuis are genomically diverse, low-virulence, opportunistic pathogens for both animals and humans. The majority of human M. avium subsp. hominissuis infections occur in HIV-immunocompromised people, immunocompetent persons with underling pulmonary disease, and children with cystic fibrosis (2, 12, 17). Considered ubiquitous in the environment (the most likely source of infection for humans), M. avium subsp. hominissuis has been isolated from water, soil, and dust (9). Domestic water distribution systems have been reported as possible sources of M. avium subsp. hominissuis infections in hospitals, homes, and commercial buildings (26, 27). In animals, M. avium subsp. hominissuis is found as a cause of lymphadenitis of the head and mesenteric lymph nodes of swine recognized at slaughter.
Mycobacterium avium subsp. avium has long been recognized as a primary pathogen causing avian tuberculosis in wild and domestic birds (37, 38). Members of this subspecies also sporadically cause disease in other animals (6, 15, 30).
For veterinarians, the MAC member of greatest importance is M. avium subsp. paratuberculosis. This MAC member causes a chronic granulomatous enteritis called Johne's disease or paratuberculosis, most often in ruminants (16, 22, 31). Mycobacterium avium subsp. paratuberculosis is capable of infecting and causing disease a wide array of animal species, including nonhuman primates, without need of immunosuppressive coinfections. The herd-level prevalence of M. avium subsp. paratuberculosis infections in dairy cattle exceeds 50% in most major dairy product-producing countries (29, 31). Two systematic reviews and meta-analyses report a consistent association of M. avium subsp. paratuberculosis with Crohn's disease, and the zoonotic potential of M. avium subsp. paratuberculosis continues to be a controversial subject discussed in the literature (1, 11). Unlike for most other M. avium subspecies, isolation of M. avium subsp. paratuberculosis requires the addition of the siderophore mycobactin to culture media and prolonged culture incubation for successful isolation from a tissue, soil, or fecal samples (43). After this lengthy incubation period with special media, resultant acid-fast organisms then need to be accurately identified.
Unlike the M. avium subspecies, whose type strains were obtained from nonhuman hosts, the type strain of M. intracellulare (ATCC 13950) was isolated from a human, specifically a child who died from disseminated disease. Recently, numerous isolates considered to be M. intracellulare were reclassified as M. chimaera sp. nov. as part of the MAC (35). Few of these isolates were found to be clinically relevant, suggesting that this MAC species has low pathogenicity, and this factor is crucial to therapeutic decision making. Mycobacterium intracellulare appears to have a distinct environmental niche, more prevalent in biofilms and at significantly higher CFU numbers than M. avium (10, 36). It accounts for more documented human infections than M. avium subsp. hominissuis in several countries, including South Korea and Japan (19, 20, 23).
Contemporary methods for MAC identification, e.g., high-performance liquid chromatography (HPLC) of cell wall mycolic acids, and genetic probes based on rRNA targets, e.g., AccuProbe, cannot discriminate among M. avium subspecies (2, 9). Given the differences in pathogenicity among M. avium subspecies and the implications regarding the infection source, a practical and accurate method of simply identifying M. avium subspecies is needed (13, 25, 35). In this study, we describe the specificity, discrimination capacity, and sensitivity of a novel five-target PCR, called the MAC multiplex, using a wide array of reference and clinical MAC isolates and numerous nonmycobacterial organisms.
MATERIALS AND METHODS
Primer design and MAC multiplex PCR assay.
