Abstract
Helicobacter pylori is a Gram-negative bacterial pathogen with a small genome of 1.64–1.67 Mb. More than 20 putative DNA restriction-modification (R-M) systems, comprising more than 4% of the total genome, have been identified in the two completely sequenced H. pylori strains, 26695 and J99, based on sequence similarities. In this study, we have investigated the biochemical activities of 14 Type II R-M systems in H. pylori 26695. Less than 30% of the Type II R-M systems in 26695 are fully functional, similar to the results obtained from strain J99. Although nearly 90% of the R-M genes are shared by the two H. pylori strains, different sets of these R-M genes are functionally active in each strain. Interestingly, all strain-specific R-M genes are active, whereas most shared genes are inactive. This agrees with the notion that strain-specific genes have been acquired more recently through horizontal transfer from other bacteria and selected for function. Thus, they are less likely to be impaired by random mutations. Our results also show that H. pylori has extremely diversified R-M systems in different strains, and that the diversity may be maintained by constantly acquiring new R-M systems and by inactivating and deleting the old ones.
Helicobacter pylori is one of the most common bacterial pathogens that colonizes the gastric mucosa of humans. H. pylori is implicated in a wide range of gastroduodenal diseases (1, 2). H. pylori is commonly believed to be a very diverse species. It is believed that, in addition to genetic recombination, de novo mutation could have a role in generating the high level of genetic variation in H. pylori (3). MutH and MutL homologues cannot be found in H. pylori genomes, which suggests H. pylori may not have a functional mismatch repair system (4, 5). Recent analysis of the complete genomic sequences of two unrelated H. pylori isolates reveals that although intraspecies variation does exist, the overall genomic organization, gene order, and predicted proteins of the two strains are quite similar (5, 6). Approximately 6–7% of the genes are specific to each strain (5). The 26695 and J99 strains have a relatively small genome size of 1.67 and 1.64 megabase pairs (4, 5). However, more than twenty DNA restriction-modification (R-M) systems can be identified in each strain based on sequence similarities. The biological significance of this large complement of R-M systems is not clear. The majority of the H. pylori R-M systems are of Type II, which consist of two separate enzymes: the restriction endonucleases, which are responsible for degrading unmodified foreign DNA, and the modification DNA methyltransferases (methylase or M), which protect endogenous DNA from endonucleolytic digestion by methylating them at the endonuclease recognition sites (7).
Interesting observations have been reported regarding H. pylori R-M genes. A novel H. pylori gene, iceA (induced when the bacteria contact the host epithelium), was identified recently (8). DNA sequences have revealed two alleles of the iceA locus, iceA1 and iceA2, existing in different H. pylori strains. Strains containing iceA1 were found to be significantly associated with peptic ulceration. Increased mucosal concentrations of interleukin-8 were also found in these strains. Surprisingly, iceA1 shares significant sequence similarity with a Type II restriction endonuclease gene, r.nlaIII (9).
Second, an interesting phenomenon of phase variation has been linked to the R-M genes in H. pylori. Short tandem repeat sequences are subject to loss or gain of a repeat unit, presumably through slipped-strand mispairing during replication. This results in frameshifting, which can alternatively activate or inactivate genes (10). Tetranucleotide repeats were found in a Type III DNA methylase gene and the length of the repeat tract determined the phase variation rate (11). In the case of the H. pylori 26695 genome, 27 putative genes that contain simple sequence repeats and that may be subject to phase variation have been identified. These putative phase-variable elements can be divided into three groups: lipopolysaccharide (LPS) biosynthesis, cell-surface-associated proteins and DNA R-M systems (12). For example, the putative Type II R-M system encoded by HP1471–1472 contains a string of 14 G-residues in the HP1471 gene.
Third, H. pylori R-M genes are one of the major components of the strain-specific genes. The strain-specific genes are believed to be involved in drug resistance (13) and bacterial surface structure (14), as well as restriction-modification (15). A PCR-based subtractive hybridization method was used to investigate genes that are unique to individual H. pylori strains (16, 17). Among the 18 strain-specific genes identified by this method, seven are R-M genes (16). In addition, genome sequence comparison of two H. pylori strains showed that R-M system genes account for 15–20% of the strain-specific genes. We reported (18) a biochemical analysis of the Type II R-M systems in H. pylori J99. We now report a similar analysis of H. pylori 26695 and comparison of these R-M systems with the 16 Type II R-M systems present in strain J99.
Materials and Methods
Bacterial Strains and Growth.
Escherichia coli DB24 is derived from E. coli GM4714 (19), with an additional mutation at the dcm locus introduced via P1 transduction (obtained from E. Raleigh and M. Sibley, New England Biolabs). E. coli ER2566-pLysP is an E. coli ER2566 derivative that contains a mutant T7 lysozyme gene in the plasmid pACYC184. E. coli ER2566-pLysP cells carrying pLT7K with cloned endonuclease genes were grown on LB plates containing 100 μg/ml ampicillin and 20 mM glucose at 42°C overnight. Cells were then removed from the plate and grown in LB media containing 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 20 mM glucose at 37°C for 2 h. The cell suspension was equilibrated at 30°C for half an hour before induction with 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG). Following 30 min induction, 200 μg/ml of rifampicin was added to the cell suspension to reduce the background nuclease activity by inhibiting the host E. coli RNA polymerase (19). Induced cultures were grown at 30°C for 2 more hours, and then the E. coli cells were collected by centrifugation.
