Abstract
Members of the genus Borrelia are among the most common infectious agents causing tick-borne disease in humans worldwide. Here, we developed a Light Upon eXtension (LUX) real-time PCR assay that can detect and quantify Borrelia species in ticks that have fed on humans, and we applied the assay to 399 such ticks. Borrelia PCR-positive ticks were identified to species level by sequencing the products of conventional PCR performed using Borrelia group-specific primers. There was a 19% prevalence of Borrelia spp. in the detached ticks, and the number of spirochetes per Borrelia PCR-positive tick ranged from 2.0 × 102 to 4.9 × 105, with a median of 7.8 × 103 spirochetes. Adult ticks had a significantly larger number of spirochetes, with a median of 8.4 × 104 compared to the median of nymphs of 4.4 × 104. Adult ticks also exhibited a higher prevalence of Borrelia (33%) than nymphs (14%). Among the identified species, Borrelia afzelii was found to predominate (61%) and was followed by B. garinii (23%), B. valaisiana (13%), B. burgdorferi sensu stricto (1%), B. lusitaniae (1%), and B. miyamotoi-like (1%). Also, 3% of the ticks were coinfected with multiple strains of B. afzelii. Notably, this is the first report of B. lusitaniae being detected in ticks in Sweden. Our LUX real-time PCR assay proved to be more sensitive than a corresponding TaqMan assay. In conclusion, the novel LUX real-time PCR method is a rapid and sensitive tool for detection and quantification of Borrelia spp. in ticks.
Lyme borreliosis (LB) is the most common tick-borne disease in humans in Europe (26), and it is caused by spirochetes belonging to the Borrelia burgdorferi sensu lato complex. That group comprises the species B. burgdorferi sensu stricto, B. afzelii, and B. garinii, which are usually transmitted by the vector Ixodes ricinus. Furthermore, there have been reports of B. valaisiana, B. lusitaniae, and B. spielmanii being detected in samples of human skin and cerebrospinal fluid (5, 7, 30), which suggests that those three species can also give rise to LB. It is often hard to distinguish the clinical symptoms of LB from those of other diseases (10), and hence, it can be difficult to establish a correct diagnosis, especially if the patient is unable to recall having a tick bite.
Today, diagnosis is based mainly on serological tests, although some PCR-based approaches, such as the TaqMan real-time PCR assay (3, 12), have been developed to detect Borrelia species in clinical samples. Even if real-time PCR is not yet considered to be a routine method in clinical practice, it can nonetheless provide valuable information about Borrelia infections, with regard to species type and the number of spirochetes present. Additional major advantages of PCR in this context are its simplicity, sensitivity, robustness, and speed. Other assays besides the TaqMan assay include a method based on SYBR green dye chemistry (37) and another using Light Upon eXtension (LUX) (Invitrogen Corporation). Compared to the SYBR green real-time PCR assay, the LUX assay offers the benefit of using a self-quenched primer with a hairpin loop structure, which makes it more specific; that is, it entails less unspecific binding and primer-dimer formation. Furthermore, the fluorophore is attached to the hairpin loop in the LUX setup, and thus, in contrast to the TaqMan assay, this PCR technique does not need an internal probe and is therefore a better choice if broader specificity is required. The LUX assay also has the capacity for melting curve analysis, which offers the possibility of discriminating between PCR products with different base pair compositions (23) and thereby revealing false-positive samples.
Ixodes ricinus has been found in 23 of the 25 provinces in Sweden (9), but it is most common in the southern and central parts of the country and along the northeastern coast (14). Various investigators have described the prevalence and diversity of Borrelia in ticks collected in the field in Sweden (4, 8, 9, 14), and to date, five species of these bacteria have been recorded: B. afzelii, B. garinii, B. valaisiana, B. burgdorferi sensu stricto, and also one that is closely related to B. miyamotoi, which is known to be associated with relapsing fever. According to the cited studies, the prevalence of Borrelia spp. in Sweden varies between 3% and 23%. However, detection was not achieved by real-time PCR in those investigations, and thus, no attempts were made to quantify the Borrelia spirochetes in the ticks. To our knowledge, no quantification of Borrelia spirochetes in ticks detached from humans has ever been performed.
Our aim was to study the prevalence of Borrelia and to quantify Borrelia cells in ticks that had fed on humans, and we developed a LUX real-time PCR assay for that purpose. In addition, we examined possible geographical differences in prevalence, and we also studied the temporal and spatial distribution of Borrelia species.
MATERIALS AND METHODS
Study sites and collection of ticks.
The ticks analyzed in the present study were also used in an investigation focused on the clinical outcome in the humans involved (L. Fryland, P. Wilhelmsson, P.-E. Lindgren, D. Nyman, C. Ekerfelt, and P. Forsberg, submitted for publication), and more detailed information about collection of the specimens is to be published by the group conducting the latter investigation. In short, we used a total of 399 ticks that had been attached to humans in nine areas in Östergötland County, Sweden, between June 2007 and January 2008. The specimens were obtained from eight primary health care centers (PHCs) located in the towns/communities of Ekholmen, Johannelund, Linghem, Kisa, Skärblacka, Söderköping, Valdemarsvik, and Åtvidaberg, and some were also acquired from the Department of Infectious Diseases at Linköping University Hospital. The ticks were available at those facilities because people had been asked to bring detached ticks to their local PHCs. The subjects also completed a health questionnaire and provided a blood sample during the initial visit made to donate the ticks. A second blood sample was obtained 3 months later, and both samples were analyzed for anti-Borrelia antibodies to determine seroconversion or increase in antibody titer. The ticks that people provided were kept in plastic tubes at room temperature and were transported to the Division of Medical Microbiology, Linköping University, within 3 days. They were photographed to determine species type and developmental stage, based on size and color of the dorsal shield. This study was approved by the Ethics Committee of the Faculty of Medicine, Linköping University (no. M132-06).
DNA extraction from ticks.
