Abstract
The objectives of this study were to assess the performance of genotypic algorithms for predicting CXCR4-using virus, with enhanced sensitivity Trofile HIV coreceptor tropism assay (ES Trofile) as the reference, and to compare the concordance/accuracy of genotypic tests with ES Trofile and with the original Trofile assay. Paired phenotypic and genotypic determinations of HIV-1 coreceptor usage were compared in plasma samples from HIV-1-infected patients. Sequencing of the third hypervariable (V3) loop of the viral gene and phenotypic assays were performed for each sample. Genotypic rules used to predict tropism were Geno2pheno (false-positive rate at 1 to 20%), position-specific scoring matrix X4R5 (PSSMX4R5) and PSSMsinsi (where “sinsi” stands for syncytium inducing and non-syncytium inducing), and the 11/25, 11/24/25, and net charge rules. Two hundred forty-four phenotypic and genotypic samples were tested. Coreceptor usage was obtained from ES Trofile for 145 (59%) samples and from Trofile for 99 (41%) samples. The highest concordance (82.6%) was obtained with PSSMX4R5 when ES Trofile was used as the reference. Geno2pheno at a 20% false-positive rate showed the highest sensitivity (76.7%) for CXCR4-using virus detection with ES Trofile. Samples from naïve subjects and those with CD4 cell counts between 200 and 500 cells/mm3 showed the best predictive performance. Overall, the accuracy of the bioinformatics tools to detect CXCR4-using virus was similar for ES Trofile and Trofile; however, the negative predictive values for genotypic tools with ES Trofile were slightly higher than they were with Trofile. The accuracy of genotypic algorithms for detecting CXCR4-using viruses is high when using ES Trofile as the reference. Results are similar to those obtained with Trofile. The concordance with ES Trofile is better with higher CD4 cell counts and nonexposure to antiretroviral therapy.
The determination of HIV-1 tropism is now of clinical interest because the chemokine coreceptors CCR5 and CXCR4 are targets for drugs that block HIV-1 entry. Maraviroc, the first CCR5 antagonist approved for clinical use, specifically inhibits the replication of R5-tropic HIV-1 variants; therefore, viral tropism testing is mandatory before using this drug. Several assays have been developed to determine HIV-1 coreceptor usage (1, 10, 18). Phenotypic assays using either HIV primary isolates or recombinant viruses are considered the gold standard for HIV-1 tropism assessment. Among them, the assay from Trofile (Monogram Biosciences, South San Francisco, CA) is the only clinically proven, commercially available diagnostic test to determine HIV-1 coreceptor usage and therefore the most widely used phenotypic test worldwide. In spite of their accuracy, phenotypic methodologies have the inconvenience of their complexity, expensiveness, and the requirement of special facilities and expertise, which makes them unfeasible to be used as a routine part of clinical diagnosis. An alternative method for tropism determination consists of the genotypic prediction of HIV-1 coreceptor usage through bioinformatics tools based on third hypervariable (V3) loop viral sequences. These genotypic methods have demonstrated good correlation with phenotypic tests, including the Trofile assay, in different studies (5, 8, 14, 16, 19), and preliminary data from prospective clinical studies suggest that they may predict clinical response to maraviroc (12, 21, 24). However, a number of factors are thought to reduce the ability of genotyping to predict HIV-1 tropism, including the presence of minority CXCR4-using variants (9, 16).
Because of the low sensitivity of the original Trofile assay to detect minority CXCR4 variants when present, an enhanced version (ES Trofile) has been launched by Monogram Biosciences that has significantly improved the ability to identify low levels of CXCR4-using variants, allowing a 30-fold increase in analytical sensitivity for detecting CXCR4-using variants in env clone mixtures (23). This new test constitutes the current gold standard for tropism determination and has replaced the original version of Trofile, which is currently not available. To date, the performance of genotypic algorithms for the prediction of HIV-1 tropism using ES Trofile as a reference has not yet been explored. The objectives of this study were to evaluate the accuracy of genotypic algorithms for detecting CXCR4-using virus when measured against ES Trofile and to compare the concordance/accuracy of genotypic tests with ES Trofile and with the original Trofile assay.
(This work was accepted as a late breaker in the 12th European AIDS Conference. 11 to 14 November 2009, Cologne, Germany [abstract LBPE1.2/10].)
MATERIALS AND METHODS
Study population.