Identification of Putative R-M Genes.
Putative Type II R-M genes were identified based on the original annotations (4, 5) and our own sequence search (18). First, sequence analysis was used to identify all of the putative methylases in the H. pylori 26695 genome. Next, the search for additional endonuclease candidates is based on the principle of “guilt by association.” Endonuclease and methylase genes in a given R-M system are usually located next to each other. Therefore, an unknown gene with no homolog in GenBank next to a putative methylase gene is a candidate endonuclease gene.
Cloning Potential Type II R-M Systems.
DNA sequences coding for all 14 potential Type II R-M systems of H. pylori 26695 were obtained from the TIGR web site (http://www.tigr.org/). Those candidate genes were first amplified by using PCR, and then cloned into plasmid vectors. However, the subsequent expression of those restriction endonuclease genes in E. coli can be very challenging because the gene products normally are cytotoxic unless the host genomic DNA is completely methylated by the corresponding MTase. To maximize the success rate of expressing these lethal endonucleases, two cloning strategies were used in this study. Putative endonuclease and MTase genes were amplified by PCR using H. pylori 26695 genomic DNA and a pair of primers corresponding to the 5′ and 3′ sequences of each pair of R-M genes. The amplified genes were cloned into plasmid pUC19 between the BamHI and EcoRI sites. The recombinant plasmid DNA was transformed into E. coli ER2683, which lacks endogenous DNA R-M systems (E. Raleigh and M. Sibley, New England Biolabs). These target genes were sequenced to verify that no mutations were introduced during the PCR and cloning processes. Expression of these R-M genes was under the control of a constitutive lac promoter. E. coli ER2683 cells carrying the pUC19 vector with both R and M genes were grown in LB broth containing 100 μg/ml ampicillin. Cells were harvested after the OD590 reached 0.8–0.9.
The pLT7K system was used to clone potential Type II restriction endonuclease genes directly in the absence of an adjacent gene coding for the cognate MTase (18). Potential restriction endonuclease genes were cloned into pLT7K and sequenced to check for mutations during PCR and cloning. Recombinant plasmids containing the wild-type putative endonuclease genes were then transformed into E. coli ER2566 pLysP, which encodes a T7 RNA polymerase. E. coli (ER2566pLysP) cells carrying the pLT7K vector with cloned restriction endonuclease genes were grown on LB plates containing 100 μg/ml ampicillin and 20 mM glucose at 42°C overnight. Cells were then removed from the plate and grown in LB media containing 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 20 mM glucose at 37°C for 2 h. The cell suspension was equilibrated to 30°C for 30 min before induction with 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG). Following 30 min induction, 200 μg/ml of rifampicin was added to the cell suspension to reduce the background nuclease activity. Two hours later, cells were harvested at 6000 × g for 20 min.
Characterization of Restriction Endonuclease Activity.
Restriction endonuclease activity was assayed by using phage lambda and T7 DNA as substrates. The recognition sequences of all active endonucleases were determined by DNA mapping and computer analysis using the programs mapsort and gap (Genetics Computer Group, Madison, WI). The locations of the cleavage sites by the endonuclease were mapped by double digestion of plasmid DNA (pBR322 or pUC19) with known restriction endonucleases and the endonuclease of interest.
Characterization of Methylation Activity.
The methylation activity of each potential M gene was determined by a restriction enzyme digestion assay and/or dot blot assay using two rabbit primary antibodies raised against DNA with N6-methyladenine (m6A) and N4-methylcytosine (m4C) (18).
Results
We have searched the H. pylori DNA sequence looking for the following: a potential DNA MTase (M) gene containing conserved MTase sequence motifs (20, 21) and an adjacent putative restriction endonuclease (R) gene, which either shares sequence similarity with the existing Type II endonuclease or shares no similarity with any known gene in the GenBank. Fourteen potential Type II R-M systems were found by using these criteria and are summarized in Table 1. Type II restriction endonucleases usually do not share sequence similarities with each other (7). Among the 14 putative Type II endonuclease genes, only three (HP91, HP1209, and HP1366) share significant sequence similarities with genes for MboI, NlaIII (IceA), and MboII, respectively (Table 1). In the case of the DNA MTases, they were further annotated into three groups based on the methylation products: m4C, N5-methylcytosine (m5C), and m6A (7, 20).
Table 1.