The ticks were washed in 70% ethanol and then in phosphate-buffered saline (PBS), and they were subsequently sectioned longitudinally into two equal parts using a sterile scalpel. One half of each tick was subjected to DNA extraction using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany) and the supplementary protocol designated “purification of total DNA from ticks,” according to the manufacturer's instructions, which gave 50 μl of DNA in the supplied elution buffer. The DNA concentration in each sample was determined using a spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific, Wilmington, DE). The extracted DNA was stored at −20°C pending further analysis.
Reference bacterial strains and samples used to develop the real-time PCR assay.
A panel comprising DNA from 16 bacterial species, three human blood samples, and one human skin surface sample was used to develop a real-time PCR assay as described below. DNA from one strain of each of the following reference Borrelia species served as positive controls: B. burgdorferi sensu stricto B31 ATCC 35210, B. afzelii ACA-1 (2), B. garinii IP90 (18), B. valaisiana VS116 (35), B. japonica H014 (16), B. duttonii 1120 (obtained from the strain collection of Guy Baranton, Institut Pasteur), B. hispanica CR1 (obtained from the strain collection of Guy Baranton, Institut Pasteur), B. persica (obtained from the strain collection of Eduard Korenberg, Gamaleya Research Institute, Moscow, Russia), B. coriaceae (obtained from the strain collection of Alan G. Barbour, UC Irvine), B. anserina (obtained from the strain collection of Alan G. Barbour, UC Irvine), and B. turicatae (obtained from the strain collection of Alan G. Barbour, UC Irvine). These strains were cultivated for 12 days at 35°C in 8 ml of Barbour-Stoenner-Kelly (BSK) medium supplemented with 9% rabbit serum (Sigma-Aldrich Sweden, Stockholm, Sweden) and then harvested by centrifugation at 8,000 × g for 10 min at 20°C. DNA was extracted from the bacterial pellets using a DNeasy blood and tissue kit (Qiagen) according to the instructions of the manufacturer. DNA from one strain of each of the five bacterial species that can be members of human skin flora (i.e., Escherichia coli C-1467, Staphylococcus aureus ATCC 3359, Staphylococcus epidermidis CCUG 21989, Streptococcus pyogenes CCUG 33061, and Propionibacterium acnes CCUG 1794) was used as a negative control. These strains were cultivated on blood agar plates at 37°C overnight. One colony of each strain was then transferred to LB medium and incubated overnight at 37°C, after which bacterial DNA was isolated using a DNeasy blood and tissue kit (Qiagen) according to the manufacturer's protocol. DNA from human blood and skin surface samples was used as additional negative controls. The human blood and skin surface samples were collected from staff at the Division of Medical Microbiology, Linköping University. A skin surface sample was taken by gently scraping a scalpel on the lower arm. An aliquot (200 μl) of each blood sample and 5 mg of the skin surface sample were used for DNA extraction, which was also done with the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's protocol.
Design of Borrelia primers for real-time PCR and for conventional PCR.
All sequences of the 16S rRNA gene available for different strains of Borrelia spp. in GenBank were obtained from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov), and these sequences were aligned using BioEdit software (Tom Hall, Ibis Therapeutics, Carlsbad, CA). The forward primer B16S_FL, 5′-GAC TCG TCA AGA CTG ACG CTA AGT C-3′, and reverse primer B16S_R, 5′-GCA CAC TTA ACA CGT TAG CTT CGG TAC TAA C-3′, were designed to target a conserved, 131-bp-long Borrelia-specific region of the 16S rRNA gene (bases in bold at the 5′ end of the B16S_FL primer correspond to additional bases added to create the hairpin loop structure). According to the BLAST search (1), the designed primers matched 100% with the sequences of strains of the following species: B. burgdorferi sensu stricto, B. garinii, B. afzelii, B. valaisiana, B. lusitaniae, B. spielmanii, B. andersonii, B. hispanica, B. miyamotoi, B. turdi, B. parkeri, B. crocidurae, B. tanukii, B. duttonii, B. hermsii, B. theileri, B. persica, B. anserina, B. turicatae, B. turcica, B. japonica, B. coriaceae, B. recurrentis, and B. lonestari. The LUX primer pair was designed and evaluated using OligoAnalyzer 3.0 (Integrated DNA Technologies, Coralville, IA). The forward primer B16S_FL was labeled with the reporter dye 6-carboxyfluorescein (FAM) at the first thymine base from the 3′ end (Invitrogen Corporation, Paisley, United Kingdom).
PCR primers for species identification were based on the 5S-23S rRNA intergenic spacer (IGS). We used the same set of primers as that reported by Postic et al. (28): 5′-CTG CGA GTT CGC GGG AGA-3′ and 5′-TCC TAG GCA TTC ACC ATA-3′, which amplify a genetically diverse region within the IGS in a conventional PCR assay. To increase the sensitivity of the assay, we applied a nested PCR approach with an additional set of primers designed to target the PCR product obtained from the first amplification: B5S-23S_Fn, 5′-GAG TTC GCG GGA GAG TAA G-3′, and B5S-23S_Rn, 5′-TAG GCA TTC ACC ATA GAC TCT T-3′. According to the BLAST search, the designed primers for the 5S-23S IGS matched 100% with sequences of strains belonging to the following Borrelia species, all of which are present in Europe (26, 29): B. burgdorferi sensu stricto, B. afzelii, B. garinii, B. valaisiana, B. spielmanii, and B. lusitaniae. However, it is not known whether the primers targeting the 5S-23S rRNA IGS can detect the B. miyamotoi-like species, which has previously been found in Sweden (8). All tick samples positive for Borrelia in the LUX real-time PCR assay, which failed to produce PCR products with primers targeting the 5S-23S IGS, were instead analyzed with primers targeting the 16S-23S IGS (4): F, 5′-GTA TGT TTA GTG AGG GGG GTG-3′; R, 5′-GGA TCA TAG CTC AGG TGG TTA G-3′; Fn, 5′-AGG GGG GTG AAG TCG TAA CAA G-3′; and Rn, 5′-GTC TGA TAA ACC TGA GGT CGG A-3′. These primers were employed to detect the B. miyamotoi-like spirochete (4), and, according to the BLAST search, they matched 100% with sequences belonging to the following species: B. miyamotoi, B. burgdorferi sensu stricto, B. afzelii, B. garinii, B. recurrentis, B. duttonii, B. turicatae, B. hermsii, and B. japonica.
