Abstract
To improve the diagnosis of classical rabies virus with molecular methods, a validated, ready-to-use, real-time reverse transcription-PCR (RT-PCR) assay was developed. In a first step, primers and 6-carboxyfluorescien-labeled TaqMan probes specific for rabies virus were selected from the consensus sequence of the nucleoprotein gene of 203 different rabies virus sequences derived from GenBank. The selected primer-probe combination was highly specific and sensitive. During validation using a sample set of rabies virus strains from the virus archives of the Friedrich-Loeffler-Institut (FLI; Germany), the Veterinary Laboratories Agency (VLA; United Kingdom), and the DTU National Veterinary Institute (Lindholm, Denmark), covering the global diversity of rabies virus lineages, it was shown that both the newly developed assay and a previously described one had some detection failures. This was overcome by a combined assay that detected all samples as positive. In addition, the introduction of labeled positive controls (LPC) increased the diagnostic safety of the single as well as the combined assay. Based on the newly developed, alternative assay for the detection of rabies virus and the application of LPCs, an improved diagnostic sensitivity and reliability can be ascertained for postmortem and intra vitam real-time RT-PCR analyses in rabies reference laboratories.
Rabies, the oldest and one of the most feared lethal zoonotic diseases known to humanity, is caused by different Lyssavirus species of the family Rhabdoviridae (31). The classical rabies virus (RABV) is also referred to as genotype 1 and is widely distributed across the globe, with only certain regions and countries (e.g., western Europe, the United Kingdom, New Zealand, Hawaii, Australia, Japan, and Antarctica) being free from the disease, either historically or through successful eradication programs. The transmission of RABV is maintained in most parts of the world by omni- or carnivorous mammals, such as dogs, foxes, jackals, mongoose, skunks, raccoon dogs, and raccoons; in the Americas, RABV also is transmitted by a wide variety of bat species (24). However, most human rabies cases occur in Africa and Asia, with an estimated 55,000 deaths per year (40). Definitive rabies diagnoses in both human and animal samples rely on postmortem laboratory findings. The gold standard of both the WHO and the International Office of Epizootics (OIE) is the detection of lyssavirus antigen by the fluorescent antibody test (FAT) (8). In animal samples from human contacts or with epidemiological relevance, virus isolation should be used as a confirmatory test for FAT-inconclusive and -negative samples (40). With the advent of modern molecular techniques, reverse transcription-PCR (RT-PCR) was applied for virus characterization and the diagnosis of lyssaviruses based on the detection of virus-specific genetic material (28). Classical PCR assays proved to be a sensitive and specific tool for routine diagnostic purposes (32, 33), particularly in decomposed samples (7, 1, 39) or archival specimens (22, 3). The rapidity and high sensitivity of the PCR technique also has offered new prospects of the intra vitam diagnosis of humans being superior to conventional techniques (6). Viral RNA can be detected in several biological fluids and samples (e.g., saliva, cerebrospinal fluid, tears, skin biopsy, and urine), but serial sampling is needed because of the intermittent shedding of virus.
Various conventional, gel-based PCR protocols for the amplification of lyssavirus RNA fragments are available (11). In generic pan-lyssavirus approaches, heminested (15) or fully nested assays are used (9, 13, 35). However, gel-based systems used for the detection of amplified PCR products bear the risk of cross-contamination, especially when using (hemi)nested PCRs and do not allow the exact quantification of genome copies. Furthermore, there is no test for specificity included in the assay (2). To overcome this, several methods have been developed for rabies diagnosis, including hybridization (28), restriction fragment length polymorphism (RFLP), PCR-enzyme-linked immunosorbent assay (ELISA) (5), in situ hybridization (10), and sequencing. The latter has become widely used, as the sequences obtained can be used for further genetic characterization (21).
Only the use of fluorogenic probes allows the detection of sequence-specific templates in real time. Also, hybridization reactions provide specificity, and cross-contaminations can be largely avoided (14, 16). Consequently, TaqMan RT-PCR assays were developed for various lyssavirus genotypes other than RABV (13, 19, 27, 37).
In North America, TaqMan PCR assays to detect RABV either were comparable (29) or had a considerably reduced detection limit compared to that of heminested PCR (20). A SYBR green real-time PCR also has been used as a method to improve intra vitam rabies diagnosis by testing the saliva of diseased persons (26). Similarly, a real-time RT-PCR assay with TaqMan technology was developed by using sequences of rabies variants prevalent in Thailand, Myanmar, Cambodia, Indonesia, and India (36). Orlowska and coworkers (27) compared a real-time PCR for RABV to a heminested RT-PCR method and found comparable sensitivity and specificity. The broadest approach was a real-time TaqMan RT-PCR for the detection and differentiation of several lyssavirus genotypes (4); however, there were limitations in the rapidity and specificity and the need for separate reactions. The technical constraints of this assay were overcome by a later approach using generic primers and genotype-specific probes in one reaction. This assay is able to detect and distinguish RABV and European bat lyssaviruses 1 and 2 (37).
In this study, a different modified assay solely targeting RABV was developed. The designed assay was subject to validation as a single and combined assay with the protocol described by Wakeley and coworkers (37).
To respond to demands in quality assurance and to test the functionality and robustness of the assays, different innovative internal process controls were included in this study. Artificial labeled positive controls (LPC) were designed, generated, and assessed to replace conventional positive controls that bring the risk of cross-contamination.
MATERIALS AND METHODS
Design of primers and probes.
The alignment of the different RABV sequences was undertaken using the Genetics Computer Group software package (GCG, Wisconsin). Alignment-based primer and probe selection with a consensus sequence of 203 different RABV variants published in the NCBI GenBank was supported by the software package Beacon Designer 2.06 (Premier Biosoft, Palo Alto, CA). A total of 20 primers were selected, and 13 resulting primer pairs were screened for sensitivity on a limited panel of RABV (data not shown). Based on the CT values and the absence of comigrating bands after visualization in agarose gels, the assay R14 was selected as the most promising. All oligonucleotides (Table 1) were synthesized by Eurofins MWG, GmbH (Ebersberg, Germany), and stored at −20°C until use.
TABLE 1.
