Abstract
Lactic acid bacteria (LAB) are responsible for olfactory changes in wine during malolactic fermentation (MLF). A side characteristic of MLF is the release of grape derived aroma compounds from their glycosylated precursors by β-glycosidase activities of these bacteria. Apart from Oenococcus oeni, which is regarded as the most promising species for MLF, glycosidic activities have also been observed in wine related members of the genera Lactobacillus and Pediococcus. Nevertheless, information on the involved enzymes including their potential use in winemaking is limited. In this study we report that β-glucosidases with similar protein sequences can be identified in the genomes of Lactobacillus brevis, O. oeni and Leuconostoc mesenteroides. TTG serves as start codon for the glucosidase gene of O. oeni. The β-glucosidase of O. oeni ATCC BAA-1163 was expressed in E. coli and partially characterized. The enzyme displayed characteristics similar to β-glucosidases isolated from L. brevis and L. mesenteroides. A pH optimum between 5.0 and 5.5, and a Km of 0.17 mmol L−1 pNP-β-D-glucopyranoside were determined. A glycosyltransferase activity was observed in the presence of ethanol. The enzyme from O. oeni was capable to hydrolyze glycosides extracted from Muskat wine. This study also contains a report on glycosidase activities of several LAB species including Oenococcus kitaharae.
Introduction
Although the majority of aroma and flavor compounds of wine are produced during the alcoholic fermentation, the varietal bouquet is determined mainly by volatile constituents derived from the grapes. Apart from the free flavor compounds found in the berries, a significant amount is present as odorless non-volatile glycosides (Maicas and Mateo 2005; Ribéreau-Gayon et al. 2006). Many of these aroma compounds develop in wine during fermentation and ageing (Mateo and Jimenez 2000; Ribéreau-Gayon et al. 2006). This is especially obvious in grape varieties like Chardonnay, Semillon, Pinot Gris, Trebbiano and most of the red varieties where the typical varietal aroma is only formed during yeast fermentation (Ugliano 2009).
Enzymatic hydrolysis of these glycosides could enhance the natural aroma spectrum of wine or fruit juices and has therefore often been of considerable interest in wine research (Günata et al. 1998; Mateo and Jiménez 2000).
Recent studies revealed that lactic acid bacteria (LAB) can hydrolyze glycosylated aroma precursors in the course of malolactic fermentation (MLF). Several studies have shown that Oenococcus oeni possesses such enzymatic activities (Grimaldi et al. 2000; Ugliano et al. 2003; D'Incecco et al. 2004). However, the precursor hydrolysis during MLF does not result in stoichiometric increase in the concentration of the corresponding volatile compounds (Boido et al. 2002; Ugliano and Moio 2006), implying that other processes cause partial loss of the liberated aglycons during fermentation (Boido et al. 2002; D'Incecco et al. 2004). Glycosidase activities have also been reported for wine related strains of Lactobacillus spp. and Pediococcus spp. (Grimaldi et al. 2005). The study of glycosidases from LAB for aroma release could be of interest because most fungal glycosidases present in pectolytic preparations were reported to be strongly inhibited under wine conditions, especially by glucose and the high acidity (Maicas and Mateo 2005). Nevertheless, previous studies on glycosidase activities of wine LAB mainly considered the potential of the intact cells, little information exists on the nature of the responsible enzymes.
β-glucosidases (EC 3.2.1.21) have already been isolated from Lactobacillus casei (intracellular; Coulon et al. 1998) and Lactobacillus plantarum (extracellular; Sestelo et al. 2004). In our recent study (Michlmayr et al. 2009) we identified an intracellular enzyme from Lactobacillus brevis which displayed functional similarities to a β-glucosidase previously isolated from Leuconostoc mesenteroides (Gueguen et al. 1997). Based on the protein sequence of the enzyme from L. brevis we report the presence of a similar sequence in the genome of O. oeni. Further, the heterologous expression of this enzyme in E. coli and its initial characterization are described. Finally, we discuss the glycosidase activities of several wine related species under different growth conditions, including the first report on glycosidic activity of the newly described species Oenococcus kitaharae (Endo and Okada 2006).