Optimization of primers designed for detection and quantification of Borrelia spp. by conventional PCR.
Optimization and evaluation of assay specificity were performed using the designed primers B16S_FL (without a fluorophore) and B16S_R in a conventional PCR assay with DNA templates from the reference panel, as described above. Different annealing temperatures (55 to 60°C) were tested. The reaction mixture in the optimized assay (final volume of 50 μl) contained 5 μl of 10× PCR buffer (Amersham Biosciences, Uppsala, Sweden), 1 μl of deoxynucleoside triphosphate (dNTP) (10 mM), 1 μl of each primer (10 μM), 0.25 μl of Taq DNA polymerase (5 U μl−1; Amersham Biosciences), 5 μl of template DNA (2 to 4 ng μl−1), and 36.75 μl of RNase-free water. The amplification program comprised 95°C for 2 min, followed by 95°C for 15 s, 58°C for 30 s, 72°C for 30 s in 40 cycles, and finally 72°C for 7 min. The reactions were performed in a PTC-100TM programmable thermal controller (M. J. Research, Inc., Waltham, MA), and products were analyzed by agarose gel electrophoresis.
Conventional PCR assays used for species identification.
A nested PCR assay was performed to amplify the 5S-23S rRNA IGS for species identification. Specificity of the assay was determined using the same reference panel as that employed to develop the LUX assay described above. The reaction mixture (final volume of 50 μl) contained the following: 5 μl of 10× PCR buffer (Amersham Biosciences), 1 μl of dNTP (10 mM), 1 μl each of the primers targeting the 5S-23S IGS (28) (10 μM), 0.38 μl of high-fidelity polymerase (3.5 U μl−1; Amersham Biosciences), 5 μl of template DNA (2 to 4 ng μl−1), and 36.62 μl of RNase-free water. The amplification program comprised 95°C for 5 min, followed by 95°C for 15 s, 57°C for 30 s, 39 cycles of 72°C for 30 s, and finally 72°C for 7 min. An aliquot (5 μl) of the PCR product obtained in this assay was added to the second PCR mixture, which was prepared using the same volumes, concentrations, and amplification program as those used for the first mixture, except with a different primer pair (B5S-23S_Fn and B5S-23S_Rn), and the number of cycles was increased to 42. The nested PCR assay used to amplify the 16S-23S rRNA IGS for B. miyamotoi-like identification was performed as described by Bunikis et al. (4).
All reactions were conducted in a PTC-100TM programmable thermal controller (M. J. Research, Inc., Waltham, MA), and PCR products were analyzed by agarose gel electrophoresis.
LUX real-time PCR assay.
Each PCR amplification was carried out in a 96-well reaction plate (Applied Biosystems, Warrington, United Kingdom), using a 20-μl aliquot of reaction mixture containing the following: 10 μl of Platinum qPCR SuperMix uracil-d-glycosylase (UDG) (Invitrogen), 0.04 μl of Rox reference (Invitrogen), 0.4 μl of LUX B16S_FL primer (10 μM), 0.4 μl of B16S_R primer (10 μM; Invitrogen), 4.16 μl RNase-free water, and 5 μl of template DNA. Thereafter, the plate was centrifuged at 900 × g for 5 min.
The reactions were performed on an ABI PRISM 7500 fast real-time PCR system (Applied Biosystems). The reaction mixture was preheated at 50°C for 2 min (carry-over prevention step, activation of the enzyme UDG) and 95°C for 2 min (denaturation of UDG and activation of Platinum Taq DNA polymerase) and then subjected to 45 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s. Immediately after completion of PCR, melting curve analyses were performed by heating to 95°C for 15 s, followed by cooling to 60°C for 1 min, and subsequent heating to 95°C at 0.8°C min−1 with continuous fluorescence recording. The real-time PCR and melting curve results were analyzed using Sequence Detection Software version 1.3.1 (Applied Biosystems).
Determination of the detection/quantification limit and efficiency of the LUX real-time PCR assay.
A 10-fold serial dilution ranging from 101 to 107 gene copies of the B. burgdorferi B31 was used as a standard to determine the detection limit and efficiency of the real-time PCR assay. The gene copy numbers were calculated by converting the concentration of total double-stranded DNA (dsDNA) of B. burgdorferi B31 (measured spectrophotometrically) to the number of genome copies based on the molecular weight of the genome. According to Ornstein and Barbour (25), there is a mean of 10 genomes per B. burgdorferi B31 cell and one 16S rRNA gene copy per genome. Therefore, calculations included the assumption that all Borrelia species have 10 16S rRNA gene copies per cell.
To check for possible inhibition, we used five ticks in different developmental stages (i.e., five larvae, five nymphs, and five adults), which were collected in the field and kindly provided by Jan Landin, Department of Physics, Chemistry and Biology, Linköping University. These specimens were processed as described above. B. burgdorferi B31 cells cultivated in BSK medium were washed with PBS and counted in a phase-contrast microscope. One half of each tick was spiked with a known number of B. burgdorferi B31 cells, and the other half served as a negative control for Borrelia. A serial dilution was prepared in PBS to represent Borrelia concentrations of 104, 103, 102, 101, and 100 spirochetes per tick sample and then incubated for 30 min at 37°C. The tick samples were subsequently used for DNA extraction as described above. Real-time PCR amplification was performed using LUX primer pairs targeting the 16S rRNA gene to verify the efficiency and the quantification limit of the assay.
Validation of the LUX real-time PCR assay.