Putative Type II R-M systems in H. pylori 26695 strain
| HP | Annotation | Putative function | Activity confirmed isoschiz./sequence | Name |
|---|---|---|---|---|
| 49 | None | ENase | − | |
| 50 | M.DpnIA, 55%, m6A | MTase | +, m6A | M.HpyAVIA |
| 51 | M.DdeI, 60%, m5C | MTase | +, m5C | M.HpyAVIB |
| 52 | None | ENase | − | |
| 53 | None | ENase | +, ? | HpyAV |
| 54 | M.HgaI, 55%, m6A | MTase | +, m6A | M.HpyAV |
| 91 | R.MboI, 68% | ENase | +, MboI | HpyAIII |
| 92 | M.MboI, 75%, m6A | MTase | +, m6A | M.HpyAIII |
| 262 | None | ENase | − | |
| 263 | M.ScaI, 28%, m4C | MTase | Inactive MTase | |
| 368 | None | ENase | − | |
| 369 | M.CviQI, 31%, m6A | MTase | Inactive MTase | |
| 478 | M.VspI, 62%, m6A | MTase | +, m6A | M.HpyAVII |
| 479 | None | ENase | − | |
| 481 | M.FokI, 49%, m6A | MTase | Inactive MTase | |
| 482 | None | ENase | − | |
| 483 | M.BspRI, 64%, m5C | MTase | Inactive MTase | |
| 484 | None | ENase | − | |
| 909 | None | ENase | − | |
| 910 | M.HincII, 52%, m6A | MTase | +, m6A | |
| 1120 | None | ENase | − | |
| 1121 | M.BsuFI, 58%, m5C | MTase | +, m5C | M.HpyAVIII |
| 1208 | M.NlaIII, 60%, m6A | MTase | +, m6A | HpyAI |
| 1209 | R.NlaIII, 98%, IceA | ENase | − | |
| 1351 | None | ENase | +, HinfI | HpyAIV |
| 1352 | M.HinfI, 78%, m6A | MTase | +, m6A | M.HpyAIV |
| 1366 | R.MboII, 49% | ENase | +, MboII | HpyAII |
| 1367 | M.MboII, 64%, m6A | MTase | +, m6A | M.HpyAIIA |
| 1368 | M.MjaI, 41%, m4C | MTase | +, m4C | M.HpyAII |
| 1471 | BcgI B, 29%, | S | − | |
| 1472 | BcgI A, 32%, m6A | R-M | – |
HP, ORF number of H. pylori 26695; %, percent of sequence identity; MTase, DNA methylase; ENase, restriction endonuclease; S, specificity subunit. Fully active R-M systems are in bold type. Inactive endonuclease or methylase indicates an ORF with similarity to a known endonuclease or methylase but no function was detected. +, activity detected; −, no activity detected.
The restriction endonuclease activity of each potential R gene was determined by cloning the endonuclease gene and its adjacent methylase gene as a pair into pUC19 vector and then assaying for the presence of DNA cleavage activity in E. coli cells expressing the target gene. Sometimes, introducing both endonuclease and methylase genes into a new host cell at the same time could cause problems, because the endogenous DNA may not be sufficiently modified by the incoming MTase if the endonuclease gene is expressed immediately (22). To reduce the possibility of obtaining false-negative results, those putative H. pylori endonuclease genes whose gene products showed no detectable endonuclease activity in the pUC19 system were cloned into pLT7K. pLT7K is a cloning/expression vector that features repressor and antisense control elements so that toxic genes can be expressed in E. coli under extremely tight control (18).
The DNA methylation activity of each potential M gene was determined by a restriction enzyme digestion assay and/or antibody detection assay. The DNA methylase component of a Type II R-M system usually recognizes the same sequence as the corresponding endonuclease. Thus, after a restriction endonuclease is characterized, the methylation activity of its corresponding methylase can easily be determined by digesting the DNA, isolated from cells containing the relevant methylase gene, with the identified endonuclease. Because the majority of the examined H. pylori R-M systems showed no detectable endonuclease activity, it was impossible to determine the DNA MTase activity by using the endonuclease digestion method described above. Recently, two polyclonal antibodies raised against m6A or m4C have been developed, and these two antibodies were found to bind specifically to DNA containing these two methylated nucleotides (18).
HP49–50-51.
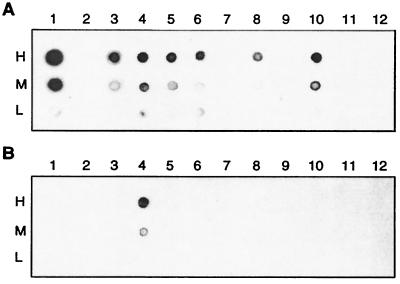
These three ORFs encode two functional MTases and one nonfunctional restriction enzyme-related gene that form part of a Type IIs R-M system. By using antibody analysis we find that the HP50 MTase is an m6A enzyme (Fig. 1A, lane 6). The gene sequence is similar to that encoding M.MnlIB (A. Lubys and A. Janulaitis, personal communication), which is a component of a known system that recognizes the sequence 5′-CCTC-3′ (23). The HP50 product, M.HpyAVIA, must methylate the A-residue on the complementary strand of the recognition sequence. Clones expressing M.HpyAVIA are resistant to digestion by MnlI. HP51 is an m5C MTase, M.HpyAVIB, that is also functional. When HP51 is cloned separately into pUC19, it too protects DNA against digestion by MnlI and the gene shows strong sequence similarity to the gene encoding M.MnlIA—the other MTase of the MnlI system. HP52 is almost certainly a remnant of a gene that may have once encoded a functional isoschizomer of MnlI, but in which we can no longer detect activity.
Figure 1.