Oligonucleotides used in this study
| Assay or function | Name | Role | Length (nt) | Sequencec | Positiona | PCR or in vitro RNA fragment size (bp or nt) |
|---|---|---|---|---|---|---|
| R13b | JW12 | Primer | 19 | ATGTAACACCYCTACAATG | 55-73 | 110 |
| N165-146 | Primer | 20 | GCAGGGTAYTTRTACTCATA | 165-146 | ||
| LysGT1-FAM | Probe | 29 | 6-FAM-ACAAGATTGTATTCAAAGTCAATAATCAG-TAMRA | 81-109 | ||
| LacZ-Cy5 | Probe | 25 | Cy5-TCC AGT CGG GAA ACC TGT CGT GCC A-BHQ3 | 56-80 | ||
| R14 | RV-N_F | Primer | 23 | GATCCTGATGAYGTATGTTCCTA | 266-288 | 87 |
| RV-N_R | Primer | 19 | RGATTCCGTAGCTRGTCCA | 353-335 | ||
| RabGT1-B-FAM | Probe | 25 | 6-FAM-CAGCAATGCAGTTYTTTGAGGGGAC-TAMRA | 297-321 | ||
| LacZ-Cy5 | Probe | 25 | Cy5-TCC AGT CGG GAA ACC TGT CGT GCC A-BHQ3 | 56-80 | ||
| Generation of LPC R13 | oLPC-R13 | Template for in vitro transcription | 130 | GCA GCA GGG TAC TTG TAC TCA TAT GAC TGA TTA TTG ACT TTG AATACA ATC TTG TAG ATG GCA CGA CAG GTT TCC CGA CTG GAT CTCATT GTA GAG GTG TTA CAT TCG CCC TAT AGT GAG TCG TAT TAC A | Artificial | 111 |
| Generation of LPC R14 | oLPC-R14 | Template for in vitro transcription | 129 | GCA GGA TTC CGT AGC TGG TCC ATG AGT CCC CTC AAA GAA CTGCAT TGC TGA GAT GGC ACG ACA GGT TTC CCG ACT GGA TCT TAG GAACAT ACG TCA TCA GGA TCT CGC CCT ATA GTG AGT CGT ATTACA | Artificial | 110 |
According to the sequences of SAD B19 (GenBank accession no. M31046) and the cloning vector pUC13 (L09130), respectively.
The RABV genotype 1 assay was described by Wakeley et al. (37) and modified by the LacZ-Cy5 probe.
The T7 promoter sites in the oLPC are depicted in boldface. Primer and probe binding sites specific for RABV are underlined, and the binding site for the LacZ-Cy5 probe are shown in italics. TAMRA, 6-carboxytetramethylrhodamine; BHQ3, black hole quencher 3.
Generation of LPC and testing of analytical sensitivity.
For the construction of the labeled positive controls (LPC), long oligonucleotides were synthesized by Eurofins MWG, GmbH (Ebersberg, Germany), and used as the template for the generation of the artificial RNA (Table 1). The in vitro-transcribed LPC RNA was produced using the Riboprobe T7 system (Promega, GmbH, Mannheim, Germany) and purified using the miRNeasy mini kit (Qiagen, Hilden, Germany), including a second on-column DNA digestion (RNase-free DNase set; Qiagen, Hilden, Germany). The concentration of the LPC in vitro RNA was determined by spectrophotometry (Nanodrop; PEQLAB Biotechnologie GmbH, Erlangen, Germany), and the exact number of RNA molecules was calculated using the formula (X g/μl RNA/[transcript length in nucleotides × 340]) × 6.022 × 1023 = Y molecules/μl.
The stock solutions of the in vitro-transcribed RNA were stored at −80°C, and the diluted working solutions were stored at −20°C. Using the working solutions the analytical sensitivity of the real-time RT-PCR assays was determined by testing R13 and R14 LPC standard RNA diluted serially 10-fold.
Samples, viruses, and cells.
RABVs, Lagos bat virus (LBV), Mokola virus (MOKV), Duvenhage virus (DUVV), European bat lyssavirus 1 and 2 (EBLV-1 and EBLV-2), and Australian bat lyssavirus (ABLV) were obtained from the archive of the National Reference Laboratory for Rabies, located at the Friedrich-Loeffler-Institut, Wusterhausen, Germany. Brain homogenates from original samples or from mice after inoculation were used. For a few archived viruses, tissue culture supernatant was used (Table 2 ). For the latter, viruses were cultured using murine neuroblastoma cells (NA-cells; cat. no. 229; Collection of Cell Lines in Veterinary Medicine [CCLV], Insel Riems, Germany) as described previously (38). Brain samples from 12 different species that tested negative by FAT were used as negative controls.
TABLE 2.
Diagnostic sensitivity and specificity using the R13, R14, and combined assay for the detection of RABV with prediluted RNAa
| Lab ID | Material | Yr | Host | Continent | Country | R13 assay (CT) | R14 assay (CT) | R13-R14 combination assay (CT) |
|---|---|---|---|---|---|---|---|---|
| 13118 | BS* | 1983 | Dog | Africa | Algeria | 39.20 | 30.96 | 30.73 |
| 20827 | BS | 1991 | Duiker | Africa | Botswana | 27.77 | 23.28 | 24.29 |
| 13130 | BS | 1974 | Africa | Egypt | 29.27 | 28.24 | 26.81 | |
| 20829 | BS | 1998 | Dog | Africa | Egypt | 22.57 | 20.75 | 21.35 |
| 20830 | BS | 1998 | Dog | Africa | Egypt | 28.39 | 26.97 | 27.34 |
| 13136 | BS | 1989 | Africa | Nigeria | 22.79 | 22.91 | ||
| 13137 | BS | 1988 | Dog | Africa | Nigeria | 25.30 | 24.41 | |
| 20825 | BS | 2005 | Dog | Africa | Nigeria | 28.86 | 28.94 | |
| 20826 | BS | 1996 | Human | Africa | Nigeria | 22.79 | 21.32 | |
| 20828 | BS | 1990 | Cat | Africa | South Africa | 26.79 | 22.41 | 23.7 |
| 13176 | BS | 1992 | Dog | Africa | Sudan | 32.17 | 32.42 | |
| 13116 | BS | Dog | Africa | Tunesia | 30.00 | 25.21 | 24.44 | |