Considering the aspects of sensitivity and specificity, we compared the LUX assay with the TaqMan-based real-time PCR method reported by Ornstein and Barbour (25). The primer pair and probe in the TaqMan assay are designed to target the same 136-bp region of the 16S rRNA gene as the primer pair designed for the LUX assay. We also applied an internal control to all extracted tick samples to check for PCR inhibition and thereby prevent false-negative results. A modified real-time PCR assay for the mitochondrial tick housekeeping gene 16S Ixodes DNA was run as previously described by Schwaiger and Cassinotti (32). The same primers (F-16sIxodes and R-16sIxodes), but no TaqMan probe, were employed in a SYBR green assay. The PCR amplification was carried out in 96-well reaction plates (Applied Biosystems), and the reaction mixture (20 μl) contained 10 μl of FastStart Universal SYBR green Master (ROX) (Roche, Mannheim, Germany), 0.4 μl of each primer (10 μM) (Sigma-Aldrich Sweden AB, Stockholm, Sweden), 7.2 μl of RNase-free water, and 2 μl of template DNA. The reactions were performed in an ABI Prism 7500 fast real-time PCR system (Applied Biosystems) using the same reaction conditions as those described by Schwaiger and Cassinotti (32). Melting analyses of all reactions were performed as reported above.
Nucleotide sequencing of the PCR products and species identification by sequence analysis.
Macrogen, Inc. (Seoul, South Korea), performed nucleotide sequencing of the PCR products that we obtained from primers targeting the following: the 16S rRNA gene, 5S-23S rRNA IGS, 16S-23S rRNA IGS, and the 16S Ixodes DNA. The sequencing reactions were based on BigDye chemistry.
In addition to the 5S-23S IGS sequences acquired in this study, IGS sequences from Borrelia spp. in GenBank were used in the phylogenetic analysis (n = 41). By including a representative selection of IGS sequences from Borrelia spp. that are common in and around Europe (i.e., B. afzelii, B. garinii, B. valaisiana, B. burgdorferi sensu stricto, and B. lusitaniae), we were able to identify the species found in our investigation. Sequence alignment was performed using Clustal W2 (European Bioinformatics Institute, Cambridge, United Kingdom). Phylogenetic analyses were conducted using MEGA version 4 (17, 31), and the phylogenetic tree was constructed by applying neighbor-joining and Kimura-2-parameter methods with pairwise deletion. The significance of the relationship was ascertained by bootstrap analysis (500 replicates).
Cloning of PCR products of 5S-23S IGS from ticks carrying more than one Borrelia strain.
PCR products containing different Borrelia sequences, determined as dual peaks in the sequences analysis, were separated by cloning of the PCR products from amplification of the 5S-23S IGS. The PCR products were purified using a GeneJET PCR purification kit (Fermentas, Glen Burnie, MD) according to the manufacturer's protocol. For bacterial transformation and cloning procedures, a TransformAid bacterial transformation kit and a CloneJET PCR cloning kit (both from Fermentas) were used as stipulated in the protocols provided by the manufacturer. DNA was extracted from transformants with plasmids containing PCR products as inserts and purified using a GeneJET plasmid miniprep kit (Fermentas) according to the manufacturer's instructions. Sequencing of the inserted PCR products was performed by Macrogen, Inc. (Seoul, South Korea).
Statistical analysis.
Fisher's exact test and chi-square test were applied to compare the distribution of Borrelia PCR-positive ticks and Borrelia species among the different PHCs (i.e., geographic sampling locations). The Mann-Whitney test was used to compare adults and nymphs, as well as different months, regarding the number of Borrelia spirochetes per tick. Median values and 95% confidence intervals were determined. Statistical analyses were performed, and graphs were drawn using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). All P values of <0.05 were considered significant.
Nucleotide sequence accession number.
Sequences obtained in this investigation have been deposited in GenBank with accession numbers HM173532 to HM173598.
RESULTS
Development of a broad-range and sensitive LUX real-time PCR assay.
We designed a LUX real-time PCR primer pair to target a highly conserved 131-bp region of the 16S rRNA gene. Without a fluorophore and at a primer annealing temperature of 58°C, this pair could detect all of the tested Borrelia reference strains, as indicated by conventional PCR followed by sequence analysis of the PCR products (data not shown). As expected, when analyzing the DNA samples that served as negative controls, human blood, human skin surface, E. coli, S. aureus, S. epidermidis, S. pyogenes, and P. acnes, no PCR products were detected.
Five independent LUX real-time PCR runs, including a 10-fold serial dilution of gene copies from B. burgdorferi B31, were performed, exhibiting a dynamic range in the interval of 101 to 107 gene copies per reaction. The slope as a mean of the standard curves was −3.64 ± 0.08 (r2 = 0.99). The melting temperature of the PCR products was 80.3 ± 0.2°C. Using the known copy numbers of reference DNA, the lower quantification limit was 101 gene copies per PCR. However, it was possible to detect, but not to accurately quantify, fewer than 101 gene copies. Ten gene copies of the 16S rRNA is equivalent to the number of copies that exists in one Borrelia cell (25). The lower quantification limit was similar in the PCR assay using the Borrelia spiked tick samples; thus, no inhibition of the PCR amplification was detected. The assay did not show any unspecific binding or primer-dimer formation. The primers targeting the 5S-23S IGS were able to amplify PCR products from all the Borrelia strains used as references.
Borrelia in every fifth tick detected with a novel, sensitive LUX real-time PCR assay.
All 399 ticks that we analyzed were identified as I. ricinus; 101 (25.3%) were adult females, 296 (74.2%) were nymphs, and two (0.5%) were in the larval stage (Table 1). The LUX-based real-time PCR assay showed the presence of Borrelia spp. in 75 ticks (19%; Table 1), whereas the TaqMan assay detected Borrelia in only 72 of those 75 and in no other ticks (data not shown). No obvious seasonal trend in the number of Borrelia-positive ticks was detected during the collection period.
TABLE 1.