Based on the sequences of IS900 (GenBank accession no. X16293), IS901 (accession no. X59272), IS1311 (accession no. U16276), DT1 (accession no. L04543), and the 16S rRNA gene (34), primer sets were designed to amplify specific portions of each target gene (Table 1). Assay conditions were optimized in previous studies (41). For amplification, an aliquot (2 μl) of the DNA sample was added to 48 μl of PCR mixture containing 10 mM Tris-HCl (pH 8.8), 2.5 mM MgCl2, 50 mM KCl, 1 M betaine, 0.2 mM each deoxynucleoside triphosphate (Promega, Madison, WI), 10 pmol of each primer, and 2.5 U of HotStar Plus Taq polymerase (Qiagen, Gaithersburg, MD). After an initial denaturation step (8 min at 95°C) to activate the HotStar Plus Taq polymerase, 29 cycles of amplification were performed as follows: denaturation at 95°C for 60 s, annealing at 60°C for 40 s, and DNA extension at 72°C for 35 s. A final extension was performed at 72°C for 10 min. Amplification was carried out in a DNA 9700 thermocycler (TGradient; Biometra, Germany). After amplification, PCR products were analyzed on a 2.0% agarose gel, stained with ethidium bromide, and visualized on a UV transilluminator. Multiplex PCR products of 484, 398, 753, 608, and 296 bp resulted from amplification of the 16S rRNA gene, IS900, IS901, IS1311, and DT1 targets, respectively (Fig. 1). DNA isolated from M. avium subsp. paratuberculosis ATCC 19698, M. avium subsp. avium ATCC 35712, M. intracellulare ATCC 13950, and M. terrae ATCC 15755 were used as controls for each primer set in each PCR run.
TABLE 1.
Oligonucleotide primers used for MAC multiplex PCR
| Genetic construct | Product size (bp) | Orientationa | Oligonucleotide sequence | Target organism(s) |
|---|---|---|---|---|
| IS900 | 398 | F | 5′ TGGACAATGACGGTTACGGAGGTGG 3′ | M. avium subsp. paratuberculosis |
| R | 5′ CGCAGAGGCTGCAAGTCGTGG 3′ | |||
| IS901 | 753 | F | 5′ GAACGCTGCTCTAAGGACCTGTTGG 3′ | M. avium subsp. avium |
| R | 5′ GGAAGGGTGATTATCTGGCCTGC 3′ | |||
| DT1 | 296 | F | 5′ CGTTGGCTGGCCATTCACGAAGGAGT 3′ | M. avium subsp. avium and M. intracellulare |
| R | 5′ GCTAGTTGGATCGCGCCGAACACCGG 3′ | |||
| IS1311 | 608 | F | 5′ GCGTGAGGCTCTGTGGTGAA 3′ | All MAC members |
| R | 5′ ATGACGACCGCTTGGGAGAC 3′ | |||
| 16S rRNA gene | 484 | F | 5′ ATAAGCCTGGGAAACTGGGT 3′ | All mycobacterial species |
| R | 5′ CACGCTCACAGTTAAGCCGT 3′ |
F, forward; R, reverse.
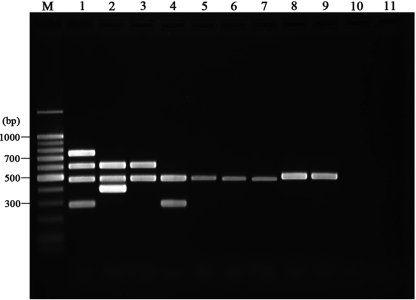
FIG. 1.
Representative gel resulting from the MAC multiplex. Lane M, molecular size marker; lane 1, M. avium subsp. avium ATCC 35712; lane 2, M. avium subsp. paratuberculosis ATCC 19698; lane 3, M. avium subsp. hominissuis 104; lane 4, M. intracellulare ATCC 13950; lane 5, M. terrae ATCC 15755; lane 6, M. phlei ATCC 11758; lane 7, Mycobacterium abscessus ATCC 19977; lane 8, M. tuberculosis Ra; lane 9, Mycobacterium bovis (BCG); lane 10, E. coli ATCC 35218; lane 11, negative control.
A priori MAC multiplex interpretation criteria, consistent with current MAC nomenclature (39), are listed in Table 2. Simultaneous amplification of the three targets IS1311, the 16S rRNA gene, and IS900, was interpreted as corresponding to M. avium subsp. paratuberculosis; amplification of DT1, IS1311, the 16S rRNA gene, and IS901 was interpreted as corresponding to M. avium subsp. avium serotype 2 or 3 (“bird type”) or M. avium subsp. silvaticum. Amplification of both IS1311 and the 16S rRNA gene but neither IS900 nor IS901 was interpreted as corresponding to M. avium subsp. hominissuis. Amplification of only the DT1 and 16S rRNA gene targets indicated M. intracellulare. Mycobacterial species outside the MAC were indicated when 16S rRNA gene amplification alone was observed.