A total of 145 plasma samples were collected during a 15-month period (March 2008 through June 2009) from HIV-infected naïve and treatment-experienced patients who were recruited at the outpatient HIV clinic of a university hospital (Hospital General Universitario de Elche, Elche, Alicante, Spain). Eligible patients were all viremic HIV-infected adults who were ≥18 years old. The study also included a set of 99 plasma samples phenotypically characterized using the original Trofile version obtained from the same institution from the following sources: 80 samples from HIV-infected naïve and treatment-experienced patients from the clinical cohort and 19 samples from HIV-infected, treatment-experienced patients who had been enrolled in the maraviroc phase 2b/3 development program (MOTIVATE 2; recruitment began in February 2005 and halted in July 2005) and in the maraviroc expanded access program (EAP; recruitment began in October 2007 and ended in January 2008). Written informed consent was obtained from the patients before participation in the study. Patients included in the MOTIVATE 2 and EAP studies gave specific informed consent and fulfilled the inclusion criteria to be enrolled in the programs.
Procedures.
The coreceptor phenotype was determined in the 145 plasma samples using ES Trofile by Monogram Biosciences (South San Francisco, CA). The V3 genotypes were determined from a different aliquot of the same single blood draw in our local laboratory. Viral RNA was extracted from frozen plasma samples using the QIAamp RNA blood kit according to the manufacturer's instructions (Qiagen). An ∼105-bp fragment of the envelope gene encoding the V3 loop was amplified by a previously described nested PCR protocol (4, 14). Completed sequences were aligned, assembled, and compared with Chromas and Winstar programs. Nucleotide mixtures were defined by a second-highest peak in the electropherogram (≥25% of the highest peak). Codons containing mixtures were translated into each possible amino acid, considering all possible combinations of V3 sequences for each sample. V3 loop sequences were interpreted using bioinformatic algorithms, which were developed to infer the HIV-1 tropism based on the nucleotide or amino acid sequences of the V3 region obtained after amplification from plasma HIV RNA, taking into account the key amino acids at determined positions plus other sites in V3 that differ between CCR5-using and CXCR4-using strains (16). The following Web-available genotypic algorithms were used: Geno2pheno, a bioinformatic tool based on the V3 sequence plus additional host-specific features, selecting false-positive rates (FPR) at 1%, 2.5%, 5%, 10%, 15%, and 20% (http://coreceptor.bioinf.mpi-sb.mpg.de/cgi-bin/coreceptor.pl); position-specific scoring matrix (PSSM), in which a sequence can be assigned a score based on the comparison with a sequence of known X4 viruses (http://indra.mullins.microbiol.washington.edu/webpssm/) (PSSMX4R5 and PSSMsinsi); the 11/25 rule, which is based on the presence of positively charged amino acids at positions 11 and/or 25 of the third hypervariable (V3) loop of the envelope glycoprotein gp120 (9); and the 11/24/25 rule and net charge rule, where the overall net charge of V3 is used to predict HIV-1 tropism from the V3 genotype. HIV-1 strains were classified as CCR5-using virus (viral population using exclusively coreceptor CCR5) or CXCR4-using virus, the latter including both pure CXCR4 (viral population using exclusively coreceptor CXCR4) and dual/mixed virus (viral population harboring virus that can use both coreceptors [dual] and/or CCR5-using virus plus CXCR4-using virus [mixed]). HIV-1 subtype was determined with the Geno2pheno bioinformatic tool. Previous studies have shown that HIV-1 rapid subtyping tools using online websites instead of phylogenetic analysis can be useful for differentiating clade B from non-clade B sequences (6).
Statistical analyses.
Statistical analysis was performed using the SPSS package, version 15.0 (SPSS). Descriptive analyses of demographic and baseline characteristics of the patients were expressed as medians and ranges. Sensitivities, specificities, positive (PPV) and negative predictive values (NPV) with 95% confidence intervals (CI), and accuracy of the bioinformatics tools for CXCR4-using virus prediction taking ES Trofile as a reference were calculated from 2-by-2 contingency tables.
RESULTS
Demographic characteristics.
A total of 145 phenotypic and genotypic samples were initially included. The baseline characteristics of the study population are summarized in Table 1.
TABLE 1.