Prevalence of Borrelia species in I. ricinus ticks that had been removed from humans and obtained at primary health care centers in Östergötland County, Sweden
| Stage | Total no. of ticks examined by the LUX assay | No. of Borrelia-positive ticks (%) | No. of ticks containing indicated Borrelia speciesa |
||||||
|---|---|---|---|---|---|---|---|---|---|
| B. afzelii | B. garinii | B. valaisiana | B. burgdorferi sensu stricto | B. lusitaniae | B. miyamotoi-like | UT | |||
| Nymph | 296 | 42 (14) | 27 | 6 | 2 | 1 | 6 | ||
| Adult | 101 | 33 (33) | 13* | 9 | 6 | 1 | 1 | 3 | |
| Larva | 2 | 0 (0) | |||||||
| Total | 399 | 75 (19) | 40 | 15 | 8 | 1 | 1 | 1 | 9 |
Abbreviations: UT, untypeable; *, includes the coinfected ticks.
The SYBR green real-time PCR assay detected the tick-specific extraction control 16S Ixodes DNA in all samples (data not shown). The threshold cycle (CT) value range for this DNA was 11 to 23 (median of 16) for adult ticks and 14 to 26 (median of 18) for nymph ticks.
Higher number of Borrelia cells in adults than in nymphs.
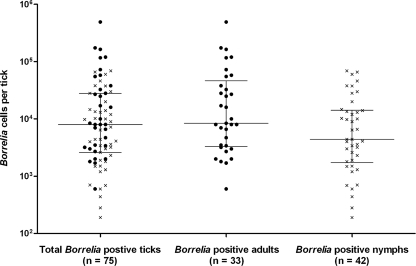
According to the LUX real-time PCR assay, the number of Borrelia cells per Borrelia PCR-positive tick ranged from 2.0 × 102 to 4.9 × 105 (Fig. 1), with a median of 7.8 × 103. The number of Borrelia cells in adults ranged from 6.0 × 102 to 4.9 × 105 and in nymphs from 2.0 × 102 to 7.0 × 104. The number of Borrelia cells was significantly higher in adult ticks than in nymphs (median per tick of 8.4 × 103 versus 4.4 × 103). Furthermore, the prevalence of Borrelia was greater in adult ticks than in nymphs, and no Borrelia was detected in larvae. The prevalence varied from 11% to 33% in the ticks obtained at the PHCs in Östergötland County (data not shown). Comparison of the PCR-positive ticks from all collection sites indicated that the prevalence of Borrelia was significantly lower in those from one PHC (Kisa) than in those from the other PHCs. However, no seasonal trend in Borrelia cell number was observed over the collection period. Moreover, no significant seasonal difference could be detected for the number of developmental stages of the ticks provided over the study period.
FIG. 1.
Total number of ticks, adult ticks (•), and nymphs (×) PCR positive for Borrelia plotted against the number of Borrelia cells per tick. Horizontal lines indicate the median, with upper and lower quartiles.
Six different Borrelia species detected in the detached ticks.
It was possible to determine the Borrelia species in 66 of the 75 ticks that were positive for Borrelia in the LUX real-time PCR (Table 1) using the primer pairs targeting the 5S-23S IGS and 16S-23S IGS regions. Six different species were recorded (Table 1), among which B. afzelii (n = 40) predominated and was followed by B. garinii (n = 15), B. valaisiana (n = 8), B. burgdorferi sensu stricto (n = 1), B. lusitaniae (n = 1), and B. miyamotoi-like (n = 1). Notably, B. lusitaniae was identified for the first time in Sweden. B. afzelii dominated at both the adult (39%) and the nymph (64%) stages. Considering the diversity of Borrelia species in relation to the developmental stages of the ticks, we found that B. afzelii occurred more often in nymphs than in adults, whereas the opposite pattern was observed for B. garinii. Three times more adult ticks (n = 6) than nymphs (n = 2) were positive for B. valaisiana. Two adult ticks were coinfected with different strains of B. afzelii (Table 1), and both of those specimens were obtained at the same PHC (Åtvidaberg). B. afzelii and B. garinii were also found in ticks from all of the collection sites.
Nine LUX real-time PCR-positive samples contained species that were not typeable, possibly because the primers targeting the 5S-23S and 16S-23S IGS do not amplify these Borrelia sequences. However, both the LUX and TaqMan real-time PCR assays successfully amplified the correct length of PCR products from these nine samples, as confirmed by electrophoresis. Nucleotide sequencing also verified that the LUX PCR products originated from Borrelia.
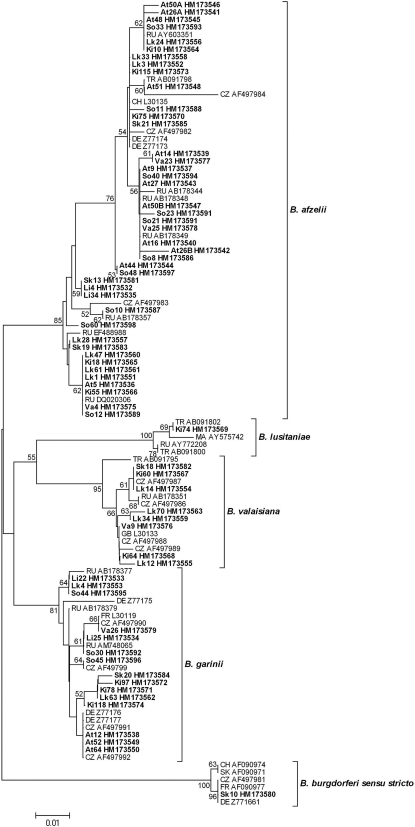
Neighbor-joining was used to construct a phylogenetic tree based on the 5S-23S rRNA IGS sequences of Borrelia species (Fig. 2). Sixty-seven sequences from the current study (i.e., from the coinfected ticks included) and 41 reference sequences retrieved from GenBank were gathered into clusters. A cluster represented a group of sequences within the same Borrelia species that displayed more than 93% sequence similarity, and we found that some sequences within the same cluster showed 100% similarity, even though they had disparate origins (e.g., the ticks came from different sampling sites). The B. afzelii sequences of the two coinfected ticks included in Fig. 2 are denoted At26A, At26B, At50A, and At50B.