Antibody assay to measure methyltransferase activities by using rabbit primary antibodies raised specifically against m6A or m4C. Dot blot assays were performed by spotting total DNAs, isolated from clones expressing individual H. pylori methylase genes, onto a nitrocellulose BA85 membrane (18). Positive signals were detected by using a secondary anti-rabbit antibody conjugated with horseradish peroxidase (18). Lanes 1–12 represent HP1351–1352; pHKUV5 vector (a negative control); HP91–92; HP1366–1367-1368; HP54; HP50–51; HP263; HP478; HP481; HP910; HP1471–1472; and HP1121, respectively. (A) MTase dot blot assay using antibodies against m6A. (B) MTase dot blot assay using antibodies against m4C. Three dilutions of DNA samples were spotted: H (high), 0.45 μg; M (medium), 0.15 μg; and L (low), 0.05 μg.
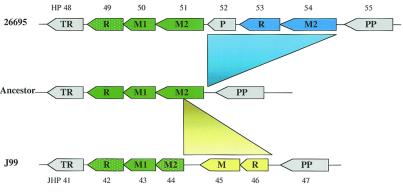
This region in H. pylori J99 (JHP43–44) is more complex and shows many rearrangements. The m6A MTase, M.Hpy99V, is the product of JHP43 (equivalent to HP50) and is the only functional gene. The adjacent ORF, JHP44, appears to be a chimera between the N terminus of HP52 and the C terminus of HP51. Finally, a new R-M system is inserted after this chimera. This is diagrammed in Fig. 2. It appears as though an earlier strain contained an intact MnlI R-M system and that in J99 it is almost completely rearranged and has lost most of the activities, whereas in 26695, only the R gene appears to have accumulated inactivating mutations (Fig. 2).
Figure 2.
Schematic diagram showing the possible evolution of HP49–54 in strain 26695 and JHP42–46 in strain J99 from a putative ancestral locus. The shared Type II R-M systems are shown in green. The strain-specific Type II R-M system in strain 26695 is shown in blue, and the strain-specific Type II R-M system in J99 is shown in yellow. Functional genes are shown in solid colors and inactive genes are patterned with white dots. Non-R-M genes are shown in gray. R, restriction endonuclease gene; M, DNA methylase gene; P, putative gene; TR, transcriptional regulator gene; PP, proline permease gene. HP numbers are marked above the genes and JHP numbers are marked below the genes.
HP53–54.
These two ORFs encode a fully functional Type II R-M system called HpyAV. HP54 is the gene for an m6A/m5C MTase and has many similar sequences in GenBank. However, it contains an unusually long C terminus of unknown function. The M gene is active because adenine methylation could be detected by m6A antibodies (Fig. 1, lane 5). HP53 has no similar sequences in GenBank, which is typical of an R gene. Restriction endonuclease activity was detectable when the HP53 gene was expressed in E. coli cell premodified by the M gene of HP54. The endonuclease activity of HpyAV was purified through several columns. However HpyAV cleaves DNA very frequently and we have not been able to achieve complete digestion on DNA substrates. It probably recognizes a 4- or 5-bp asymmetric sequence. In J99, a totally different R-M system (Hpy99II, a Tsp45I isoschizomer) is found at this locus (Fig. 2).
HP91–92.
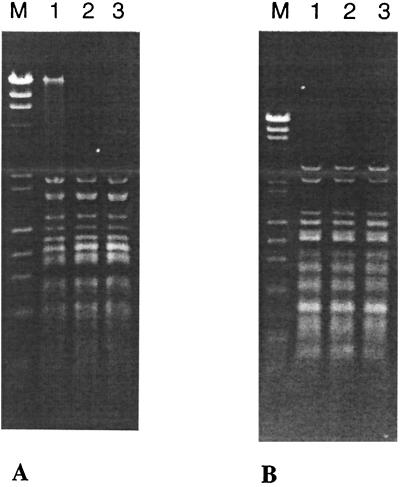
HP91 is a fully functional R gene and encodes the restriction enzyme HpyAIII. The gene shows similarities to those of several known isoschizomers, such as MboI, MjaIII, and DpnII, all of which recognize GATC. The digestion patterns produced by HpyAIII are identical to those produced by these other enzymes (Fig. 3A). The cognate MTase is encoded by HP92 and shows sequence similarities to other known GATC MTases. Antibody tests show that this is an m6A MTase (Fig. 1, lane 3). These genes have a corresponding pair in J99, but there only the M gene is active. The J99 R gene contains multiple frameshifts. However, after correcting the frameshifts the similarity in sequence is very high, which identifies the J99 system as also recognizing GATC.
Figure 3.
Determination of the recognition specificities of H. pylori endonucleases by double digestion. DNA endonucleotic cleavage activity of isoschizomers of MboI and MboII, encoded by HP91–92 and HP1366–1367-1368, respectively. (A) Bacteriophage lambda DNA was digested with MboI (lane 1), MboI and purified endonuclease HpyAIII (lane 2), or HpyAIII (lane 3). (B) Lambda DNA was digested with MboII (lane 1), MboII and purified endonuclease HpyAII (lane 2), or HpyAII alone (lane 3).
HP262–263.