TABLE 2.
Interpretation criteria for MAC multiplex results
| Identification | Criterion |
||||
|---|---|---|---|---|---|
| Panmycobacterium 16S rRNA gene | IS900 | IS901 | IS1311 | DT1 | |
| M. avium subsp. avium (serotypes 2 and 3) or M. avium subsp. silvaticuma | + | − | + | + | + |
| M. avium subsp. avium (serotype 1) | + | − | + | + | − |
| M. avium subsp. paratuberculosis | + | + | − | + | − |
| M. avium subsp. hominissuis | + | − | − | + | − |
| M. intracellulare | + | − | − | − | + |
| Mycobacteria other than MAC | + | − | − | − | − |
| Nonmycobacterial species | − | − | − | − | − |
An M. avium subspecies rarely isolated and of uncertain taxonomic status.
Bacterial strains, cultures, and preparation of mycobacterial single-cell suspensions.
The origins, sources and/or strain names, descriptions, and reference identifications of mycobacterial strains used to evaluate the assay are listed in Table 3. Reference strains of each Mycobacterium species were obtained from the American Type Culture Collection (ATCC). All nonmycobacterial organisms were obtained from the ATCC or the State Laboratory of Hygiene, Madison, WI (WSLH). Mycobacteria were cultivated in 7H9 broth supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; Difco Laboratories, MD) supplemented with 2 μg/ml of mycobactin J (Allied Monitor, Fayette, MO) for 2 weeks (rapid growers) or 4 weeks (slow growers) at 37°C. Nonmycobacterial organisms, such as Escherichia coli, were grown in Luria-Bertani (LB) liquid medium for 2 days. All bacteria were washed 3 times with 10 mM phosphate-buffered saline (PBS; pH 7.4), bacterial cell pellets were collected after centrifugation, and small aliquots were stored at −80°C until use.
TABLE 3.
Type strains used in the specificity test for multiplex PCR
| Bacterial species | No. of isolates tested | Strain name(s)/sourcea | Amplification target in PCR |
||||
|---|---|---|---|---|---|---|---|
| 16S rRNA gene | IS900 | IS901 | IS1311 | DT1 | |||
| M. avium complex | |||||||
| M. avium subsp. paratuberculosis | 6 | K-10, ATCC 19698, ATCC 43015, ATCC 43544, ATCC 43545, ATCC 49164 | + | + | − | + | − |
| M. avium subsp. hominissuis | 2 | ATCC 700898, 104 | + | − | − | + | − |
| M. avium subsp. avium | 2 | ATCC 35712, ATCC 25291 | + | − | + | + | + |
| M. intracellulare | 2 | ATCC 13950, ATCC 25122 | + | − | − | − | + |
| M. avium subsp. silvaticum | 1 | ATCC 49884 | + | − | + | + | + |
| Mycobacteria other than MAC | |||||||
| M. abscessus | 1 | ATCC 19977 | + | − | − | − | − |
| M. bovis | 1 | ATCC 19210 | + | − | − | − | − |
| M. bovis BCG | 1 | BCG Pasteur 1173P2 | + | − | − | − | − |
| M. celatum | 2 | ATCC 51130, ATCC 51131 | + | − | − | − | − |
| M. chelonae | 1 | ATCC 35749 | + | − | − | − | − |
| M. flavescens | 1 | ATCC 14474 | + | − | − | − | − |
| M. fortuitum | 2 | ATCC 49403, ATCC 49404 | + | − | − | − | − |
| M. gastri | 1 | ATCC 15754 | + | − | − | − | − |
| M. gordonae | 1 | ATCC 14470 | + | − | − | − | − |
| M. kansasii | 1 | ATCC 12478 | + | − | − | − | − |
| M. malmoense | 1 | ATCC 29571 | + | − | − | − | − |
| M. marinum | 1 | ATCC 927 | + | − | − | − | − |
| M. nonchromogenicum | 1 | ATCC 19530 | + | − | − | − | − |
| M. peregrinum | 1 | ATCC 14467 | + | − | − | − | − |
| M. phlei | 1 | ATCC 11758 | + | − | − | − | − |
| M. scrofulaceum | 1 | ATCC 19981 | + | − | − | − | − |
| M. smegmatis | 2 | ATCC 14468, mc2155 | + | − | − | − | − |