| 13230 | BS | Skunk | Americas | Canada | 34.36 | 30.09 | 30.21 | |
| 13231 | BS | Fox | Americas | Canada | 30.02 | 32.02 | 26.61 | |
| 13239 | BS | Bat | Americas | Canada | 24.11 | 23.97 | ||
| 13240 | BS* | 1986 | Americas | Canada | 25.37 | 25.5 | ||
| 13250 | BS | 1973 | Human | Americas | Chile | 28.79 | 28.76 | |
| 13254 | BS* | 1979 | Human | Americas | Chile | 20.45 | 20.46 | |
| 13196 | BS | 1981 | Dog | Americas | Mexico | 26.21 | 22.88 | 23.21 |
| 13242 | BS* | 1966 | Bat | Americas | South America | 27.45 | 23.64 | 24.49 |
| 13199 | BS* | 1980 | Skunk | Americas | United States | 35.46 | 32.76 | 33.68 |
| 13213 | TCS | 1981 | Skunk | Americas | United States | 27.98 | 25.75 | 25.16 |
| 13216 | TCS | Bat | Americas | United States | 28.55 | 28.73 | ||
| 20823 | BS | 1988 | Bat | Americas | United States | 27.59 | 28.9 | 28.46 |
| 20824 | BS | 1988 | Raccoon | Americas | United States | 22.31 | 24.76 | 22.77 |
| 13091 | BS | 1994 | Camel | Asia | Abu Dhabi | 17.47 | 16.69 | 16.73 |
| 20277 | BS | 2005 | Dog | Asia | Afghanistan | 18.10 | 19.58 | |
| 20278 | BS | 2005 | Pig | Asia | Afghanistan | 22.91 | 24.59 | |
| 20280 | BS | 2006 | Dog | Asia | Afghanistan | 15.72 | 17.04 | |
| 20281 | BS | 2006 | Dog | Asia | Afghanistan | 14.87 | 17.26 | |
| 20282 | BS | 2006 | Dog | Asia | Afghanistan | 26.32 | 23.75 | |
| 20287 | BS | 2008 | Dog | Asia | Afghanistan | 17.85 | 18.81 | |
| 20292 | BS | 2008 | Dog | Asia | Afghanistan | 14.55 | 15.79 | |
| 5989 | BS | 2002 | Dog | Asia | Aserbaidshan | 29.12 | 24.21 | 24.76 |
| 13081 | TCS | 1985 | Asia | China | 26.09 | 22.99 | 22.68 | |
| 20833 | BS | 1989 | Dog | Asia | China | 29.01 | 22.23 | 19.29 |
| 20834 | BS | 1989 | Cow | Asia | China | 28.57 | 25.41 | 22.61 |
| 13181 | BS | 1982 | Dog | Asia | India | 26.33 | 24.51 | |
| 13184 | BS | 1993 | Monkey | Asia | India | 27.89 | 22.44 | 23.18 |
| 13101 | BS | 1988 | Dog | Asia | Indonesia | 31.09 | 25.77 | 26.64 |
| 13151 | BS | 1991 | Wolf | Asia | Iran | 30.80 | 30.19 | 29.52 |
| 13160 | TCS | 1991 | Sheep | Asia | Iran | 18.39 | 16.54 | 16.34 |
| 13161 | TCS | 1991 | Sheep | Asia | Iran | 19.86 | 19.51 | 19.26 |
| 13164 | TCS | 1991 | Hyena | Asia | Iran | 25.01 | 26.3 | 24.34 |
| 20276 | BS | 2004 | Dog | Asia | Iraq | 16.81 | 18.25 | 22.59 |
| 20279 | BS | 2005 | Dog | Asia | Iraq | 15.43 | 12.82 | 12.61 |
| 20283 | BS | 2007 | Dog | Asia | Iraq | 17.55 | 13.83 | 12.99 |
| 20284 | BS | 2007 | Dog | Asia | Iraq | 16.31 | 13.67 | 13.76 |
| 20285 | BS | 2007 | Mongoose | Asia | Iraq | 17.62 | 17.9 | 16.78 |
| 20288 | BS | 2008 | Cow | Asia | Iraq | 21.77 | 21.49 | 20.82 |
| 20289 | BS | 2008 | Cow | Asia | Iraq | 18.85 | 18.25 | 17.81 |
| 20290 | BS | 2008 | Cow | Asia | Iraq | 24.20 | 25.18 | 24.28 |
| 20291 | BS | 2008 | Dog | Asia | Iraq | 22.56 | 18.47 | 19.1 |
| 20293 | BS | 2008 | Dog | Asia | Iraq | 16.55 | 12.62 | 12.97 |
| 20294 | BS | 2008 | Dog | Asia | Iraq | 16.58 | 12.48 | 12.5 |
| 20295 | BS | 2009 | Dog | Asia | Iraq | 17.41 | 12.86 | 13.38 |
| 20296 | BS | 2009 | Dog | Asia | Iraq | 18.84 | 15.01 | 13.9 |
| 20297 | BS | 2009 | Horse | Asia | Iraq | 14.90 | 11.92 | 11.81 |
| 20298 | BS | 2009 | Cow | Asia | Iraq | 20.05 | 15.73 | 15.99 |
| 20299 | BS | 2008 | Dog | Asia | Iraq | 17.52 | 13.68 | 13.62 |
| 13109 | BS | 1974 | Dog | Asia | Malaysia | 26.91 | 21.91 | 22.19 |
| 13141 | BS | Asia | Oman | 30.15 | 29.56 | 28.15 | ||
| 13088 | BS* | 1979 | Dog | Asia | Pakistan | 28.69 | 28.22 | |
| 13042 | BS* | 1987 | Fox | Asia | Saudi Arabia | 26.98 | 22.35 | 22.52 |
| 13044 | TCS | 1990 | Fox | Asia | Saudi Arabia | 20.48 | 19.42 | 19.5 |
| 13096 | BS | 1974 | Dog | Asia | Singapore | 29.81 | 25.1 | 25.21 |
| 13099 | BS | 1974 | Dog | Asia | Taiwan | 33.56 | 29.02 | 29.23 |
| 20831 | BS | Asia | Thailand | 14.07 | 25.86 | 23.65 | ||
| 20832 | BS | Asia | Thailand | 25.87 | 26.8 | 25.28 | ||
| 13056 | BS* | 1984 | Dog | Asia | Turkey | 26.45 | 23.47 | 23.89 |
| 13077 | BS | 1995 | Fox | Europe | Bulgaria | 27.02 | 24.28 | 24.34 |
| 13078 | BS | 1995 | Human | Europe | Bulgaria | 13.8 | 12.68 | 12.77 |
| 13079 | BS | 1995 | Fox | Europe | Bulgaria | 29.66 | 26.9 | 26.74 |
| 12952 | BS | 2001 | Fox | Europe | Estonia | 31.01 | 27.39 | 27.8 |
| 13001 | TCS | 1991 | Europe | Finland | 30.87 | 27.25 | 27.7 | |
| 13000 | BS | 1990 | Raccoon dog | Europe | Finland | 33.05 | 29.39 | 30.0 |
| 1390 | TCS | 1998 | Fox | Europe | Germany | 29.98 | 30.98 | 29.26 |
| 11164 | BS | 2005 | Fox | Europe | Germany | 23.43 | 23.9 | |
| 11240 | BS | 2005 | Human | Europe | Germany | 16.9 | 18.27 | 16.66 |
| 11251 | BS | 2005 | Human | Europe | Germany | 20.03 | 21.71 | 20.28 |
| 12542 | BS | 2005 | Human | Europe | Germany | 24.99 | 24.53 | 24.2 |
| 13057 | BS | 2005 | Human | Europe | Germany | 17.01 | 17.25 | 16.64 |