These two genes do not encode functional proteins, but they do show similarities to known genes. HP263 is most similar to M.MjaVI, an m4C MTase that methylates the sequence CCGG. In particular, there is strong similarity within the variable region that encodes sequence specificity. HP262 also shows similarity to the ORF in Methanococcus jannaschii that lies next to that encoding M.MjaVI, which is probably an ancestral R gene of the CCGG specificity. We conclude that these two genes are inactive versions of an R-M system that once recognized CCGG. Another strain of H. pylori (ATCC 49503) encodes an active restriction endonuclease of this same specificity (24).
In J99 this same pair of genes exists (JHP247–248) and the MTase is an active m4C MTase. These observations suggest that the J99 gene, encoding M.Hpy99VIII, also has the specificity CCGG. It is unknown which C residue is modified.
HP368–369.
These two genes are also inactive. They have homologues in J99, but those genes are in the opposite orientation on the chromosome and at a very different location (JHP1012–1013). All of these genes show strong similarities to the sequences of the Hpy188III R-M system, which recognizes the sequence TCNNGA (24). It is likely that these genes in both 26695 and J99 are derived from a previously functional system that also recognized TCNNGA.
HP478–479.
By antibody testing, HP478 encodes an active m6A MTase, M.HpyAVII (Fig. 1, lane 8). Its counterpart in J99, JHP430, is inactive and there are 33 predicted amino acid changes within the 545 amino acid coding region. The M genes, HP478 and JHP430, both show over 60% sequence similarity to the gene for VspI, which recognizes ATTAAT (25). DNA isolated from cells expressing HP478 gene was partially resistant to VspI endonuclease digestion, suggesting that it is possible that these systems also recognize ATTAAT or a closely related sequence. Because HP477 is likely to encode an outer membrane protein, HP479 is the candidate R gene, although we found no activity when it was cloned into E. coli. A similar situation is encountered in J99, where the corresponding, closely related gene, JHP431, is also inactive.
HP481–482.
These two genes are inactive, as are their counterparts (JHP433–434) in J99. However, both sets of sequences show strong similarities to other R-M sequences present in GenBank. HP481 and JHP433 are closely related to the gene for M.Hpy188I, which recognizes the sequence TCNGA and forms m6A (24) and another system from H. pylori strain A17–2 (GenBank accession no. AJ278104) that also recognizes TCNGA (R. Sapranauskas and A. Janulaitis, personal communication). Similarly, the R genes also show similarities and so it is likely that in both 26695 and J99 these genes are derived from a once-functional R-M system recognizing TCNGA.
HP483–484.
Both of these genes are inactive, but in J99 the corresponding M gene, JHP435, encodes an active m5C MTase. The corresponding inactive R gene is JHP436. Both M gene sequences show strong similarities to the M gene from the HpyCH4IV R-M system, which recognizes ACGT (24). Thus, we conclude that these genes are another defunct R-M system that once recognized ACGT. Expression of the J99 gene (JHP435) protects against HpyCH4IV confirming its specificity.
HP909–910.
This system encodes an active m6A MTase based on antibody tests (Fig. 1, lane 10) and an inactive endonuclease because no activity is found in clones expressing HP909. Both genes show strong similarities to the genes encoding the characterized Hpy8I R-M system, which recognizes GTNNAC (A. Lubys, J. Minkute, and A. Janulatis, personal communication; GenBank accession no. AF283672). Thus, this R-M system contains an active M gene and an inactive R gene recognizing GTNNAC. In J99, both genes of the corresponding system JHP845/846 are inactive.
HP1120–1121.
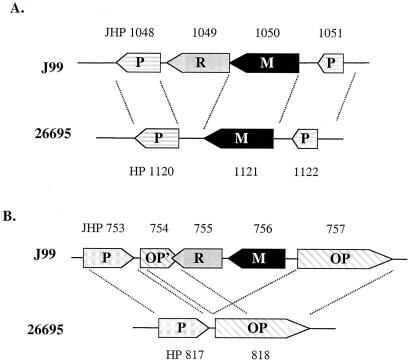
HP1121 encodes a functional m5C MTase, M.HpyAVIII, that recognizes the sequence GCGC. In J99 there is a corresponding system, Hpy99III (JHP1049–1050), with a fully functional pair of genes. The R gene in J99 is JHP1049, but it has no counterpart in 26695. Instead, the adjacent gene, HP1120, corresponds to JHP1048. Because the flanking genes on the other side of the M gene are conserved between J99 and 26695, we conclude that the R gene from the precursor R-M system has been deleted in 26695 (Fig. 4A).
Figure 4.
Schematic diagram showing the alignment of homologous loci. (A) The alignment of homologous loci where the strain-specific R-M system genes of JHP755–756 are inserted in strain J99. (B) The deletion of an endonuclease gene (homologue of JHP1049) in strain 26695. R, restriction endonuclease gene (shown in gray); M, DNA methylase gene (shown in black). Non-R-M genes are shown in patterned boxes. P, putative gene; OP, osmoprotection protein gene; OP', 5′ end of a truncated OP gene. JHP numbers are marked above the genes and HP numbers are marked below the genes.
HP1208–1209.