MATERIAL AND METHODS
In silico studies
The identity of the β-glucosidase from L. brevis (Michlmayr et al. 2009) was determined by LC-ESI-QTOF-MS/MS analysis of its tryptic peptides as described by Mattanovich et al. (2009). The resulting peptide sequences (YPATLMTSYNK and AVNTGELDEGTLNK) closest matched a sequence annotated as “beta glucosidase related glycosidase” (GenBank accession no. YP_796051) from L. brevis strain ATCC 367. A search for similar sequences was conducted in the NCBI Database (GenBank) using the BLASTp algorithm. Sequences were aligned with T-Coffee (Notredame et al. 2000; http://www.ebi.ac.uk/Tools/t-coffee) using the Blosum matrix. A distance-matrix (neighbour-joining) tree was constructed with the Phylip package (Protdist, Neighbor; Felsenstein 1993; http://bioweb2.pasteur.fr/phylogeny). The software used to visualize the tree was iTOL (Letunic and Bork 2007). The source for information on membrane transporters was TransportDB (Ren et al. 2007; http://www.membranetransport.org).
Construction of expression vectors for the β-glucosidase from O. oeni
PCR was performed with the Phusion Master Mix (New England Biolabs, Ipswich, MA, USA) according to the suppliers specifications. PCR products were purified with the Wizard SV Gel and PCR Clean-UP Kit (Promega, Madison, WI, USA). The restriction enzymes NdeI and XhoI and the Quick ligation kit were also purchased from New England Biolabs. Plasmid purification was done with the GenElute Plasmid Miniprep Kit from Sigma (St. Louis, MO).
All primer sequences are listed in table 1.
Table 1.
Primers used for the construction of expression vectors
| Primer | Primer sequence | Restriction site |
Vector | Construct |
|---|---|---|---|---|
| OnbglF | 5′- CTTGCTTTATTCGCCAATTT -3′ | - | ||
| OnbglR | 5′- TCTGCCGTTTCAAATAACGA -3′ | - | ||
| Onbgl2F | 5′ - GATATACATATGTCTAAGATTACTTCAATTA -3′ | NdeI | pET-16b | pHM1 |
| Onbgl2R | 5′- CGGATCCTCGAGTTAACTTTGATTGGCGAG -3′ | XhoI | ||
| Onbgl3F | 5′ - CATATGTCTAAGATTACTTCAATTA - 3′ | NdeI | pET-21a | pHM2ATG |
| Onbgl3R | 5′ - CTCGAGACTTTGATTGGCGAG - 3′ | XhoI | ||
| TTGF | 5′ - AGATATACATTTGTCTAAGATTACTTC - 3′ | - | pET-21a | pHM2TTG |
| TTGR | 5′ - ATGTATATCTCCTTCTTAAAGTTAAAC - 3′ | - | ||
| Onbgl4F | 5′ - CATATGATGGTTTCCGATGGA - 3′ | NdeI | pET-21a | pHM3 |
| LbbglF | 5′ - GATATACATATGGACATCGAACGAACGC - 3′ | NdeI | pET-21a | pHM4 |
| LbbglR | 5′ - GTGGTGCTCGAGTTGACGTAATAAGGTGTTTGC - 3′ | XhoI |
The coding region for the putative β-glucosidase (GenBank Accession No. ZP_01543735) from O. oeni ATCC BAA-1163 was amplified with the primers OnbglF and OnbglR and sequenced. The fragment was then used as template for a PCR with primers Onbgl2F and Onbgl2R, designed with restriction sites for NdeI and XhoI, respectively. Primer Onbgl2F was designed to change the original start codon (TTG) into ATG. The fragment was cloned into the expression vector pET-16b (Novagen, Madison, WI, USA) with the N-terminal His10-tag in frame (construct pHM1).