Baseline characteristics of the study population
| Characteristic | No. (%) of patientsa |
P value | ||
|---|---|---|---|---|
| Total | Naïve | Treatment experienced | ||
| No. of samples | 145 | 65 | 80 | |
| Median age in years (range) | 39.2 (22-75) | 37.4 (22-71) | 41.7 (22-75) | 0.177 |
| Female | 35 (24.1) | 14 (21.5) | 21 (26.3) | 0.510 |
| Transmission route | 0.010 | |||
| Intravenous drug use | 55 (37.9) | 15 (23.1) | 40 (50.0) | |
| Male homosexual activity | 41 (28.3) | 26 (40.0) | 15 (18.8) | |
| Heterosexual activity | 40 (27.6) | 19 (29.2) | 21 (26.3) | |
| Other | 4 (2.8) | 2 (3.1) | 2 (2.5) | |
| Not known | 5 (3.4) | 3 (4.6) | 2 (2.5) | |
| Median no. of years from HIV infection diagnosis (range) | 5.88 (0-26.3) | 0.63 (0-23.8) | 11.5 (0.2-26.3) | <0.001 |
| Median no. of CD4 cells/mm3 (range) | 300 (0-820) | 375 (10-820) | 270 (0-786) | 0.027 |
| Median HIV-1 RNA load, log10 copies/ml (range) | 4.5 (1.5-7.0) | 4.7 (2.0-6.9) | 4.0 (1.5-7.0) | <0.001 |
| CDC stage | <0.001 | |||
| A | 82 (56.6) | 55 (84.6) | 27 (33.8) | |
| B | 18 (12.4) | 3 (4.6) | 15 (18.8) | |
| C | 45 (31.1) | 7 (10.8) | 38 (47.6) | |
| Hepatitis C virus coinfection | 56 (39.2) | 17 (26.2) | 39 (48.8) | 0.004 |
| HIV-1 subtypeb | 0.035 | |||
| B | 125 (86.2) | 57 (87.5) | 68 (85) | |
| Other | 6 (4.1) | 5 (7.7) | 1 (1.3) | |
| Not tested | 14 (9.7) | 3 (4.6) | 11 (13.8) | |
Values represent the numbers and percentages of patients exhibiting each characteristic unless otherwise noted in the first column.
Subtype of the V3 loop predicted by the Geno2pheno (coreceptor) bioinformatic tool.
Concordance between V3 genotypic tools and ES Trofile.
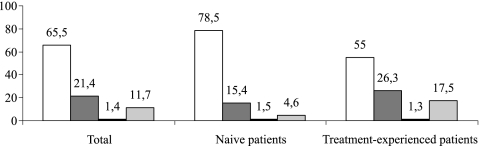
Phenotypic tropism determination was successful in 128 of 145 samples (88.3%), while genotypic testing could be performed in 128 of the 145 samples (88.3%). In 8 cases, nonreportable results were obtained from both phenotypic and genotypic testing. Thus, coreceptor usage was finally determined in 119 paired phenotypic and genotypic samples by the genotypic algorithms previously described, taking ES Trofile as the reference. Overall, there was a prevalence of 52.1% for CCR5 viruses, 32% for dual/mixed viruses, and 1.8% for CXCR4 viruses according to ES Trofile. The proportion of CXCR4 and dual/mixed virus was significantly higher in treatment-experienced patients than in naïve patients (27.6% versus 17%, respectively; P = 0.039) (Fig. 1).
FIG. 1.
Prevalence of HIV-1 tropism in naïve and treatment-experienced patients (n = 145). The white, dark-gray, black, and light-gray bars represent R5, dual/mix, X4 variant, and nonreportable percentages, respectively.
Among bioinformatics tools, PSSMX4X5 showed the best concordance with ES Trofile (82.6%; 95% CI, 76.7 to 86.4%). Geno2pheno (false-positive rate, 20%) showed the highest sensitivity to predict CXCR4 (76.7%; 95% CI, 61.8 to 87.5%), with a specificity of 73.6% (95% CI, 68.7 to 77.2%) (Tables 2 and 3). Although there were no overall significant differences in the accuracy of the bioinformatics tools according to antiretroviral experience, Geno2pheno 1% and 2.5% did perform better in assessing HIV tropism in samples from naïve patients than from treatment-experienced patients (Table 4). When samples were stratified in relation to the CD4 cell counts, those from patients with CD4 cell counts between 200 and 500 cells/mm3 showed the best predictive performance, and among the algorithms, again the PSSMX4R5 algorithm displayed the highest concordance with ES Trofile. In contrast, patients with CD4 cell counts of less than 200 cells/mm3 exhibited the least accurate results (Table 5).
TABLE 2.