| SAD B19 | TCS | Vaccine strain | Europe | Germany | 18.22 | 19.00 | 18.32 | |
| GRA 11/04 | BS | 2004 | Polar fox | Europe | Greenland | 15.67 | 16.20 | 14.56 |
| GRA 16/06 | BS | 2006 | Polar fox | Europe | Greenland | 13.90 | 14.71 | |
| GRA 2/07 | BS | 2007 | Polar fox | Europe | Greenland | 12.58 | 12.68 | 10.72 |
| GRA 2/08 | BS | 2008 | Polar fox | Europe | Greenland | 15.33 | 16.41 | 15.24 |
| GRA 4/07 | BS | 2007 | Polar fox | Europe | Greenland | 12.14 | 12.54 | 12.54 |
| GRA 5/03 | BS | 2003 | Dog | Europe | Greenland | 15.31 | 16.76 | 14.67 |
| GRA 5/04 | BS | 2004 | Polar fox | Europe | Greenland | 15.15 | 17.06 | 13.94 |
| GRA 6/05 | BS | 2005 | Polar fox | Europe | Greenland | 16.71 | 16.99 | 15.70 |
| GRA 6/08 | BS | 2008 | Polar fox | Europe | Greenland | 12.79 | 12.25 | 12.71 |
| GRA 7/05 | BS | 2005 | Polar fox | Europe | Greenland | 17.93 | 18.50 | 17.13 |
BS, brain suspension; BS*, brain suspension from inoculated mice; TCS, tissue culture supernatant.
RNA extraction.
Viral RNA was extracted from cell culture using the QIAamp viral RNA kit (Qiagen, Hilden Germany) or RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After the column was washed twice with the appropriate buffer, RNA was eluted using 50 μl elution buffer and stored at −80°C until use.
TaqMan real-time RT-PCR.
To minimize the risk of cross-contamination, a one-step reverse transcription-PCR (RT-PCR) (RT-PCR) protocol was carried out using the commercially available QuantiTect probe RT-PCR kit (Qiagen, Hilden, Germany) or the RNA UltraSense one-step quantitative RT-PCR system (Invitrogen, Carlsbad, CA). The real-time RT-PCR assay with the QuantiTect probe RT-PCR kit was optimized using a total volume of 25 μl. Briefly, for a single well, 5.25 μl RNase-free water, 12.5 μl 2× QuantiTect probe RT-PCR master mix, 0.25 μl RT mix, 5.0 μl of prediluted RNA, and 2.0 μl primer-probe mix R13 or R14 was mixed. In combined assays, 1 μl of each primer-probe mix R13 and R14 was used. Based on stock solutions of the primer and probes of 100 pmol/μl (100 μM), the two different primer-probe mixes were created. For the newly developed assay R14, 20 μl of the relevant forward and reverse primer, 2.5 μl of the 6-carboxyfluorescein (FAM)-labeled R14 rabies probe, and 2.5 μl of the Cy5-labeled positive-control probe were mixed in 155 μl 0.1× Tris-EDTA (TE) buffer. Thus, a final concentration of 10 μM each primer and 1.25 μM each probe was combined in the R14 primer-probe mix. For the R13 primer-probe mix, identical concentrations with the relevant primers and probes were established.
The reactions were carried out either on an Mx3000P/Mx3005P multiplex quantitative PCR system (Stratagene, La Jolla) or on a LightCycler (Roche Diagnostics, Mannheim, Germany). The RNAs were reverse transcribed and amplified according to the following heating and cooling program: 1 cycle of 50°C for 30 min and 95°C for 15 min, followed by 42 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. In the LightCycler, while keeping the same temperature regimen, the number of cycles was set at 45 and the duration time for each cycle step was 20 s. For each RT-PCR, a critical threshold cycle number (CT) was determined corresponding to the PCR cycle number at which the fluorescence of the reaction exceeded a value determined to be statistically higher than the background by the respective software associated with each system.
To estimate the reproducibility of the assays, most samples (N = 69) were run twice with R13, R14, and the combined assay on an Mx3000P (FLI, Riems, Germany), LightCycler 1.0 (FLI, Wusterhausen, Germany), and Mx3005P (DTU, Lindholm, Denmark). The data were transferred into an Excel spreadsheet (Microsoft), and the resulting graphs and correlation coefficients (of determination) were calculated.
Sequencing.
A selection of samples that tested negative in real-time RT-PCR with either the developed assay (R14) or the previously described one, R13, with the RABV protocol from Wakeley et al. (37), were retested with a conventional RT-PCR as described before (15). Resulting amplicons were subject to the sequence analysis of the partial N gene essentially as described previously (21).
RESULTS
Development and validation of the new R14 real-time RT-PCR for RABV. (i) Primers and probe.
In a first step, 203 sequences of the conserved N gene of different rabies virus strains were used for the creation of a consensus sequence and the identification of highly conserved sequence regions. Besides the genome region used by Wakeley et al. (37), only a few nucleotides downstream make a suitable region for the design of alternative primers, and a respective hybridization probe was chosen. The newly selected primers amplified a fragment of 87 bp in size (Table 1).
(ii) Construction of the LPC.