FIG. 2.
Phylogenetic tree based on the 5S-23S rRNA intergenic spacer region of different Borrelia species, constructed by neighbor-joining using Kimura 2-parameter and pairwise deletion with a bootstrap value of 500 replicates. Strains found in Östergötland County, Sweden, are shown in bold. Brackets denote Borrelia species clusters with more than 93% sequence similarity. The scale bar corresponds to the expected number of substitutions per nucleotide site. The reliability of the tree was tested by 500 bootstrap replicate analyses; only values greater than 50% are shown. The source of each reference sequence is indicated by an accession number preceded by a country code: CZ, Czech Republic; DE, Germany; FR, France; GB, Great Britain; MA, Morocco; SK, Slovakia; CH, Switzerland; TR, Turkey; RU, Russia.
DISCUSSION
Using our new LUX real-time PCR assay, we found that 19% of ticks removed from humans in Östergötland County, Sweden, were positive for Borrelia. This prevalence is similar to that observed in a study conducted in the Netherlands (13) showing that 20.4% of ticks detached from humans were Borrelia positive, whereas it is twice as high as the proportion detected in an investigation performed in Switzerland (24). Another study, conducted in Texas (38), found only 1% Borrelia prevalence in ticks removed from humans. In the latter investigation, analyzed ticks were provided by individuals that had been bitten in areas where the associated Lyme borreliosis was considered to be nonendemic. Additionally, in the United States, only three species of the B. burgdorferi sensu lato complex have been described and only one of them is known to be human pathogenic (36). It should be mentioned that Borrelia spirochetes were not quantified in these three studies, because real-time PCR assays were not used. Furthermore, Borrelia species were not determined in the Swiss investigation.
Using indirect immunofluorescence to detect Borrelia in field-collected ticks, Gustafson et al. (9) found positive specimens in 20 of the 23 Swedish provinces where I. ricinus was encountered, with an average prevalence of 10% in nymphs and 15% in adults. However, the prevalence of Borrelia varied greatly between the provinces, as exemplified by 0% and 13% found in nymph and adult ticks, respectively, in Östergötland. It is plausible that the higher prevalences that we observed were due to the increased occurrence of Borrelia in ticks since 1995. The Borrelia prevalence in adults that we recorded (33%) is also three times higher than that noted by Fraenkel et al. (8) in a study of field-collected adult ticks from the south and east coasts of Sweden. This discrepancy might be the result of dissimilarities in climate and ecosystem conditions, but it may also be explained by the use of different PCR assays. When a Borrelia-positive tick bites a host, dramatic changes occur in the expression pattern of the Borrelia population, seen as rapid multiplication of the spirochetes in the tick midgut, leading to the overall higher density of Borrelia cells in the tick (27). If the PCR assay applied is not sensitive enough, a lower density of Borrelia spirochetes in field-collected ticks may give false-negative results.
In our study, the prevalence of Borrelia was significantly higher in adult ticks than in nymphs. This was probably the case because adult ticks require an extra blood meal from a host that may be infected with the bacteria, an assumption that is supported by the results of an investigation of field-collected ticks conducted in Switzerland in 2004 (15). We observed geographical differences in Borrelia prevalence in Östergötland County, and these local disparities may be related to factors such as the presence/density of reservoir hosts, forest structure, and types of biotope.
Considering both adult and nymph ticks, we found that B. afzelii was the dominating species in Östergötland County, followed by B. garinii, B, valaisiana, B. burgdorferi sensu stricto, B. lusitaniae, and B. miyamotoi-like (Table 1). Furthermore, B. afzelii and B. garinii were identified at all collecting sites. Those two species have also been described to predominate among ticks detached from humans in the Netherlands (13), and the same pattern has been seen in field-collected ticks from the south and east coasts of Sweden (8). Moreover, B. afzelii and B. garinii are the most abundant Borrelia species in Europe (11). The diversity of reservoir hosts is likely to have an impact on the geographic distribution of Borrelia species. It is well known that small mammals (e.g., rodents) frequently serve as intermediate hosts for strains of B. afzelii and that strains of B. garinii and B. valaisiana are associated with a variety of bird species (19). The fact that we also identified B. lusitaniae for the first time in Sweden may be important, because it is possible that some strains of this species give rise to human LB (6). In addition, there is evidence that B. lusitaniae is becoming established in the northern part of Europe (33).
In our investigation, two adult ticks coinfected with two different strains of B. afzelii were obtained from the same PHC. In comparison, other studies have shown various prevalences of coinfections in field-collected ticks: 3% among adult ticks in England (21), 4% in both adults and nymphs in Switzerland (15), 14% in nymphs in the United States (34), 64% in nymphs in Denmark (33), and 16% among adults and nymphs in Slovakia and Poland (22). In the study conducted in Slovakia and Poland, 5% of all the positive ticks were coinfected with different strains of one particular species (i.e., B. garinii or B. valaisiana). The discrepancies in the prevalences of coinfections between our investigation and other studies might be explained by differences in the transmission pathway, that is, whether there was host-tick or tick-tick (cofeeding) transmission (20). Notably, all the coinfected ticks that we found came from the same area, and there was high sequence similarity between the Borrelia strains that they carried (Fig. 2), which seems to suggest closely related transmission pathways (e.g., these strains may cocirculate among intermediate hosts in the area).
The number of Borrelia cells ranged from 2.0 × 102 to 4.9 × 105 per tick in our study (Fig. 1), which agrees with the range and medians found in field-collected nymph and adult ticks in the northeastern United States (34). We also observed a significantly higher number of Borrelia cells in adults than in nymphs. Adults have a larger body volume than nymphs and can thus be engorged with more host blood, which should allow faster replication of Borrelia, resulting in the detection of higher numbers of the spirochetes.