Accuracy of the bioinformatic tools to predict CXCR4-using virus when measured against ES Trofile and the original Trofilea
| Bioinformatic tool | % overall correct (95% CI range) with: |
|
|---|---|---|
| ES Trofile (n = 145) | Trofile (n = 99) | |
| Geno2pheno 1% | 79.3 (75.2-80.7) | 67.4 (62.0-67.4) |
| Geno2pheno 5% | 79.3 (73.4-84.1) | 70.9 (62.5-76.3) |
| Geno2pheno 10% | 76.9 (69.7-83.2) | 73.3 (63.6-80.8) |
| Geno2pheno 20% | 74.4 (67.0-79.7) | 67.4 (57.2-76.3) |
| PSSMX4R5 | 82.6 (76.7-86.4) | 73.5 (65.6-76.9) |
| PSSMsinsi | 80.2 (74.1-84.9) | 73.5 (65.6-76.9) |
| 11/25 rule | 75.2 (69.8-80.6) | 73.8 (65.9-77.2) |
| 11/24/25 rule | 74.4 (68.9-80.0) | 75.0 (67.0-78.4) |
| Net charge rule | 73.6 (66.3-80.2) | 69.0 (59.1-77.5) |
ES Trofile, enhanced Trofile HIV-1 coreceptor tropism assay; CI, confidence interval; PSSM, position-specific score matrix. Geno2pheno 1% predicts tropism through the V3 sequence plus additional host-specific features. The rate of false-positive selection is 1% (likewise for other percentages). For PSSMX4R5, a sequence is assigned a score based on the comparison with a sequence of known X4 viruses; for PSSMsinsi, the scoring distinguishes syncytium-inducing from non-syncytium-inducing viruses. The 11/25 rule is based on the presence of positively charged amino acids at positions 11 and/or 25 of the V3 loop of the envelope glycoprotein gp120, the 11/24/25 rule is based on the presence of positively charged amino acids at positions 11 and/or 24 and/or 25 of the V3 loop of the envelope glycoprotein gp120, and the net charge rule is the overall net charge of V3 is used to predict HIV-1 tropism from the V3 genotype.
TABLE 3.
Sensitivity, specificity, and positive and negative predictive values of the bioinformatic tools to predict CXCR4 viruses against the enhanced sensitivity Trofile and original Trofile assaysa
| Bioinformatic tool | % sensitivity (95% CI) |
% specificity (95% CI) |
PPV (95% CI) |
NPV (95% CI) |
||||
|---|---|---|---|---|---|---|---|---|
| ES Trofile | Trofile | ES Trofile | Trofile | ES Trofile | Trofile | ES Trofile | Trofile | |
| Geno2pheno 1% | 20.0 (11.7-22.7) | 20.0 (13.3-20.0) | 98.9 (96.2-99.8) | 100 (95.4-100) | 85.7 (50.2-97.4) | 100 (66.4-100) | 78.9 (76.8-79.7) | 64.6 (61.6-64.6) |
| Geno2pheno 5% | 36.7 (24.7-46.2) | 40.0 (29.6-46.6) | 93.4 (89.4-96.6) | 92.2 (85.0-96.7) | 64.7 (43.5-81.6) | 77.8 (57.6-90.6) | 81.7 (73.8-84.5) | 69.1 (63.8-72.5) |
| Geno2pheno 10% | 56.7 (42.1-69.5) | 57.1 (45.3-66.5) | 83.5 (78.7-87.7) | 84.3 (76.2-90.7) | 53.1 (39.5-65.2) | 71.4 (56.6-83.1) | 85.4 (80.5-89.7) | 74.1 (67.0-79.8) |
| Geno2pheno 20% | 76.7 (61.8-87.5) | 62.9 (50.3-73.8) | 73.6 (68.7-77.2) | 70.6 (61.9-78.1) | 48.9 (39.4-55.8) | 59.5 (47.5-69.8) | 90.5 (84.5-94.9) | 73.5 (64.5-81.3) |
| PSSMX4R5 | 43.3 (31.4-51.0) | 39.4 (29.5-43.7) | 95.6 (91.7-98.1) | 96.0 (89.4-98.9) | 76.5 (55.4-90.0) | 86.7 (64.8-96.2) | 83.7 (80.2-85.9) | 70.6 (65.8-72.7) |
| PSSMsinsi | 40.0 (27.7-49.6) | 39.4 (29.5-43.7) | 93.4 (89.4-96.6) | 96.0 (89.4-98.9) | 66.7 (46.2-82.6) | 86.7 (64.8-96.2) | 82.5 (78.9-85.3) | 70.6 (65.8-72.7) |
| 11/25 rule | 26.7 (15.7-37.5) | 41.2 (31.4-45.5) | 91.2 (87.6-94.8) | 96.0 (89.3-98.9) | 50.0 (29.4-70.3) | 87.5 (66.7-96.4) | 79.0 (75.9-82.1) | 70.6 (65.7-72.7) |
| 11/24/25 rule | 26.7 (15.6-38.0) | 44.1 (34.2-48.3) | 90.1 (86.5-93.8) | 96.0 (89.3-98.9) | 47.1 (27.5-67.1) | 88.2 (68.4-96.6) | 78.8 (75.6-82.1) | 71.6 (66.6-73.8) |
| Net charge rule | 53.3 (38.7-66.8) | 52.9 (40.7-63.4) | 80.2 (75.4-84.7) | 80.0 (71.6-87.1) | 47.1 (34.2-59.0) | 64.3 (49.4-77.0) | 83.9 (78.9-88.6) | 71.4 (64.0-77.8) |
PPV, positive predictive value; NPV, negative predictive value.