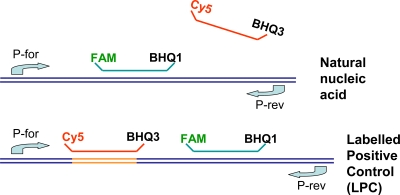
Long oligonucleotides (oLPC) were used as the template for the preparation of labeled in vitro-transcribed artificial positive control RNA (LPC). The LPC RNAs were tagged by an additional binding sequence for an extra probe (LacZ-Cy5). The LacZ-Cy5 probe was integrated into the R13 and R14 primer-probe mixes (Table 1). In the case of the amplification and detection of RABV RNA, no binding of the LacZ-Cy5 probe can be observed. In contrast, the analyses of LPC-RNA produce RABV-specific FAM signals as well as the tag-specific Cy5 fluorescence. The principle of the LPC is depicted in Fig. 1. Based on the equimolar concentration of the probe binding sites for the RABV and LacZ probes, nearly identical CT values can be observed (Fig. 2).
FIG. 1.
Graphical depiction of the principle of the labeled positive control (LPC).
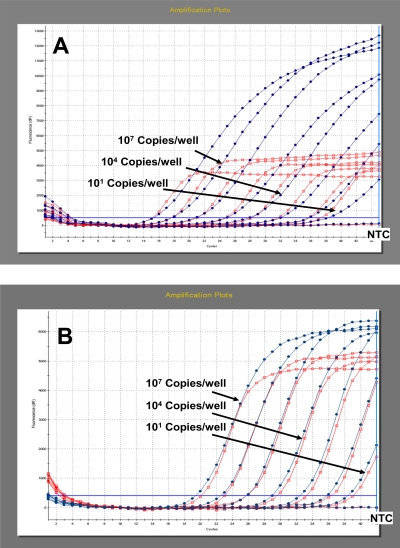
FIG. 2.
Amplification plot for the definition of the analytical sensitivity of the R13 assay (A) and the newly designed R14 system (B), based on in vitro-transcribed LPC RNA. Nearly similar CT values were observed for the FAM-labeled rabies probe (blue line with dots) as well as for the Cy5-labeled LacZ probe (red line with open square).
(iii) Analytical sensitivity and specificity.
Using a series of 10-fold dilutions of LPC RNA, it could be verified that the R13 and R14 real-time RT-PCR systems, as well as the combined assay, amplified the control RNA in a linear fashion from 107 copies per well down to less than 10 copies per well with an PCR efficiency of more than 90% (Fig. 2). Identical high PCR efficiencies were calculated using an RNA dilution series of different RABV strains (see Fig. S1a and b in the supplemental material).
The analytical specificity of the R14 assay was assessed by in silico BlastN searches. Only the forward primer of the R14 systems showed partial identity with EBLV-2, whereas the reverse primer and the R14 probe did not show any significant similarity with other pathogens or housekeeping genes. The specificity of the R14 assay was confirmed by the analysis of related lyssaviruses.
(iv) Diagnostic sensitivity and specificity.
Of 93 brain samples or virus isolations from initially FAT-positive animals, 80 tested positive by the R14 assay. Samples that tested negative originated from India, Pakistan, Afghanistan, and Nigeria (Table 2). Furthermore, none of the negative-control samples, or any of genotypes other than RABV, gave a result above the threshold indicative for the presence of RABV-specific RNA (data not shown).
(v) Reproducibility.
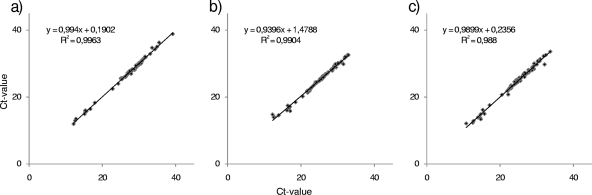
The reproducibility was very high (Fig. 3). The corresponding coefficient of determination (R2) was 0.9904.
FIG. 3.
Reproducibility shown by the CT values for each sample tested twice using the R13 assay (a), the R14 assay (b), and the combination (c). The regression line is indicated, and the coefficients of determination (R2) are provided.
(vi) Integration of internal controls.
For checking the functionality of the RNA extraction as well as the inhibition-free amplification of the R14 fragment, two different internal control (IC) systems were assessed, i.e., the beta-actin housekeeping gene system described by Wakeley et al. (37) and the external universal control system based on in vitro-transcribed enhanced green fluorescent protein RNA (18). Both IC systems used HEX (hexachloro-6-carboxyfluorescein)-labeled hydrolysis probes for the detection. The comparison of the single R14 assay to the duplex R14/IC assays showed similar analytical sensitivities for both systems (data not shown).
Comparison of the developed assay (R14) to a published assay (R13).
The specificity of the R13 assay (37) was evaluated using the same panel of samples as that used before. In this study, of 93 samples 85 tested positive by the R13 assay. Eight samples were negative or gave inconclusive results in the different laboratories (did not score as reproducibly positive), comprising samples from Europe, Chile, North America, the Sudan, and Greenland (Table 2). Overall, a high reproducibility was observed, with R2 = 0.9963. None of the negative-control samples gave a result above the threshold indicative for the presence of virus-specific RNA. In addition, the R13 assay showed reactivity with one ABLV isolate (data not shown); however, these data have to be verified using additional isolates of the same genotype.
When combining both assays in a single mix, all initially positive samples with either method also scored positive (Table 2). The reproducibility of the novel combined R13/14 assay was determined as R2 = 0.98 (Fig. 3).
Sequence comparison.
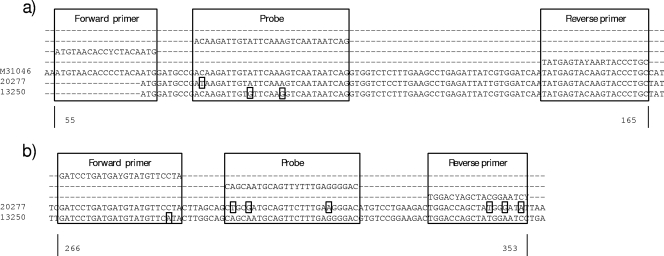
Selected sequences of samples that tested negative in either assay were aligned and compared to the sequences of primers and probes. The R14 assay did not detect strains belonging to the Arctic-like viruses from India, Pakistan, and Afghanistan (23). Also, two Nigerian RABVs belonging to the lineage Africa 2 were not detected with R14. Sequence comparison showed that Arctic-like sequences had up to three nucleotide differences compared to the sequence of the reverse primer RABV-N_R and also three nucleotide differences from the probe (Fig. 4b). For the same isolates, the primer and probe binding regions for the R13 assays revealed only one mismatch in the probe binding region (Fig. 4a).