| M. terrae | 1 | ATCC 15755 | + | − | − | − | − |
| M. tuberculosis H37Ra | 1 | ATCC 25177 | + | − | − | − | − |
| M. tuberculosis H37Rv | 1 | ATCC 25618 | + | − | − | − | − |
| M. ulcerans | 1 | ATCC 19423 | + | − | − | − | − |
| M. vaccae | 1 | ATCC 15483 | + | − | − | − | − |
| M. xenopi | 1 | ATCC 19250 | + | − | − | − | − |
| Bacteria (nonmycobacteria) | |||||||
| Enterobacter aerogenes | 1 | WSLH | − | − | − | − | − |
| Escherichia coli | 3 | ATCC 25922, ATCC 35218, DH5α | − | − | − | − | − |
| Enterococcus faecalis | 1 | ATCC 29212 | − | − | − | − | − |
| Pseudomonas aeruginosa | 1 | WSLH | − | − | − | − | − |
| Corynebacterium pseudotuberculosis | 1 | JTC | − | − | − | − | − |
| Bacteroides fragilis | 1 | ATCC 25285 | − | − | − | − | − |
| Staphylococcus aureus | 1 | ATCC 29213 | − | − | − | − | − |
| Nocardia asteroides | 1 | WSLH | − | − | − | − | − |
| Nocardiopsis dassonvillei | 1 | WSLH | − | − | − | − | − |
| Nocardia farcinica | 1 | WSLH | − | − | − | − | − |
| Streptomyces rimosus | 1 | WSLH | − | − | − | − | − |
| Rhodococcus equi | 1 | WSLH | − | − | − | − | − |
| Total | 53 | ||||||
ATCC, American Type Culture Collection, Manassas, VA; JTC, Johne's Testing Center, Madison, WI; WSLH, Wisconsin State Laboratory of Hygiene, Madison, WI.
DNA extraction.
To prepare the bacterial DNA from pure cultures, 1 ml of the bacterial pellet was resuspended in 10 mM PBS (pH 7.4) and transferred into a 1.5-ml Eppendorf tube. The DNA was extracted by using either a QIAamp DNA stool minikit (Qiagen Inc., Valencia, CA) or the conventional cetyltrimethylammonium bromide (CTAB) method (18) following procedures described previously (3) or as described by the manufacturer, with minor modifications.
Multiplex specificity and sensitivity.
To determine MAC multiplex specificity, a total of 53 reference bacterial strains consisting of 28 different mycobacterial species and 12 nonmycobacterial species were tested (Table 3). To evaluate assay sensitivity, purified DNA from M. avium subsp. avium ATCC 35712, M. avium subsp. paratuberculosis ATCC 19698, M. avium subsp. hominissuis 104, M. intracellulare ATCC 13950, M. abscessus ATCC 19977, M. tuberculosis Ra, M. bovis BCG, E. coli ATCC 35218, and M. terrae ATCC 15755 isolated from pure cultures were serially diluted from 100 ng to 5 fg, then assayed using the MAC multiplex.
MAC multiplex PCR on clinical isolates from broth culture.
A total of 223 MAC strains, consisting of 117 isolates of M. avium subsp. paratuberculosis, 80 isolates of M. avium subsp. hominissuis, 3 isolates of M. avium subsp. avium, and 23 isolates of M. intracellulare were obtained from human and veterinary diagnostic laboratories. The origins and sources of these isolates are listed in Table 4. The M. avium subsp. paratuberculosis isolate identities had been confirmed by both analysis of L1 and L9 and single-target IS900 PCR detection (28). The human-origin M. avium subsp. paratuberculosis isolates were identified by using a previously described molecular typing method based on two IS900 integration loci (28), IS1311 restriction fragment length polymorphism (21), rpoB sequence analysis (5), and hsp65 code sequevar analysis (19, 40). All clinical isolates were tested by MAC multiplex PCR on a blind basis after their propagation in modified Bactec 12B or MGIT ParaTB culture media.
TABLE 4.