TABLE 4.
Accuracy of the bioinformatic tools to predict CXCR4 using virus in naïve and treatment-experienced patients
| Bioinformatic tool | % overall correct (95% CI) |
|
|---|---|---|
| Naïve (n = 65) | Treatment experienced (n = 80) | |
| Geno2pheno 1% | 84.5 (78.5-87.3) | 73.8 (68.4-73.8) |
| Geno2pheno 2.5% | 84.5 (78.5-87.3) | 77.0 (70.3-77.0) |
| Geno2pheno 5% | 81.0 (73.6-88.1) | 77.0 (68.3-81.7) |
| Geno2pheno 10% | 75.9 (67.4-84.4) | 77.0 (66.0-85.7) |
| Geno2pheno 15% | 77.6 (68.5-85.7) | 75.4 (64.1-83.6) |
| Geno2pheno 20% | 72.4 (62.7-80.0) | 75.4 (64.4-81.6) |
| PSSMX4R5 | 84.5 (78.5-87.3) | 80.3 (70.4-86.4) |
| PSSMsinsi | 79.3 (74.5-85.4) | 80.3 (70.4-86.4) |
| 11/25 rule | 79.3 (73.0-86.4) | 70.5 (62.4-77.7) |
| 11/24/25 rule | 77.6 (71.2-85.4) | 70.5 (62.4-77.7) |
| Net charge rule | 69.0 (62.5-78.3) | 78.7 (67.6-87.0) |
TABLE 5.
Accuracy of the bioinformatic tools to predict CXCR4 using virus according to CD4 cell count
| Bioinformatic tool | % overall correct (95% CI) |
||
|---|---|---|---|
| CD4 < 200 (n = 40) | CD4 200-500 (n = 64) | CD4 > 500 (n = 33) | |
| Geno2pheno 1% | 61.8 (51.6-66.6) | 87.7 (83.2-87.7) | 70.4 (64.5-70.4) |
| Geno2pheno 2.5% | 64.7 (53.0-69.5) | 89.5 (83.8-89.5) | 70.4 (64.5-70.4) |
| Geno2pheno 5% | 67.6 (53.5-75.9) | 86.0 (78.6-92.0) | 66.7 (60.7-72.6) |
| Geno2pheno 10% | 67.6 (51.7-79.9) | 78.9 (70.0-85.4) | 70.4 (60.4-76.4) |
| Geno2pheno 15% | 73.5 (57.2-85.2) | 73.7 (64.6-80.2) | 74.1 (61.5-80.1) |
| Geno2pheno 20% | 76.5 (60.2-86.8) | 68.4 (59.3-74.9) | 77.8 (61.8-87.7) |
| PSSMX4R5 | 73.5 (58.3-81.7) | 87.7 (80.6-92.5) | 70.4 (64.5-70.4) |
| PSSMsinsi | 76.5 (61.0-84.6) | 80.7 (75.8-87.7) | 70.4 (64.5-70.4) |
| 11/25 rule | 67.6 (53.5-75.9) | 77.2 (74.3-84.6) | 70.4 (64.5-70.4) |
| 11/24/25 rule | 67.6 (53.5-75.9) | 75.4 (72.6-83.3) | 74.1 (64.6-74.1) |
| Net charge rule | 73.5 (57.2-85.2) | 77.2 (69.6-85.3) | 59.3 (49.0-73.8) |
Accuracy of the bioinformatic tools comparing Trofile and ES Trofile.