FIG. 4.
Details of binding sites for two isolates, 13250 (Chile; GenBank accession no. HQ116829) and 20277 (Afghanistan; GenBank accession no. HQ116830), that were not detected by the R13 assay (a) or by the R14 assay (b). Differences are indicated. The sequence of SAD B19 (M31046) was included for better visualization.
An isolate from Chile (13250) that did not yield a CT value with the R13 assay had two mismatches in the probe binding region (Fig. 4a), while the nucleotide sequence of the same isolate only has one mismatch within the forward primer of the R14 assay (Fig. 4b).
DISCUSSION
In this study, we developed and validated a specific and sensitive real-time RT-PCR for the detection of RABV, compared it to a published assay (37), and improved the performance of the assays by the combination of both primer and probe sets as well as by the introduction of labeled positive controls (LPC).
Generally, the use of fluorogenic probes as detection systems for PCRs have improved the analytical sensitivity due to very short amplification products, which result in increased specificity and the prevention of cross-contaminations through the absence of post-PCR handling. The rapidity of the test, with results obtained in real time by obviating gel electrophoresis, offers the possibility to provide timely information to the relevant authorities.
As the R14 diagnostic assay can detect less than 10 genome copies/well, it represents a valuable tool for intra vitam diagnosis. A further advantage of real-time PCR techniques is the possibility to quantify viral RNA in real time, giving a relatively quick and reliable method for measuring the levels of viral RNA. Since the assay has proven reproducibility (Fig. 3), it is suitable for experimental infection studies with RABV investigating the spread of the virus and its accumulation in various tissues.
Both investigated assays had limits to detect some of the RABVs in the panel. Specifically, the newly developed RT-PCR (R14) did not detect RABVs of the Arctic-like lineage from Asia and viruses from Nigeria (Africa 2) (Table 2). It was demonstrated before that the number of sequence mismatches between primer and probe sets and target sequences of rabies viruses significantly affects amplification and detection (20, 37, 25). In this case, sequence analysis showed that both probe and reverse primer sequences were different from the sequences of this lineage (Fig. 4). In contrast, the R13 assay had only one nucleotide difference from the Arctic-like viruses and hence resulted in good performance. The opposite is the case for RABVs from Chile. These samples exemplify that the assays complement each other in combination, which could be shown practically with the combined R13/14 assay. Overall, all FAT-positive samples were confirmed as positive using the combined real-time RT-PCR assay. During the development and validation of the combined assay, an RABV and sequence panel with wide geographical and phylogenetic variation were used. Also, with the previously published R13 RT-PCR being revalidated with a different virus panel, in total more than 130 virus isolates were successfully tested. In conclusion, it appears likely that this combined assay will detect RABV strains occurring worldwide.
Generally, a combination of two assays targeting different locations on the genome will limit the chance of false negatives tremendously. Furthermore, by choosing a similar fragment size and similar specifications for primers and probes (Table 1), a combination as multiplex or parallel assays gives diagnosticians many options for implementation on various PCR machines.
The approach of a combined version of primer and probe sets to overcome the diversity among RABVs previously has been applied to improve intra vitam human rabies diagnosis (25). Using a collection of representatives of the world-wide diversity of RABV, all three primer-probe sets were shown to detect a wide range of RABV strains with very few detection failures (25). In fact, the most successful primer-probe combination was a slightly modified version of the Wakeley protocol for RABV (25), indicating that assays can and should be improved and revalidated when more sequence data are available.
The diversity of RABV is important, especially for reference laboratories for rabies. International travel leads to possibilities for people to have contact with various RABV variants (12). Also, companion animals often accompany passengers, thus there is a need for reference laboratories to provide fast and reliable results for subsequent decision-making by public and veterinary health authorities. For this purpose, sensitive, specific-but-broad-spectrum assays such as the one described here should be applied. Other published real-time RT-PCR assays have been validated using small numbers of RABV strains only (29, 20, 27, 36), thus their use for international reference laboratories needs additional evaluation.
A further advantage of RT-PCR is that RABV RNA can be detected in a range of biological fluids and samples (e.g., saliva, cerebrospinal fluid, tears, and skin biopsy samples). Generally, RT-PCR has been reported to confirm rabies diagnosis intra vitam in suspect human cases, when conventional diagnostic methods have failed and postmortem material is not available (11). Owing to the intermittent shedding of virus, negative results should not be used to exclude a diagnosis of rabies. However, diagnostic sensitivity is of eminent importance for intra vitam diagnosis. Because of the high sensitivity of the real-time PCR described here, this technique offers a potential use in this diagnostic scheme, as demonstrated earlier with a SYBR green real-time PCR assay (26).
At present, no recommended standard protocol for rabies diagnosis using RT-PCR has been published by the OIE or WHO. Since both generic approaches as well as specific PCR assays targeting lyssavirus genotypes or even variants have advantages and limitations, a cascade-type diagnostic procedure for rabies PCRs would be preferable, as described for the diagnosis of other viral diseases (17). A pan-lyssavirus or even a pan-rhabdovirus PCR assay could be combined with more-specific and -sensitive genotype-specific real-time PCRs or even variant specific real-time PCRs to confirm each other, allow genotyping, and obtain epidemiologically relevant information in real time. In this respect each single assay must be validated.
Besides the fully validated target assays, the functionality and robustness of molecular genetic tests can be increased by the application of innovative control systems. Internal process controls (e.g., beta-actin gene) check the efficiency of total RNA extraction as well as the inhibition of free amplification (37, 34). The disadvantage of such housekeeping gene systems is the inconsistent RNA load in different sample materials. Thus, a reduced sensitivity of the target assay based on partial inhibition is unlikely to be identified. In such cases the amplification of an external control using a defined amount of heterologous in vitro-transcribed RNA (18) can be helpful. The newly designed R14 assay was successfully combined with two published control assays (37, 18), and the combined systems can be used routinely. Furthermore, the R14 system seems sufficiently robust for the implementation of alternative internal control systems generally used in the different laboratories (e.g., commercial IC).