Identification of clinical isolates by MAC multiplex PCR (100% congruence with source methods)
| MAC organism | No. of isolates tested | Isolate type/origin, source |
|---|---|---|
| M. avium subsp. paratuberculosis | 109 | Ruminant clinical sample isolates; Johne's Testing Center, WI |
| 8 | Crohn's disease patients; University of Central Florida, FL | |
| M. avium subsp. hominissuis | 3 | Human pulmonary infection; Korea Institute of Tuberculosis |
| 3 | Water sample; Environmental Protection Agency, USA | |
| 4 | TJ-1, -2, -3, and -4, human infection; Division of Cellular and Molecular Medicine, St. George's University of London, Cranmer Terrace, London SW17 0RE, United Kingdom | |
| 1 | JSH-1, human sputum; Sequella Inc., Rockville, MD | |
| 18 | Human infection; Wisconsin State Laboratory of Hygiene, WI | |
| 9 | Human infection, Department of Biological Sciences, Virginia Polytechnic Institute and State University, Blacksburg, VA | |
| 42 | Human pulmonary infection; Samsung Medical Center, South Korea | |
| M. avium subsp. avium | 3 | Bovine fecal sample; Johne's Testing Center, WI |
| M. intracellulare | 1 | Water sample; Wisconsin State Laboratory of Hygiene |
| 3 | Water sample; Environmental Protection Agency, USA | |
| 2 | Human infection; Department of Biological Sciences, Virginia Polytechnic Institute and State University, Blacksburg, VA | |
| 15 | Human pulmonary infection; Samsung Medical Center, South Korea | |
| 2 | Human pulmonary infection; Korea Institute of Tuberculosis | |
| Total | 223 |
RESULTS
Specificity and sensitivity of MAC multiplex PCR.
The MAC multiplex PCR's five products were readily visualized on agarose gels: these products were a 484-bp product specific for the mycobacterial 16S rRNA gene, a 398-bp product from IS900 specific for M. avium subsp. paratuberculosis, 753 bp from IS901 specific for M. avium subsp. avium, 296 bp from DT found in M. avium subsp. avium and M. intracellulare, and a 608-bp product from IS1311 found in all M. avium subspecies members (Fig. 1). During testing of the 12 nonmycobacterial species, no amplification was seen for any of the MAC multiplex targets. The MAC multiplex correctly positioned the ATCC mycobacterial species within or outside the M. avium complex and identified all species within the MAC correctly (Table 1).
Based on dilution trials with purified DNA from M. avium subsp. paratuberculosis ATCC 19698, M. avium subsp. avium ATCC 35712, M. intracellulare ATCC 13950, and M. bovis (BCG), the analytical sensitivities of the MAC multiplex PCR were approximately 2 to 5 fg for the 16S rRNA gene, 10 fg for IS900, 10 fg for IS901, 5 fg for IS1311, and 50 fg for DT1 (data not shown). This is roughly equivalent to 101 to 102 CFU/ml of mycobacteria in liquid culture. The MAC multiplex was able to detect M. avium subsp. paratuberculosis from a mixed pure culture containing a hundredfold excess of non-MAC mycobacteria (data not shown).
Performance of MAC multiplex with clinical isolates.
Complete identity agreement was found between the MAC multiplex and the independent methods used by source institutions providing the 223 different acid-fast strains isolated from humans and a wide variety of domestic and nondomestic animals (Table 4).
DISCUSSION
Insertion elements found in MAC subspecies provide useful markers for their identification through genotyping (2, 4, 21, 39). Moreover, the markers appear to be associated with the subspecies' epidemiologies and pathogenicities, making the use of these markers clinically relevant. With rare exceptions, IS900 is a reliable genetic marker defining M. avium subsp. paratuberculosis (4, 16), the cause of paratuberculosis (Johne's disease) in animals. IS901 is strongly associated with MAC infections in avian species, and IS901-positive strains are thus often called bird-type MAC members (7, 8, 39). IS1311 is consistently associated with the MAC, with only rare exceptions reported (2, 21). Inclusion of primers for the 16S rRNA gene target common to all mycobacteria in the MAC multiplex serves as an internal control for confirmation of mycobacterial identification (34).