The concordance and accuracy of the bioinformatics tools to predict CXCR4-tropic virus using ES Trofile as the reference were compared with the concordance/accuracy when using the original Trofile (Table 2 and 3). Overall, the concordance of the genotypic algorithms was similar when the results of the two phenotypic assays were compared, although PSSMX4R5 and Geno2pheno 1% had significantly higher concordance with the ES Trofile than with the original version (Table 2). Likewise, although the accuracy of the bioinformatics tools was not different between the two phenotypic assays, all negative predictive values of the bioinformatics algorithms were higher when ES Trofile was used as the reference than when the original assay was used (Table 3).
DISCUSSION
The new gold standard for HIV tropism determination is ES Trofile, an enhanced phenotypic test that has replaced the original Trofile version, which has not been available since June 2008. Genotypic methods are an alternative to phenotypic methods and have demonstrated a satisfactory correlation with the original Trofile assay but have not yet been validated using ES Trofile as the reference. Our study shows that the correlation of the genotype-based bioinformatics tools with ES Trofile is good and at least comparable to, and in some aspects might even be better than, the correlation with the original Trofile version.
The study of the genotypic-phenotypic correlation might be complicated by the presence of minority CXCR4-using variants, since it has been argued that genotypic tests lack sensitivity for predicting CXCR4 viruses (9). The new ES Trofile has significantly improved the ability to identify minority variants, being able to detect CXCR4-using viruses representing only 0.3% of the viral population with 100% sensitivity (17, 23), compared with the 10% lower limit of detection of the original Trofile (25). This might elicit doubts about the performance of the genotypic tests compared to the much more sensitive ES Trofile version. Our study is the first to evaluate genotypic algorithms with ES Trofile as the reference and to compare the performance of the two phenotypic methods in a sample of patients from the same cohort during a consecutive period of time. We found that the correlation of the bioinformatics models with the ES Trofile was similar to that found in previous studies when the original Trofile version was used as the reference method (14, 22). We also stated that the performance of the genotypic tests differed according to certain patients' clinical data. Samples from individuals with a CD4 cell count of 200 to 500 cells/mm3 showed the highest accuracy for tropism prediction, whereas the lowest accuracy was observed in those with <200 cells/mm3. Additionally, some of the bioinformatics tools also demonstrated higher accuracy in samples from naïve patients than samples from treatment-experienced patients. This is in agreement with the better performance of the genotypic tests in naïve patients, in whom the probability of CXCR4 variants is lower (7). In our study, treatment-experienced patients had significantly lower CD4 cell counts than naïve patients and also a higher duration of HIV infection. These findings are consistent with previous studies reporting a higher prevalence of CXCR4-using viruses in treatment-experienced patients (7) and an association of duration of HIV infection and the emergence of CXCR4 variants (13). The probability of missing CXCR4 variants might increase as the number of these species increases, thus explaining the poorer performance of genotypic tests. Finally, our results are also supported by a retrospective substudy of the MERIT clinical trial, in which sequencing of the V3 loop was shown to be comparable to ES Trofile in predicting clinical response to maraviroc in treatment-naïve patients (11).
When the concordance and accuracy of the bioinformatics tools using ES Trofile were compared with the original Trofile version in samples from our cohort of patients, the results were similar or even slightly better when ES Trofile was used as the reference. We also found higher negative predictive values when genotypic methods were compared to ES Trofile than when they were compared to the original test, which might represent an advantage when prescribing maraviroc, a selective CCR5 coreceptor antagonist with no activity against CXCR4-using viruses.
A limitation of the study is the lack of determination of both phenotypic tests, the original and ES Trofile assays, on the same plasma samples for the comparison of both methods. Unfortunately, the availability of the two procedures at the same time and their high cost were hampering factors against such an approach. The low number of non-B subtype HIV-1 strains precluded us from analyzing the accuracy of genotype within this subgroup of patients. In contrast, the high number of samples, the availability of clinical data, and the wide spectrum of patients included allowed us to explore the performance of the genotypic tests compared to the new version of the reference phenotypic method in the real-world scenario and to contribute to characterizing potential candidates for a more accurate prediction. It remains to be confirmed whether, as previously stated, the predictive capacity of genotype also improves when the bioinformatics algorithms are used in combination or when clinical variables are added (2, 3, 15, 20) when ES Trofile is used as the reference, as shown in a recent study (19).
In conclusion, we found that genotypic tests predict accurately viral tropism when using the ES Trofile assay as the reference and that this accuracy is at least comparable to or slightly higher than that observed with the original Trofile assay. The predictive capacity of genotype differs according to the antiretroviral status and CD4 cell count of the patients. Our results support that genotypic tools might also constitute suitable alternative methods to the enhanced phenotypic assays for tropism prediction.