An important advantage of the real-time RT-PCR technology is the minimizing of cross-contamination based on the amplicon detection without opening the lid of the reaction tube. Nevertheless, the accidental removal of the lid or seal can be responsible for the spreading of amplicons. Furthermore, during pipetting, handling positive controls or standards is necessary. Therefore, an easy way to generate a labeled positive control without any cloning steps was developed. Based on a synthetic oligonucleotide, including the primers and probe for the target assay and a T7 promoter site, extensive quantities of in vitro-transcribed RNA was produced. To tag the in vitro transcript, an additional probe binding site was introduced into the LPC. The tag-specific probe integrated in the master mix can hybridize only with the LPC, and only then can the tag-specific fluorophore be detected during amplification. A similar approach has been described by Snow et al. (30). In general these authors used the same strategy, but they cloned the tagged positive control prior to in vitro transcription. Here, a simplified protocol for the more-rapid generation of LPC without a cloning procedure is presented. This simple protocol should support the construction and application of LPC for increasing the diagnostic safety of molecular rabies diagnostics. In general, the implementation of LPC can play an important role for the further harmonization and standardization of real-time PCR assays.
The lack of standardization, quality issues like contamination or false-negative results, and the varying reliability of PCR results in many laboratories, especially in developing countries, have been obstacles to the general use of PCR for rabies diagnosis. Therefore, the WHO does not currently recommend this technique for the routine postmortem diagnosis of rabies (40). However, with the accreditation of quality control measures being implemented in a growing number of laboratories worldwide, recommended PCR methods may become available. Such quality controls for diagnostic rabies PCRs should encompass several measures, e.g., the inclusion of appropriate positive (LPC), negative, and inhibition controls in assay runs. The consistency and the interassay reproducibility also should be ensured over time by monitoring the performance of the assay rigorously. If PCR is applied for epidemiological surveys, positive results should be confirmed by conventional virological techniques. In most national and international legislatures, only the detection of virus or viral antigen leads to the confirmation of a rabies case and thus to subsequent control measures. Also, the isolation of viable virus provides further possibilities to study the characteristics of this particular strain.
Only if laboratories meet the required standard (40) can the PCR and, especially, real-time PCR offer its full potential as a confirmatory diagnostic test, especially in decomposed or intra vitam samples.
Supplementary Material
Acknowledgments
We thank Christian Korthase and Jeanette Kliemt at the Friedrich-Loeffler-Institute for excellent technical assistance.
This work was partially supported by EPIZONE, the EU Network of Excellence for Epizootic Disease Diagnosis and Control (FOOD-CT-2006-016236), under work package 4.1, designated real-time PCR diagnostics, and was partially funded from the project European Management Platform for Emerging and Re-emerging Infectious Disease (EMPERIE) (EC grant agreement 223.498).
Footnotes
Published ahead of print on 25 August 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Araújo, D. B., H. Langoni, M. F. Almeida, and J. Megid. 2008. Heminested reverse-transcriptase polymerase chain reaction (hnRT-PCR) as a tool for rabies virus detection in stored and decomposed samples. BMC Res. Notes 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belák, S., and P. Thoren. 2001. Molecular diagnosis of animal diseases: some experiences over the past decade. Expert. Rev. Mol. Diagn. 1:434-443. [DOI] [PubMed] [Google Scholar]
- 3.Biswal, M., R. Ratho, and B. Mishra. 2007. Usefulness of reverse transcriptase-polymerase chain reaction for detection of rabies RNA in archival samples. Jpn. J. Infect. Dis. 60:298-299. [PubMed] [Google Scholar]
- 4.Black, E. M., J. P. Lowings, J. Smith, P. R. Heaton, and L. M. McElhinney. 2002. A rapid RT-PCR method to differentiate six established genotypes of rabies and rabies-related viruses using TaqMan technology. J. Virol. Methods 105:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, E. M., L. M. McElhinney, J. P. Lowings, J. Smith, P. Johnstone, and P. R. Heaton. 2000. Molecular methods to distinguish between classical rabies and the rabies-related European bat lyssaviruses. J. Virol. Methods 87:123-131. [DOI] [PubMed] [Google Scholar]
- 6.Crepin, P., L. Audry, Y. Rotivel, A. Gacoin, C. Caroff, and H. Bourhy. 1998. Intravitam diagnosis of human rabies by PCR using saliva and cerebrospinal fluid. J. Clin. Microbiol. 36:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David, D., B. Yakobson, D. Rotenberg, N. Dveres, I. Davidson, and Y. Stram. 2002. Rabies virus detection by RT-PCR in decomposed naturally infected brains. Vet. Microbiol. 87:111-118. [DOI] [PubMed] [Google Scholar]
- 8.Dean, D. J., M. K. Abelseth, and P. Athanasiu. 1996. The fluorescence antibody test, p. 88-93. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies. World Health Organization, Geneva, Switzerland.