Our LUX real-time PCR assay was able to reveal a wide variety of Borrelia species at a detection limit of less than 10 gene copies, which is equivalent to the number of copies that exists in one Borrelia cell (25). Furthermore, compared to a TaqMan real-time PCR assay (25), our method showed greater sensitivity, which was seen as detection of more Borrelia-positive ticks. Inasmuch as all these samples were determined to species level, the possibility of false-positive results due to carryover contamination of PCR products can probably be excluded. We also noted that the mean amplification efficiency was higher for the LUX assay than the results previously reported for the TaqMan assay (25), which is important because such efficiency is a crucial marker of the success of gene quantification. In addition, again compared to the TaqMan assay (25), our assay gave a lower standard deviation, as calculated from a set of independent real-time PCR runs. Constant amplification efficiency is an important criterion for reliable comparison between samples and between real-time PCR runs, as well as for assay reproducibility.
In summary, we found that approximately 20% of 399 ticks that had fed on humans in Östergötland, Sweden, were positive for Borrelia. Six Borrelia species were detected, and B. lusitaniae was identified for the first time in Sweden. These observations suggest that the novel LUX real-time PCR assay provides a rapid and sensitive tool for detection and quantification of Borrelia in ticks. It is also plausible that this assay can be a valuable tool in clinical diagnostics as a complement to serological tests.
Acknowledgments
We are grateful for the enthusiasm and support of staff members at the primary health care centers in Ekholmen, Johannelund, Linghem, Kisa, Skärblacka, Söderköping, Valdemarsvik, and Åtvidaberg and at the Department of Infectious Diseases, University Hospital, Linköping, Sweden.
Patricia Ödman is acknowledged for comments and linguistic revision of the manuscript. We also thank Liselott Lindvall and Mari-Anne Åkeson for excellent specimen collection logistics.
This study was supported by the Medical Research Council of Southeast Sweden and by ALF funds.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Asbrink, E., A. Hovmark, and B. Hederstedt. 1984. The spirochetal etiology of acrodermatitis chronica atrophicans Herxheimer. Acta Derm. Venereol. 64:506-512. [PubMed] [Google Scholar]
- 3.Babady, N. E., L. M. Sloan, E. A. Vetter, R. Patel, and M. J. Binnicker. 2008. Percent positive rate of Lyme real-time polymerase chain reaction in blood, cerebrospinal fluid, synovial fluid, and tissue. Diagn. Microbiol. Infect. Dis. 62:464-466. [DOI] [PubMed] [Google Scholar]
- 4.Bunikis, J., U. Garpmo, J. Tsao, J. Berglund, D. Fish, and A. G. Barbour. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741-1755. [DOI] [PubMed] [Google Scholar]
- 5.Collares-Pereira, M., S. Couceiro, I. Franca, K. Kurtenbach, S. M. Schafer, L. Vitorino, L. Goncalves, S. Baptista, M. L. Vieira, and C. Cunha. 2004. First isolation of Borrelia lusitaniae from a human patient. J. Clin. Microbiol. 42:1316-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Franca, I., L. Santos, T. Mesquita, M. Collares-Pereira, S. Baptista, L. Vieira, I. Viana, E. Vale, and C. Prates. 2005. Lyme borreliosis in Portugal caused by Borrelia lusitaniae? Clinical report on the first patient with a positive skin isolate. Wien. Klin. Wochenschr. 117:429-432. [DOI] [PubMed] [Google Scholar]
- 7.Fingerle, V., U. C. Schulte-Spechtel, E. Ruzic-Sabljic, S. Leonhard, H. Hofmann, K. Weber, K. Pfister, F. Strle, and B. Wilske. 2008. Epidemiological aspects and molecular characterization of Borrelia burgdorferi s.l. from southern Germany with special respect to the new species Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 298:279-290. [DOI] [PubMed] [Google Scholar]
- 8.Fraenkel, C. J., U. Garpmo, and J. Berglund. 2002. Determination of novel Borrelia genospecies in Swedish Ixodes ricinus ticks. J. Clin. Microbiol. 40:3308-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustafson, R., T. G. Jaenson, A. Gardulf, H. Mejlon, and B. Svenungsson. 1995. Prevalence of Borrelia burgdorferi sensu lato infection in Ixodes ricinus in Sweden. Scand. J. Infect. Dis. 27:597-601. [DOI] [PubMed] [Google Scholar]
- 10.Hengge, U. R., A. Tannapfel, S. K. Tyring, R. Erbel, G. Arendt, and T. Ruzicka. 2003. Lyme borreliosis. Lancet Infect. Dis. 3:489-500. [DOI] [PubMed] [Google Scholar]
- 11.Hubálek, Z., and J. Halouzka. 1997. Distribution of Borrelia burgdorferi sensu lato genomic groups in Europe, a review. Eur. J. Epidemiol. 13:951-957. [DOI] [PubMed] [Google Scholar]
- 12.Ivacic, L., K. D. Reed, P. D. Mitchell, and N. Ghebranious. 2007. A LightCycler TaqMan assay for detection of Borrelia burgdorferi sensu lato in clinical samples. Diagn. Microbiol. Infect. Dis. 57:137-143. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, J. J., G. T. Noordhoek, J. M. Brouwers, P. R. Wielinga, J. P. Jacobs, and A. H. Brandenburg. 2008. [Small risk of developing Lyme borreliosis following a tick bite on Ameland: research in a general practice.] Ned. Tijdschr. Geneeskd. 152:2022-2026. (In Dutch.) [PubMed] [Google Scholar]