Reliance on a single target for a collection of organisms so closely related can be perilous and may result in inaccurate diagnoses of mycobacterial infections (9). The MAC multiplex has good analytical sensitivity (5 to 10 fg DNA), especially considering that it simultaneously amplifies five separate mycobacterial genomic targets. The assay is designed specifically to discriminate among MAC subspecies and is particularly good at identifying M. avium subsp. paratuberculosis, aided in all likelihood by the high copy number of IS900 (1). M. avium subsp. paratuberculosis identification based solely on the detection of IS900 should be evaluated with caution, since some environmental mycobacteria have been reported to contain IS900-like elements (1, 14, 42).
Despite the specificity of PCR primers and the full isolate identification concordance shown in this study's data set, the presence of more than one mycobacterial species in a sample can perturb the diagnosis. False-positive PCR results are not unheard of (33). Primer inhibition due to sample components or competition for primer targets in the assay also may occur and should be monitored.
The data provided from multitarget PCR allow discrimination among MAC subspecies, increase diagnostic confidence in the identity of isolates, and help resolve discrepancies. This technique is not only very promising for clear characterization of clinical MAC infections but is useful for epidemiologic studies as well.
Acknowledgments
We acknowledge financial support by the Johne's Testing Center, the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant R01-2007-000-10702-0), and the Johne's Disease Integrated Program (award no. 2008-55620-18710) from the USDA-NIFA program.
Footnotes
Published ahead of print on 1 September 2010.
REFERENCES
- 1.Abubakar, I., D. Myhill, S. H. Aliyu, and P. R. Hunter. 2008. Detection of Mycobacterium avium subspecies paratuberculosis from patients with Crohn's disease using nucleic acid-based techniques: a systematic review and meta-analysis. Inflamm. Bowel Dis. 14:401-410. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, J., I. G. Garcia, A. Aranaz, J. Bezos, B. Romero, L. de Juan, A. Mateos, E. Gomez-Mampaso, and L. Dominguez. 2008. Genetic diversity of Mycobacterium avium isolates recovered from clinical samples and from the environment: molecular characterization for diagnostic purposes. J. Clin. Microbiol. 46:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaro, A., E. Duarte, A. Amado, H. Ferronha, and A. Botelho. 2008. Comparison of three DNA extraction methods for Mycobacterium bovis, Mycobacterium tuberculosis and Mycobacterium avium subsp. avium. Lett. Appl. Microbiol. 47:8-11. [DOI] [PubMed] [Google Scholar]
- 4.Bartos, M., P. Hlozek, P. Svastova, L. Dvorska, T. Bull, L. Matlova, I. Parmova, I. Kuhn, J. Stubbs, M. Moravkova, J. Kintr, V. Beran, I. Melicharek, M. Ocepek, and I. Pavlik. 2006. Identification of members of Mycobacterium avium species by Accu-Probes, serotyping, and single IS900, IS901, IS1245 and IS901-flanking region PCR with internal standards. J. Microbiol. Methods 64:333-345. [DOI] [PubMed] [Google Scholar]
- 5.Ben Salah, I., T. Adekambi, D. Raoult, and M. Drancourt. 2008. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology 154:3715-3723. [DOI] [PubMed] [Google Scholar]
- 6.Biet, F., M. L. Boschiroli, M. F. Thorel, and L. A. Guilloteau. 2005. Zoonotic aspects of Mycobacterium bovis and Mycobacterium avium-intracellulare complex (MAC). Vet. Res. 36:411-436. [DOI] [PubMed] [Google Scholar]
- 7.Dvorska, L., T. J. Bull, M. Bartos, L. Matlova, P. Svastova, R. T. Weston, J. Kintr, I. Parmova, D. Van Soolingen, and I. Pavlik. 2003. A standardised restriction fragment length polymorphism (RFLP) method for typing Mycobacterium avium isolates links IS901 with virulence for birds. J. Microbiol. Methods 55:11-27. [DOI] [PubMed] [Google Scholar]