Acknowledgments
We acknowledge the contributions of Victoria Ortiz de la Tabla and José María Cuadrado from Hospital Universitario de San Juan Alicante, Spain, Juan Molina and Vicenta Fenoll from Hospital de la Marina Baixa, Spain, who participated in data collection and sample handling and storing, and Daniel Ciprián from Hospital General Universitario de Elche, Spain, who contributed to the statistical analysis.
This work was financially supported by the ISCIII-RETIC RD06/0006/0027, FIBELX 19/2007, and Conselleria de Sanitat Valenciana AP-089/07 grants.
We have no conflicts of interest to report.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Braun, P., and F. Wiesmann. 2007. Phenotypic assays for the determination of coreceptor tropism in HIV-1 infected individuals. Eur. J. Med. Res. 12:463-472. [PubMed] [Google Scholar]
- 2.Chueca, N., C. Garrido, M. Alvarez, E. Poveda, J. De Dios Luna, N. Zahonero, J. Hernández-Quero, V. Soriano, C. Maroto, C. De Mendoza, and F. García. 2009. Improvement in the determination of HIV-1 tropism using the V3 gene sequence and a combination of bioinformatic tools. J. Med. Virol. 81:763-767. [DOI] [PubMed] [Google Scholar]
- 3.Delobel, P., M. T. Nugeyre, M. Cazabat, C. Pasquier, B. Marchou, P. Massip, F. Barre-Sinoussi, N. Israël, and J. Izopet. 2007. Population-based sequencing of the V3 region of env for predicting the coreceptor usage of human immunodeficiency virus type 1 quasispecies. J. Clin. Microbiol. 45:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Mendoza, C., C. Rodríguez, F. García, J. M. Eiros, L. Ruiz, E. Caballero, A. Aguilera, P. Leiva, J. Colomina, F. Gutierrez, J. Del Romero, J. Aguero, and V. Soriano on behalf of the Spanish HIV Seroconverter Study Group. 2007. Prevalence of X4 tropic viruses in patients recently infected with HIV-1 and lack of association with transmission of drug resistance. J. Antimicrob. Chemother. 59:698-704. [DOI] [PubMed] [Google Scholar]
- 5.Garrido, C., V. Roulet, N. Chueca, E. Poveda, A. Aguilera, K. Skrabal, N. Zahonero, S. Carlos, F. García, J. L. Faudon, V. Soriano, and C. de Mendoza. 2008. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J. Clin. Microbiol. 46:887-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holguín, A., M. López, and V. Soriano. 2008. Reliability of rapid subtyping tools compared to that of phylogenetic analysis for characterization of human immunodeficiency virus type 1 non-B subtypes and recombinant forms. J. Clin. Microbiol. 46:3896-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt, P. W., P. R. Harrigan, W. Huang, M. Bates, D. W. Williamson, J. M. McCune, R. W. Price, S. S. Spudich, H. Lampiris, R. Hoh, T. Leigler, J. N. Martin, and S. G. Deeks. 2006. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J. Infect. Dis. 194:926-930. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer, T., O. Sander, S. Sierra, A. Thielen, and R. Kaiser. 2007. Bioinformatics predictions of HIV coreceptor usage. Nat. Biotechnol. 25:1407-1410. [DOI] [PubMed] [Google Scholar]
- 9.Low, A. J., W. Dong, D. Chan, T. Sing, R. Swanstrom, R. Jensen, S. Pillai, B. Good, and P. R. Harrigan. 2007. Current V3 genotyping algorithms are inadequate for predicting X4 coreceptor usage in clinical isolates. AIDS 21:F17-F24. [DOI] [PubMed] [Google Scholar]
- 10.Low, A. J., L. C. Swenson, and P. R. Harrigan. 2008. HIV coreceptor phenotyping in the clinical setting. AIDS Rev. 10:143-151. [PubMed] [Google Scholar]
- 11.McGovern, R., W. Dong, X. Zhong, D. Knapp, A. Thielen, D. Chapman, M. Lewis, I. James, H. Valdez, and R. Harrigan. 2010. Population-based sequencing of the V3-loop is comparable to the enhanced sensitivity Trofile assay in predicting virologic response to maraviroc of treatment-naïve patients in the MERIT trial. Abstr. 17th Conf. Retrovir. Opportunistic Infect., abstr. 92.