HP1208 is similar in sequence to several genes encoding active MTases recognizing CATG, and itself encodes an active m6A MTase, M.HpyAI, recognizing CATG. HP1209 is related to several functional genes encoding active restriction endonucleases recognizing CATG, but is itself truncated at the N terminus and is inactive when expressed in E. coli. In J99, the corresponding M gene is JHP1131 and is active, but there is no corresponding R gene. Instead it is replaced by a locus, termed iceA2, found in several different H. pylori strains (8).
HP1351–1352.
These genes encode a fully active R-M system, HpyAIV, that recognizes the sequence GANTC. The MTase is an m6A MTase. In J99, the corresponding M gene (JHP1271) is active, but the R gene (JHP1270) is not.
HP1366–1368.
These genes encode an active R-M system, HpyAII, recognizing the sequence GAAGA. Purified HpyAII cleaves lambda DNA at exactly the same sequence as the MboII endonuclease (Fig. 3B). This R gene shows great similarity to the R gene of the MboII system (26), and the adjacent gene, HP1367 encoding M.HpyAIIA, closely resembles the m6A MTase of the MboII system. In E. coli cells expressing M.HpyAIIA, overlapping GATC sites become resistant to digestion by MboI or DpnII, indicating that this MTase methylates the final A residue in its GAAGA recognition sequence. The third gene, HP1368, encodes an m4C MTase that is very similar in sequence to the second MTase of the MboII system, which has been cloned and sequenced independently (R. Morgan, personal communication). This gene was not reported earlier (26). DNA samples isolated from cells expressing HP1367 and 1368 gave positive reactions with both m6A and m4C antibodies (Fig. 1, lane 4). In J99, the two M genes have both been lost, but an inactive R gene, JHP1442, is found at a different genomic location.
HP1471–1472.
The genes of this R-M system show strong similarity to the genes of the BcgI R-M system, an unusual system that contains an M gene fused with an R gene and a separate gene encoding an S subunit of the enzyme responsible for sequence specificity (27). No restriction endonuclease and DNA methylase activities were detected. In the counterpart system in J99 (the completely inactive JHP1364–1365) the fused R-M gene is very similar to the 26695 gene, but the S gene, JH1365, differs in one of the two regions responsible for sequence recognition. We conclude that these two BcgI-like systems are derived from a precursor that recognized two different DNA sequences. It should also be noted that both HP1471 and JHP1364 contain a run of 14 G-residues that may be subject to phase variation.
In summary, four of fourteen Type II R-M systems examined were fully functional in H. pylori, strain 26695. They displayed both restriction endonuclease activity and methylation modification activity. Four others lacked detectable restriction endonuclease activity, but were positive for methylation activity. Finally, no detectable restriction enzyme activity or methylase activity was observed in the remaining six R-M systems.
Discussion
The biochemical activities of 14 Type II R-M systems from H. pylori strain 26695 have been characterized in this study. They are compared with the 16 Type II R-M systems present in the J99 strain of H. pylori (18). The results are summarized in Table 2, wherein the systems are broken down into three categories.
Table 2.
Comparison of activities of Type II R-M systems from two H. pylori strains
| 26696 (HP)
|
Sequence | J99
(JHP)
|
||
|---|---|---|---|---|
| ORF no. | Name | ORF no. | Name | |
| — | GTSAC | 45-46 | Hpy99II | |
| — | CCNNGG | 629-630 | Hpy99IV | |
| — | CGWCG | 755-756 | Hpy99I | |
| 1121 | HpyAVIII | GCGC | 1049-1050 | Hpy99III |
| 53-54 | HpyAV | ? | — | |
| 91-92 | HpyAIII | GATC | 84-85 | Hpy99VI |
| 1351-1352 | HpyAIV | GANTC | 1270-1271 | Hpy99IX |
| 1366-1367-1368 | HpyAII | GAAGA | 1442 | |
| 49-50-51 | HpyAVI | CCTC | 42-43-44 | Hpy99V |
| 262-263 | CCGG | 247-248 | Hpy99VIII | |
| 478-479 | HpyAVII | ATTAAT | 430-431 | |
| 483-484 | ACGT | 435-436 | Hpy99XI | |
| 909-910 | HpyAIX | GTNNAC | 845-846 | |
| 1208-1209 | HpyAI | CATG | 1131 | Hpy99X |
| 368-369 | TCNNGA | 1012-1013 | ||
| 481-482 | ? | 433-434 | ||
| 1471-1472 | ? | 1364-1365 | ||
Active restriction endonuclease genes are shown underlined and active methyltransferase genes are shown in italics. Fully functional R-M systems, which contain both active R and M genes, are highlighted in bold type. ?, unknown recognition sequence.