- 8.Dvorska, L., L. Matlova, W. Y. Ayele, O. A. Fischer, T. Amemori, R. T. Weston, J. Alvarez, V. Beran, M. Moravkova, and I. Pavlik. 2007. Avian tuberculosis in naturally infected captive water birds of the Ardeideae and Threskiornithidae families studied by serotyping, IS901 RFLP typing, and virulence for poultry. Vet. Microbiol. 119:366-374. [DOI] [PubMed] [Google Scholar]
- 9.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feller, L., and J. Lemmer. 2007. Aspects of immunopathogenic mechanisms of HIV infection. SADJ 62:432-434, 436. [PubMed] [Google Scholar]
- 12.Field, S. K., and R. L. Cowie. 2006. Lung disease due to the more common nontuberculous mycobacteria. Chest 129:1653-1672. [DOI] [PubMed] [Google Scholar]
- 13.Frothingham, R., and K. H. Wilson. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J. Infect. Dis. 169:305-312. [DOI] [PubMed] [Google Scholar]
- 14.Grant, I. R. 2005. Zoonotic potential of Mycobacterium avium ssp. paratuberculosis: the current position. J. Appl. Microbiol. 98:1282-1293. [DOI] [PubMed] [Google Scholar]
- 15.Haist, V., F. Seehusen, I. Moser, H. Hotzel, U. Deschl, W. Baumgartner, and P. Wohlsein. 2008. Mycobacterium avium subsp. hominissuis infection in 2 pet dogs, Germany. Emerg. Infect. Dis. 14:988-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry, M. T., L. Inamdar, D. O'Riordain, M. Schweiger, and J. P. Watson. 2004. Nontuberculous mycobacteria in non-HIV patients: epidemiology, treatment and response. Eur. Respir. J. 23:741-746. [DOI] [PubMed] [Google Scholar]
- 18.Hurley, S. S., G. A. Splitter, and R. A. Welch. 1987. Rapid lysis technique for mycobacterial species. J. Clin. Microbiol. 25:2227-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa, K., T. Yagi, M. Moriyama, T. Inagaki, T. Nakagawa, K. Uchiya, T. Nikai, and K. Ogawa. 2009. Characterization of Mycobacterium avium clinical isolates in Japan using subspecies-specific insertion sequences, and identification of a new insertion sequence, ISMav6. J. Med. Microbiol. 58:945-950. [DOI] [PubMed] [Google Scholar]
- 20.Jeong, Y. J., K. S. Lee, W. J. Koh, J. Han, T. S. Kim, and O. J. Kwon. 2004. Nontuberculous mycobacterial pulmonary infection in immunocompetent patients: comparison of thin-section CT and histopathologic findings. Radiology 231:880-886. [DOI] [PubMed] [Google Scholar]
- 21.Johansen, T. B., B. Djonne, M. R. Jensen, and I. Olsen. 2005. Distribution of IS1311 and IS1245 in Mycobacterium avium subspecies revisited. J. Clin. Microbiol. 43:2500-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson-Ifearulundu, Y., J. B. Kaneene, and J. W. Lloyd. 1999. Herd-level economic analysis of the impact of paratuberculosis on dairy herds. J. Am. Vet. Med. Assoc. 214:822-825. [PubMed] [Google Scholar]
- 23.Koh, W. J., O. J. Kwon, and K. S. Lee. 2005. Diagnosis and treatment of nontuberculous mycobacterial pulmonary diseases: a Korean perspective. J. Korean Med. Sci. 20:913-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, C. C., C. K. Tan, C. H. Chou, H. L. Hsu, C. H. Liao, Y. T. Huang, P. C. Yang, K. T. Luh, and P. R. Hsueh. 2010. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000-2008. Emerg. Infect. Dis. 16:294-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maekura, R., Y. Okuda, A. Hirotani, S. Kitada, T. Hiraga, K. Yoshimura, I. Yano, K. Kobayashi, and M. Ito. 2005. Clinical and prognostic importance of serotyping Mycobacterium avium-Mycobacterium intracellulare complex isolates in human immunodeficiency virus-negative patients. J. Clin. Microbiol. 43:3150-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matlova, L., L. Dvorska, K. Palecek, L. Maurenc, M. Bartos, and I. Pavlik. 2004. Impact of sawdust and wood shavings in bedding on pig tuberculous lesions in lymph nodes, and IS1245 RFLP analysis of Mycobacterium avium subsp. hominissuis of serotypes 6 and 8 isolated from pigs and environment. Vet. Microbiol. 102:227-236. [DOI] [PubMed] [Google Scholar]