- 9.Echevarría, J. E., A. Avellon, J. Juste, M. Vera, and C. Ibanez. 2001. Screening of active lyssavirus infection in wild bat populations by viral RNA detection on oropharyngeal swabs. J. Clin. Microbiol. 39:3678-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finnegan, C. J., S. M. Brookes, and A. R. Fooks. 2004. Detection of European bat lyssavirus mRNA in mouse brain by employing in situ hybridisation. J. Virol. Methods 121:221-227. [DOI] [PubMed] [Google Scholar]
- 11.Fooks, A. R., N. Johnson, C. M. Freuling, P. Wakeley, A. Banyard, L. M. McElhinney, D. A. Marston, A. Dastjerdi, E. Wright, R. Weiss, and T. Müller. 2009. Emerging technologies for the detection of rabies virus: challenges and hopes in the 21st century. PLoS Neglect. Trop. D 3:e530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fooks, A. R., N. Johnson, S. M. Brookes, G. Parsons, and L. M. McElhinney. 2003. Risk factors associated with travel to rabies endemic countries. J. Appl. Microbiol. 94:31-36. [DOI] [PubMed] [Google Scholar]
- 13.Foord, A. J., H. Heine, L. I. Pritchard, R. A. Lunt, K. M. Newberry, C. L. Rootes, and B. D. Boyle. 2006. Molecular diagnosis of lyssaviruses and sequence comparison of Australian bat lyssavirus samples. Aust. Vet. J. 84:225-230. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 15.Heaton, P. R., P. Johnstone, L. M. McElhinney, R. Cowley, E. O'Sullivan, and J. E. Whitby. 1997. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. J. Clin. Microbiol. 35:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, B., T. Harder, E. Starick, K. Depner, O. Werner, and M. Beer. 2007. Rapid and highly sensitive pathotyping of avian influenza A H5N1 virus by using real-time reverse transcription-PCR. J. Clin. Microbiol. 45:600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann, B., K. Depner, H. Schirrmeier, and M. Beer. 2006. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 136:200-209. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, G. J., I. V. Kuzmin, A. Schmitz, J. Blanton, J. Manangan, S. Murphy, and C. E. Rupprecht. 2006. Experimental infection of big brown bats (Eptesicus fuscus) with Eurasian bat lyssaviruses Aravan, Khujand, and Irkut virus. Arch. Virol. 151:2021-2035. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, G. J., J. S. Smith, C. A. Hanlon, and C. E. Rupprecht. 2004. Evaluation of a TaqMan PCR assay to detect rabies virus RNA: influence of sequence variation and application to quantification of viral loads. J. Clin. Microbiol. 42:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, N., L. M. McElhinney, J. Smith, P. Lowings, and A. R. Fooks. 2002. Phylogenetic comparison of the genus Lyssavirus using distal coding sequences of the glycoprotein and nucleoprotein genes. Arch. Virol. 147:2111-2123. [DOI] [PubMed] [Google Scholar]
- 22.Kulonen, K., M. Fekadu, S. Whitfield, and C. K. Warner. 1999. An evaluation of immunofluorescence and PCR methods for detection of rabies in archival Carnoy-fixed, paraffin-embedded brain tissue. Zentralbl. Veterinarmed. B 46:151-155. [DOI] [PubMed] [Google Scholar]
- 23.Kuzmin, I. V., G. J. Hughes, A. D. Botvinkin, S. G. Gribencha, and C. E. Rupprecht. 2008. Arctic and Arctic-like rabies viruses: distribution, phylogeny and evolutionary history. Epidemiol. Infect. 136:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzmin, I. V., and C. E. Rupprecht. 2007. Bat rabies, p. 259-307. In A. C. Jackson and W. Wunner. (ed.), Rabies. Academic Press, New York, NY.
- 25.Nadin-Davis, S. A., M. Sheen, and A. I. Wandeler. 2009. Development of real-time reverse transcriptase polymerase chain reaction methods for human rabies diagnosis. J. Med. Virol. 81:1484-1497. [DOI] [PubMed] [Google Scholar]
- 26.Nagaraj, T., J. P. Vasanth, A. Desai, A. Kamat, S. N. Madhusudana, and V. Ravi. 2006. Ante mortem diagnosis of human rabies using saliva samples: comparison of real time and conventional RT-PCR techniques. J. Clin. Virol. 36:17-23. [DOI] [PubMed] [Google Scholar]
- 27.Orlowska, A., M. Smreczak, P. Trêbas, and J. F. Zmudzinski. 2008. Comparison of realtime PCR and heminested RT-PCR methods in the detection of rabies virus infection in bats and terrestrial animals. Bull. Vet. Inst. Pulawy. 52:313-318. [Google Scholar]
- 28.Sacramento, D., H. Bourhy, and N. Tordo. 1991. PCR technique as an alternative method for diagnosis and molecular epidemiology of rabies virus. Mol. Cell Probes 5:229-240. [DOI] [PubMed] [Google Scholar]
- 29.Shankar, V., R. A. Bowen, A. D. Davis, C. E. Rupprecht, and T. J. O'Shea. 2004. Rabies in a captive colony of big brown bats (Eptesicus fuscus). J. Wildl. Dis. 40:403-413. [DOI] [PubMed] [Google Scholar]
- 30.Snow, M., P. McKay, and I. Matejusova. 2009. Development of a widely applicable positive control strategy to support detection of infectious salmon anaemia virus (ISAV) using Taqman real-time PCR. J. Fish Dis. 32:151-156. [DOI] [PubMed] [Google Scholar]
- 31.Tordo, N., A. Benmansour, C. Calisher, R. G. Dietzgen, R. X. Fang, A. O. Jackson, G. Kurath, S. Nadin-Davis, R. B. Tesh, and P. J. Walker. 2004. Rhabdoviridae, p. 623-644. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy, VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 32.Tordo, N., D. Sacramento, and H. Bourhy. 1996. The polymerase chain reaction (PCR) technique for diagnosis, typing and epidemiological studies, p. 157-170. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. World Health Organization, Geneva, Switzerland.
- 33.Tordo, N., H. Bourhy, and D. Sacramento. 1995. PCR technology for lyssavirus diagnosis, p. 125-145. In J. P. Clewley (ed.), Polymerase chain reaction (PCR) for human viral diagnosis. CRC, Boca Raton, FL.
- 34.Toussaint, J. F., C. Sailleau, E. Breard, S. Zientara, and K. De Clercq. 2007. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J. Virol. Methods 140:115-123. [DOI] [PubMed] [Google Scholar]
- 35.Vázquez-Morón, S., A. Avellon, and J. E. Echevarria. 2006. RT-PCR for detection of all seven genotypes of Lyssavirus genus. J. Virol. Methods 135:281-287. [DOI] [PubMed] [Google Scholar]
- 36.Wacharapluesadee, S., J. Sutipanya, S. Damrongwatanapokin, P. Phumesin, P. Chamnanpood, C. Leowijuk, and T. Hemachudha. 2008. Development of a TaqMan real-time RT-PCR assay for the detection of rabies virus. J. Virol. Methods 151:317-320. [DOI] [PubMed] [Google Scholar]
- 37.Wakeley, P. R., N. Johnson, L. M. McElhinney, D. Marston, J. Sawyer, and A. R. Fooks. 2005. Development of a real-time, TaqMan reverse transcription-PCR assay for detection and differentiation of lyssavirus genotypes 1, 5, and 6. J. Clin. Microbiol. 43:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster, W. A., and G. A. Casey. 1996. Virus isolation in neuroblastoma cell culture, p. 93-104. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies. World Health Organization, Geneva, Switzerland.
- 39.Whitby, J. E., P. R. Heaton, E. M. Black, M. Wooldridge, L. M. McElhinney, and P. Johnstone. 2000. First isolation of a rabies-related virus from a Daubenton's bat in the United Kingdom. Vet. Rec. 147:385-388. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organisation. 2005. Expert consultation on rabies, first report. World Health Organ. Tech. Rep. Ser. 931:1-121. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.