The first category shows systems that have both a functional M gene and a functional R gene. There are four such complete systems in each strain, although the recognition specificities of these systems are unique to each strain. The four systems found in J99 consist of three that have genes only found in strain J99, for example JHP755–756 is a unique R-M system, whereas the fourth has an active M gene counterpart in 26695, but no corresponding R gene. In contrast, strain 26695 has one R-M system that has no counterpart in J99 and one system in which the two M genes have been deleted; however, a remnant of an inactive R gene can be seen in J99 and two systems in which the M gene is active in J99, but the R gene is inactive. It is interesting to note that all these strain-specific R-M systems were fully functional. Genomic-sequence comparison of two unrelated H. pylori isolates showed that the (G + C) percentage is lower in strain-specific genes (35%) than the reminder of the genome (39%), indicating that they may have been acquired more recently through horizontal gene transfer (5). Analysis of the strain-specific R-M genes in H. pylori supports this notion. For example, JHP755–756 is a J99 strain-specific R-M system with a low (G + C) content of 33.9%. Comparison of corresponding loci between J99 and 26695 showed that the JHP755–756 genes were inserted into an osmoprotection protein (OP) gene and a duplication of the 5′ half of the OP gene was observed, which perhaps took place after the insertion event to retain the essential function of the OP gene (Fig. 4B).
The second group of R-M systems consists of six systems, each of which contains active methylase genes, but inactive R genes. In one case, the very common M gene encoding an m6A methylase with CATG specificity is active in both strains, but more often a methylase gene active in one strain is inactive in the other. Finally, there are remnants of three systems that are shared by the two strains, but in which neither gene is active. This diversity of R-M systems in H. pylori has been documented previously (24), although earlier the extent of active methylation was not investigated. It should also be noted that the present studies provide only a lower estimate of the total amount of methylation present because, in addition to these Type II R-M systems, there are further potentially active M genes that appear on the basis of sequence analysis to be part of Type I or Type III R-M systems. These are more difficult to assay biochemically. These findings of multiple active M genes without corresponding R genes reinforces previous warnings that restriction cannot be assumed just because site-specific modifications are found (28).
In the case of the Type II systems that have both an R gene and an M gene, but only the M gene is active, we do not presently have data to indicate whether the R gene could be reactivated in a simple fashion. For instance, if the R gene was just one or two mutations away from being active, then these systems might be expected to play a biological role, because within a clonal population active alleles would probably exist. If this were the case, then it would provide an explanation for the maintenance of activity of the M gene because only those members of the population with an active M gene could survive the reactivation of the R gene. The other formal possibility is that the R genes are many mutations away from being fully active and we are observing the slow disappearance of the system. Clearly, loss of the system must first involve the loss of the R gene, because loss of the M gene while the R gene was still active would be a lethal event, and would not be seen. In the latter case, the six systems in Table 2 with active M and inactive R genes will eventually disappear from the strains; unless, of course, the M genes convey some selective advantage outside of their potential role in R-M systems. What that role might be is presently unknown, but given the natural competence of H. pylori and the apparently widespread distribution of R-M systems within naturally occurring isolates, perhaps we are observing the selfish nature of R-M systems in action (29). In this scenario, a system that contained an active M gene and an inactive R gene could easily move into another strain and become established, awaiting either the reactivation of the R gene or the appearance of a new, fully active R gene. In this view, strains of H. pylori could be viewed as a reservoir for R-M systems, which once established in one strain would spread to other strains and remain established either as fully active systems or in latent form. Certainly, current studies of the distribution of active R-M systems in H. pylori would confirm this view, because by directly assaying for active R genes more than 30 different specificities have been discovered, including active examples of all of the identifiable dead genes in strains J99 and 26695 (23).
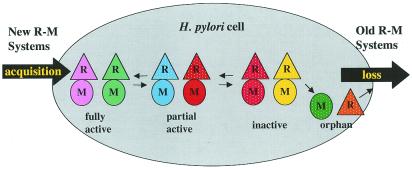
Some clues that could help decide among these possibilities can be found by examining the nature of the mutations. For instance, in the case of the GATC specificity, the R gene in J99 is inactive as a result of multiple frameshifts. In the case of the GCGC specificity, the R gene in 26695 has been lost completely. For the GANTC specificity, which is fully active in 26695, the R gene in J99 has 20 amino acid differences, but we do not know whether one, two, or more of these are responsible for the loss of activity. Finally, the GAAGA specificity, which is fully active in 26695 has only an inactive remnant of an R gene in J99 and in this case, the inactivation appears to have been caused by a sequence TAAAAAA, having been converted to a stretch of eight A residues. This is reminiscent of the phenomenon of phase variation (4, 12), although the run of A residues may be a little short to qualify as a bona fide possibility. In all of the cases above, it seems more likely that the R genes are in the process of disappearing from the strain rather than being available for reactivation. Thus, the picture that is emerging of H. pylori is one in which Type II R-M systems are widespread throughout these strains, suggesting that they are easily acquired because of the natural competence of the strain (30), but that once acquired they are also easily lost through mutation. Therefore, the set of four active systems may reflect the need for a few active systems to overcome the consequence of natural competence, combined with the propensity of R-M systems to act as selfish elements and the natural destruction of these systems by frameshift and point mutations and deletion events. As with other gene systems studies in H. pylori, the mutation rates are probably quite high and the whole genome is probably quite fluid (3). On the one hand, H. pylori might keep acquiring new R-M systems through horizontal gene transfer. On the other hand, H. pylori might lose existing R-M systems by de novo mutation and genetic recombination. The current study may provide a snapshot of the dynamic process of acquisition, mutational inactivation and loss of these numerous R-M systems (Fig. 5). Diversified R-M systems in different H. pylori strains may be maintained during this process.