- 14.Jaenson, T. G., L. Talleklint, L. Lundqvist, B. Olsen, J. Chirico, and H. Mejlon. 1994. Geographical distribution, host associations, and vector roles of ticks (Acari: Ixodidae, Argasidae) in Sweden. J. Med. Entomol. 31:240-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouda, F., J. L. Perret, and L. Gern. 2004. Density of questing Ixodes ricinus nymphs and adults infected by Borrelia burgdorferi sensu lato in Switzerland: spatio-temporal pattern at a regional scale. Vector Borne Zoonotic Dis. 4:23-32. [DOI] [PubMed] [Google Scholar]
- 16.Kawabata, H., T. Masuzawa, and Y. Yanagihara. 1993. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol. Immunol. 37:843-848. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 18.Kriuchechnikov, V. N., E. I. Korenberg, S. V. Shcherbakov, V. Kovalevskii Iu, and M. L. Levin. 1988. [Identification of Borrelia isolated in the USSR from Ixodes persulcatus Schulze ticks.] Zh. Mikrobiol. Epidemiol. Immunobiol. 12:41-44. [PubMed] [Google Scholar]
- 19.Kurtenbach, K., S. De Michelis, S. Etti, S. M. Schafer, H. S. Sewell, V. Brade, and P. Kraiczy. 2002. Host association of Borrelia burgdorferi sensu lato—the key role of host complement. Trends Microbiol. 10:74-79. [DOI] [PubMed] [Google Scholar]
- 20.Kurtenbach, K., S. De Michelis, H. S. Sewell, S. Etti, S. M. Schafer, R. Hails, M. Collares-Pereira, M. Santos-Reis, K. Hanincova, M. Labuda, A. Bormane, and M. Donaghy. 2001. Distinct combinations of Borrelia burgdorferi sensu lato genospecies found in individual questing ticks from Europe. Appl. Environ. Microbiol. 67:4926-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurtenbach, K., M. Peacey, S. G. Rijpkema, A. N. Hoodless, P. A. Nuttall, and S. E. Randolph. 1998. Differential transmission of the genospecies of Borrelia burgdorferi sensu lato by game birds and small rodents in England. Appl. Environ. Microbiol. 64:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lencakova, D., C. Hizo-Teufel, B. Petko, U. Schulte-Spechtel, M. Stanko, B. Wilske, and V. Fingerle. 2006. Prevalence of Borrelia burgdorferi s.l. OspA types in Ixodes ricinus ticks from selected localities in Slovakia and Poland. Int. J. Med. Microbiol. 296(Suppl. 1):108-118. [DOI] [PubMed] [Google Scholar]
- 23.Lowe, B., H. A. Avila, F. R. Bloom, M. Gleeson, and W. Kusser. 2003. Quantitation of gene expression in neural precursors by reverse-transcription polymerase chain reaction using self-quenched, fluorogenic primers. Anal. Biochem. 315:95-105. [DOI] [PubMed] [Google Scholar]
- 24.Nahimana, I., L. Gern, D. S. Blanc, G. Praz, P. Francioli, and O. Peter. 2004. Risk of Borrelia burgdorferi infection in western Switzerland following a tick bite. Eur. J. Clin. Microbiol. Infect. Dis. 23:603-608. [DOI] [PubMed] [Google Scholar]
- 25.Ornstein, K., and A. G. Barbour. 2006. A reverse transcriptase-polymerase chain reaction assay of Borrelia burgdorferi 16S rRNA for highly sensitive quantification of pathogen load in a vector. Vector Borne Zoonotic Dis. 6:103-112. [DOI] [PubMed] [Google Scholar]
- 26.Piesman, J., and L. Gern. 2004. Lyme borreliosis in Europe and North America. Parasitology 129(Suppl.):S191-S220. [DOI] [PubMed] [Google Scholar]
- 27.Piesman, J., B. S. Schneider, and N. S. Zeidner. 2001. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J. Clin. Microbiol. 39:4145-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postic, D., M. V. Assous, P. A. Grimont, and G. Baranton. 1994. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int. J. Syst. Bacteriol. 44:743-752. [DOI] [PubMed] [Google Scholar]
- 29.Richter, D., D. B. Schlee, R. Allgower, and F. R. Matuschka. 2004. Relationships of a novel Lyme disease spirochete, Borrelia spielmani sp. nov., with its hosts in Central Europe. Appl. Environ. Microbiol. 70:6414-6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rijpkema, S. G., D. J. Tazelaar, M. J. Molkenboer, G. T. Noordhoek, G. Plantinga, L. M. Schouls, and J. F. Schellekens. 1997. Detection of Borrelia afzelii, Borrelia burgdorferi sensu stricto, Borrelia garinii and group VS116 by PCR in skin biopsies of patients with erythema migrans and acrodermatitis chronica atrophicans. Clin. Microbiol. Infect. 3:109-116. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Schwaiger, M., and P. Cassinotti. 2003. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J. Clin. Virol. 27:136-145. [DOI] [PubMed] [Google Scholar]
- 33.Vennestrøm, J., H. Egholm, and P. M. Jensen. 2008. Occurrence of multiple infections with different Borrelia burgdorferi genospecies in Danish Ixodes ricinus nymphs. Parasitol. Int. 57:32-37. [DOI] [PubMed] [Google Scholar]
- 34.Wang, G., D. Liveris, B. Brei, H. Wu, R. C. Falco, D. Fish, and I. Schwartz. 2003. Real-time PCR for simultaneous detection and quantification of Borrelia burgdorferi in field-collected Ixodes scapularis ticks from the Northeastern United States. Appl. Environ. Microbiol. 69:4561-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, G., A. P. van Dam, A. Le Fleche, D. Postic, O. Peter, G. Baranton, R. de Boer, L. Spanjaard, and J. Dankert. 1997. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19). Int. J. Syst. Bacteriol. 47:926-932. [DOI] [PubMed] [Google Scholar]
- 36.Wang, G., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm, J., and A. Pingoud. 2003. Real-time polymerase chain reaction. Chembiochem 4:1120-1128. [DOI] [PubMed] [Google Scholar]
- 38.Williamson, P. C., P. M. Billingsley, G. J. Teltow, J. P. Seals, M. A. Turnbough, and S. F. Atkinson. 2010. Borrelia, Ehrlichia, and Rickettsia spp. in ticks removed from persons, Texas, U. S. A. Emerg. Infect. Dis. 16:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]