- 27.Miguez-Burbano, M. J., M. Flores, D. Ashkin, A. Rodriguez, A. M. Granada, N. Quintero, and A. Pitchenik. 2006. Non-tuberculous mycobacteria disease as a cause of hospitalization in HIV-infected subjects. Int. J. Infect. Dis. 10:47-55. [DOI] [PubMed] [Google Scholar]
- 28.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen, S. S., and N. Toft. 2009. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev. Vet. Med. 88:1-14. [DOI] [PubMed] [Google Scholar]
- 30.O'Toole, D., S. Tharp, B. V. Thomsen, E. Tan, and J. B. Payeur. 2005. Fatal mycobacteriosis with hepatosplenomegaly in a young dog due to Mycobacterium avium. J. Vet. Diagn. Invest. 17:200-204. [DOI] [PubMed] [Google Scholar]
- 31.Ott, S. L., S. J. Wells, and B. A. Wagner. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40:179-192. [DOI] [PubMed] [Google Scholar]
- 32.Petrini, B. 2006. Non-tuberculous mycobacterial infections. Scand. J. Infect. Dis. 38:246-255. [DOI] [PubMed] [Google Scholar]
- 33.Pithua, P., S. J. Wells, S. M. Godden, S. Sreevatsan, and J. R. Stabel. 2010. Experimental validation of a nested polymerase chain reaction targeting the genetic element ISMAP02 for detection of Mycobacterium avium subspecies paratuberculosis in bovine colostrum. J. Vet. Diagn. Invest. 22:253-256. [DOI] [PubMed] [Google Scholar]
- 34.Rossi, M. C., A. Gori, G. Zehender, G. Marchetti, G. Ferrario, C. De Maddalena, L. Catozzi, A. Bandera, A. D. Esposti, and F. Franzetti. 2000. A PCR-colorimetric microwell plate hybridization assay for detection of Mycobacterium tuberculosis and M. avium from culture samples and Ziehl-Neelsen-positive smears. J. Clin. Microbiol. 38:1772-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweickert, B., O. Goldenberg, E. Richter, U. B. Gobel, A. Petrich, P. Buchholz, and A. Moter. 2008. Occurrence and clinical relevance of Mycobacterium chimaera sp. nov., Germany. Emerg. Infect. Dis. 14:1443-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steed, K. A., and J. O. Falkinham III. 2006. Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl. Environ. Microbiol. 72:4007-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tell, L. A., L. Woods, and R. L. Cromie. 2001. Mycobacteriosis in birds. Rev. Sci. Tech. 20:180-203. [DOI] [PubMed] [Google Scholar]
- 38.Thorel, M. F., H. F. Huchzermeyer, and A. L. Michel. 2001. Mycobacterium avium and Mycobacterium intracellulare infection in mammals. Rev. Sci. Tech. 20:204-218. [DOI] [PubMed] [Google Scholar]
- 39.Turenne, C. Y., D. M. Collins, D. C. Alexander, and M. A. Behr. 2008. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J. Bacteriol. 190:2479-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turenne, C. Y., M. Semret, D. V. Cousins, D. M. Collins, and M. A. Behr. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uppal, M., S. L. McLellan, M. T. Collins, and R. S. Lambrecht. 2002. A multiplex PCR assay that discriminates Mycobacterium avium subspecies paratuberculosis from closely related Mycobacteria using primers which detect specific insertion sequences, abstr. Z-58. Abstr. 102nd Annu. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 42.Vansnick, E., P. De Rijk, F. Vercammen, D. Geysen, L. Rigouts, and F. Portaels. 2004. Newly developed primers for the detection of Mycobacterium avium subspecies paratuberculosis. Vet. Microbiol. 100:197-204. [DOI] [PubMed] [Google Scholar]
- 43.Wells, S. J., M. T. Collins, K. S. Faaberg, C. Wees, S. Tavornpanich, K. R. Petrini, J. E. Collins, N. Cernicchiaro, and R. H. Whitlock. 2006. Evaluation of a rapid fecal PCR test for detection of Mycobacterium avium subsp. paratuberculosis in dairy cattle. Clin. Vaccine Immunol. 13:1125-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]