Figure 5.
Schematic diagram showing the dynamic acquisition and loss of R-M system genes. Different R-M systems are shown in different colors. Triangles, R gene; circles, M gene. Orphan genes are not connected to each other. Functional genes are shown in solid colors and inactive genes are patterned with white dots.
Acknowledgments
We are grateful to Douglas Berg and Martin Blaser for providing H. pylori 26695 genomic DNA. We would like to thank A. Janulaitis and A. Lubys for sharing unpublished MnlI results; Shawn Stickel, Lauren Higgins, Nicole Porter, and Michael Dalton for technical help; Ira Schildkraut, Richard Morgan, and Elisabeth Raleigh for helpful discussions; and Donald Comb for his support. Part of this work was supported by National Institutes of Health Grant GM56535 (to R.J.R.).
Abbreviations
- R
restriction
- M
modification
- m6A
N6-methyladenine
- m4C
N4-methylcytosine
- m5C
N5-methylcytosine
References
- 1.Blaser M J. Sci Am. 1996;274(2):104–107. doi: 10.1038/scientificamerican0296-104. [DOI] [PubMed] [Google Scholar]
- 2.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 3.Wang G, Humayun M Z, Taylor D E. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]
- 4.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 5.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, et al. Nature (London) 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 6.Doig P, de Jonge B L, Alm R A, Brown E D, Uria-Nickelsen M, Noonan B, Mills S D, Tummino P, Carmel G, Guild B C, et al. Microbiol Mol Biol Rev. 1999;63:675–707. doi: 10.1128/mmbr.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson G G, Murray N E. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 8.Peek RM, Jr, Thompson S A, Donahue J P, Tham K T, Atherton J C, Blaser M J, Miller G G. Proc Assoc Am Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 9.Figueiredo C, Quint W G, Sanna R, Sablon E, Donahue J P, Xu Q, Miller G G, Peek RM, Jr, Blaser M J, van Doorn L J. Gene. 2000;246:59–68. doi: 10.1016/s0378-1119(00)00054-8. [DOI] [PubMed] [Google Scholar]
- 10.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Bolle X, Bayliss C D, Field D, van de Ven T, Saunders N J, Hood D W, Moxon E R. Mol Microbiol. 2000;35:211–222. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 12.Saunders N J, Peden J F, Hood D W, Moxon E R. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 13.Davies J. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 14.Stroeher U H, Manning P A. Trends Microbiol. 1997;5:178–180. doi: 10.1016/s0966-842x(97)85010-x. [DOI] [PubMed] [Google Scholar]
- 15.King G, Murray N E. Trends Microbiol. 1994;2:465–469. doi: 10.1016/0966-842x(94)90649-1. [DOI] [PubMed] [Google Scholar]
- 16.Akopyants N S, Fradkov A, Diatchenko L, Hill J E, Siebert P D, Lukyanov S A, Sverdlov E D, Berg D E. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersulyte D, Mukhopadhyay A K, Shirai M, Nakazawa T, Berg D E. Nucleic Acids Res. 2000;28:3216–3223. [Google Scholar]
- 18.Kong H, Lin L F, Porter N, Stickel S, Byrd D, Posfai J, Roberts R J. Nucleic Acids Res. 2000;28:3216–3223. doi: 10.1093/nar/28.17.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steen R, Dahlberg A E, Lade B N, Studier F W, Dunn J J. EMBO J. 1986;5:1099–1103. doi: 10.1002/j.1460-2075.1986.tb04328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson G G. Methods Enzymol. 1992;216:259–279. doi: 10.1016/0076-6879(92)16026-g. [DOI] [PubMed] [Google Scholar]
- 21.Klimasauskas S, Timinskas A, Menkevicius S, Butkiene D, Butkus V, Janulaitis A. Nucleic Acids Res. 1989;7:9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard K A, Card C, Benner J S, Callahan H L, Maunus R, Silber K, Wilson G, Brooks J E. Nucleic Acids Res. 1986;14:7939–7951. doi: 10.1093/nar/14.20.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts R J, Macelis D. Nucleic Acids Res. 2000;28:306–307. doi: 10.1093/nar/28.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q, Morgan R D, Roberts R J, Blaser M J. Proc Natl Acad Sci USA. 2000;97:9671–9676. doi: 10.1073/pnas.97.17.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degtyarev S Kh, Prikhod'ko E A, Prikhod'ko G G, Krasnykh V N. Nucleic Acids Res. 1993;21:2015. doi: 10.1093/nar/21.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bocklage H, Heeger K, Muller-Hill B. Nucleic Acids Res. 1991;19:1007–1013. doi: 10.1093/nar/19.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong H. J Mol Biol. 1998;279:823–832. doi: 10.1006/jmbi.1998.1821. [DOI] [PubMed] [Google Scholar]
- 28.Ando T, Xu Q, Torres M, Kusugami K, Israel D A, Blaser M J. Mol Microbiol. 2000;37:1052–1065. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- 29.Kusano K, Naito T, Handa N, Kobayashi I. Proc Natl Acad Sci USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]