Alternatively the gene was cloned into pET-21a with a C-terminal His6-tag. Therefore, the fragment amplified with primers Onbgl3F and Onbgl3R was cloned into the pJet1.2 blunt cloning vector (Fermentas, St. Leon-Rot, Germany). This construct was used to transform E. coli NEB 5-alpha cells (New England Biolabs). The plasmid was purified from positive transformants, the insert was cut out with NdeI and XhoI and cloned into pET-21a (pHM2ATG). Primers TTGF and TTGR were designed specifically for the construct pHM2ATG to change the start codon into TTG by site directed mutagenesis, thereby leading to loss of the restriction site NdeI.
To verify if the next (downstream from TTG, see fig. 2) ATG codon in frame can be used as initiation point, the gene was amplified with primers Onbgl4F and Onbgl3R and cloned into pET-21a (pHM3) by the same procedure as for pHM2ATG.
Fig. 2.
N-terminal nucleotide sequence of the β-glucosidase gene from O. oeni ATCC BAA-1163. A Shine-Dalgarno sequence (bold print) is located upstream of the initiation codon (TTG, position 1). The next ATG codon in frame is located at position 109.
For the construction of an expression vector for the β-glucosidase from L. brevis SK3 (pHM4) primers LbbglF and LbbglF were used to clone the gene into pET-21a.
Protein expression and purification
E. coli BL21(DE3) (Novagen) was used as expression host for all expression vectors. After electroporation positive colonies were selected on LB-Agar plates supplemented with ampicillin. Precultures were prepared by inoculating 3 mL Luria Broth containing 100 μg mL−1 ampicillin with a single colony. After 4 hours of growth at 37°C and 150 rpm, 250 mL Terrific Broth containing 100 μg mL−1 ampicillin were inoculated with this culture. After cultivation for further 4 h, lactose was added to give a final concentration of 5 g L−1. Following overnight cultivation at 25°C and 150 rpm the cells were harvested by centrifugation at 6,000 g for 20 min, the cells were resuspended in buffer A for protein purification (see below) and disrupted in a French press at 8.3 MPa in 3 cycles. The homogenate was centrifuged at 100,000 g for 30 min, the supernatant was recovered as crude extract.
The recombinant β-glucosidase was purified by immobilized metal affinity chromatography on a column (20 mL) packed with Profinity IMAC Ni-Charged Resin (Bio-Rad Inc., Hercules, CA, USA). The column was pre-equilibrated with buffer A (20 mmol L−1 KH2PO4, 500 mmol L−1 KCl, 20 mmol L−1 imidazole; pH 7.0). Then the crude extract was applied to the column at a flow rate of 1 mL min−1. After washing the column with buffer A (3 column volumes) the enzyme was eluted at a rate of 2 mL min−1 with buffer B (20 mmol L−1 KH2PO4, 500 mmol L−1 KCl, 500 mmol L−1 imidazole; pH 7.0). Active fractions were pooled, desalted and concentrated in 20 mmol L−1 citrate-phosphate buffer (pH 7.0) using Amicon Ultra Centrifugal Filter devices (Millipore, Billerica, MA) and stored at 4°C. Protein concentrations were determined using the Protein Assay kit from Bio-Rad.
The molecular weight was determined by gel filtration on a Sephacryl S-300 column (GE Healthcare) with 100 mmol L−1 citrate-phosphate buffer pH 7.0 containing 150 mmol L−1 NaCl at a flow rate of 0.1 cm min−1. The column was calibrated with the Kit for Molecular Weights 29,000-700,000 (Sigma).
SDS-PAGE including coomassie staining was performed on the PHAST System (GE Healthcare Uppsala, Sweden) with precast gels (PhastGel Gradient 8-25. The molecular weight marker was High Precision Dual Colour (Bio-Rad) with 10–250 kDa range.