- 12.Obermeier, M., A. Carganico, B. Bieniek, D. Schleehauf, S. Dupke, and K. Fischer. 2010. Tropism testing from proviral DNA analysis of a subgroup from the Berlin maraviroc cohort, abstr. 23. Abstr. 8th Eur. HIV Drug Resist. Workshop, Sorrento, Italy.
- 13.Philpott, S. M. 2003. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 1:217-227. [DOI] [PubMed] [Google Scholar]
- 14.Poveda, E., V. Briz, V. Roulet, M. González, J. Faudon, K. Skrabal, and V. Soriano. 2007. Correlation between a phenotypic assay and three bioinformatics tools for determining HIV co-receptor use. AIDS 21:1487-1490. [DOI] [PubMed] [Google Scholar]
- 15.Poveda, E., E. Seclén, M. González, F. García, N. Chueca, A. Aguilera, J. J. Rodríguez, J. González-Lahoz, and V. Soriano. 2009. Design and validation of new genotypic tools for easy and reliable estimation of HIV tropism before using CCR5 antagonists. J. Antimicrob. Chemother. 63:1006-1010. [DOI] [PubMed] [Google Scholar]
- 16.Raymond, S., P. Delobel, M. Mavigner, M. Cazabat, C. Souyris, K. Sandres-Sauné, L. Cuzin, B. Marchou, P. Massip, and J. Izopet. 2008. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS 22:11-16. [DOI] [PubMed] [Google Scholar]
- 17.Reeves, J., D. Han, P. Hunt, Y. Liu, T. Wrin, W. Huang, S. Deeks, C. Petropoulos, J. Whitcomb, E. Coakley, and N. Parkin. 2007. Enhancements to the Trofile HIV coreceptor tropism assay enable reliable detection of CXCR4-using subpopulations at less than 1%, abstr. H-1026. 47th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC). American Society for Microbiology, Washington, DC.
- 18.Rose, J. D., A. M. Rhea, J. Weber, and M. E. Quiñones-Mateu. 2009. Current tests to evaluate HIV-1 coreceptor tropism. Curr. Opin. HIV AIDS 4:136-142. [DOI] [PubMed] [Google Scholar]
- 19.Sánchez, V., M. Masiá, C. Robledano, S. Padilla, B. Lumbreras, E. Poveda, C. de Mendoza, V. Soriano, and F. Gutiérrez. 2010. A highly sensitive and specific model for predicting HIV-1 tropism in treatment-experienced patients combining interpretation of V3 loop sequences and clinical parameters. Abstr. 17th Conf. Retrovir. Opportunistic Infect., abstr. M-253. [DOI] [PubMed]
- 20.Sander, O., T. Sing, I. Sommer, A. J. Low, P. K. Cheung, P. R. Harrigan, T. Lengauer, and F. S. Domingues. 2007. Structural descriptors of gp120 loop for the prediction of HIV-1 coreceptor usage. PloS Comput. Biol. 3:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sierra, S., A. Thielen, S. Reuter, B. Jensen, S. Esser, and G. Faetkenheuer. 2010. Tropism determination and clinical outcome of 61 patients under maraviroc treatment, abstr. 20. Abstr. 8th Eur. HIV Drug Resist. Workshop, Sorrento, Italy.
- 22.Skrabal, K., A. J. Low, W. Dong, T. Sing, P. K. Cheung, F. Mammano, and P. R. Harrigan. 2007. Determining human immunodeficiency virus coreceptor use in a clinical setting: degree of correlation between two phenotypic assays and a bioinformatic model. J. Clin. Microbiol. 45:279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trinh, L., D. Han, W. Huang, T. Wrin, J. Larson, L. D. Kiss, E. Coakley, C. J. Petropoulos, N. T. Parkin, J. M. Whitcomb, and J. D. Reeves. 2008. Technical validation of an enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Antivir. Ther. 13:A128. [Google Scholar]
- 24.Van Lelyveld, S., J. Symons, A. Hoepelman, A. Stam, P. Van Ham, and M. Nijhuis. 2010. Correlation of clinical outcome of maraviroc treatment with different methods to determine HIV tropism: genotypic assay, MT-2 assay and Trofile, abstr. 41. Abstr. 8th Eur. HIV Drug Resist. Workshop, Sorrento, Italy.
- 25.Whitcomb, J. M., W. Huang, S. Fransen, K. Limoli, J. Toma, T. Wrin, C. Chappey, L. D. Kiss, E. E. Paxinos, and C. J. Petropoulos. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]