Extraction of glycosides from wine
Isolation and quantification of glycosides was performed following Williams et al. (1995). 750 mL of Muskat wine (Muskat Ottonel Classic 2006, Austria) were dealcoholised and reduced in volume to 200 mL by evaporation (Heidolph LABOROTA 4010, Schwabach, Germany). The concentrated wine was applied to a column packed with 40 mL of a C-18 reversed phase adsorbent (Amberlite XAD 16, Rohm and Haas, Chauny, France) with a flow rate of 0.3 mL min−1. The column was washed with 400 mL of water at a flow of 1.0 mL min−1, then the retained glycosides were eluted with ethanol at a flow of 0.5 ml min−1 and collected in fractions of 10 mL. The glycoside content was quantified by the glycosyl-glucose assay as described by Williams et al. (1995). 500 μL of 2.25 M H2SO4 were added to 250 μL of each fraction and incubated at 100°C for 1 h. To determine the free (nonglycosidic) glucose, 250 μL of each fraction were diluted with 500 μL HQ water and kept at room temperature. The D-glucose concentration of each sample was determined by HPLC analysis (Dionex DX500, Sunnyvale, CA, USA) with a CarboPac PA 1 column and pulsed amperometric gold electrode detection. The flow was 1.0 mL min−1, injection volume 25 μL and 15 mmol L−1 NaOH was used for elution.
The fractions with the highest glycosyl-glucose concentrations were combined and diluted in HQ water. Ethanol was removed by evaporation, and the samples were concentrated to a final volume of 10 ml. The glycosyl-glucose concentration of the final fraction was 6.6 mmol L−1.
Cultivation of lactic acid bacteria
The strains used to study their glucosidase activities were O. oeni ATCC BAA-1163, L. plantarum DSM 20174, Oenococcus kitaharae DSM 17330, L. mesenteroides DSM 20193, L. brevis DSM 20054, and the strains L. brevis SK3, L. mesenteroides SK1/2, Pediococcus. acidilacti SK 1/1 and Pediococcus pentosaceus 784a which were isolated in our laboratory. The strains identities were verified by 16S-ARDRA as previously described (Michlmayr et al., 2009). The strains were grown on de Man/Rogosa/Sharpe (MRS) broth (DSMZ medium 11: tryptone 10 g, meat extract 10 g, yeast extract 5 g, glucose 20 g, Tween 80 1 mL, K2HPO4 2g, Na-acetate 5 g, (NH4)2 citrate 2 g, MgSO4·7H2O 0.2 g, MnSO4·H2O 0.05 g, in one liter). For growth of O. oeni and O. kitaharae cysteine-HCl (0.05 %) and pantothenic acid (1 mg L−1) were added. Precultures were prepared by inoculating 10 mL MRS and incubation at 25°C for 3 to 4 days. 0.5 mL of these precultures were used to inoculate 50 mL modified medium with glucose (20 g L−1), cellobiose (20 g L−1) or apple juice (20% v/v) as carbon source. The apple juice was autoclaved separately and then added to the sterile MRS base. The sugar content of the apple juice as determined by HPLC was glucose 16 g L−1, fructose 49 g L−1, xylose 11 g L−1.
After 4 days (7 days for O. oeni, O. kitaharae) of growth the cells were harvested by centrifugation (6000 g, 5 min.), washed twice with 0.85% NaCl and resuspended in 0.85% NaCl for the determination of cell-bound glycosidase activities. The cells were kept on ice. For the determination of intracellular enzyme activities the harvested cells were resuspended in 20 mmol L−1 citrate-phosphate buffer (pH 7.0) and kept on ice. The cells were disrupted by sonication. The cell debris were separated by centrifugation (16,100 g, 30 min), and the cleared supernatants were assayed for β-glucosidase activity.
Glycosidase assays
Activity assays and kinetic measurements were performed as previously described (Michlmayr et al. 2009). The substrates used were: pNP-β-D-glucopyranoside (pNPβGlc), pNP-β-D-xylopyranoside (pNPβXyl), pNP-α-L-arabinopyranoside (pNPαAra) and pNP-α-L-rhamnopyranoside (pNPαRha). The enzyme solutions were stored in 20 mmol L−1 citrate phosphate buffer (pH 7.0). Unless otherwise mentioned, the reaction conditions were: 100 mmol L−1 citrate-phosphate buffer pH 5.5, substrate concentration 10 mmol L−1. The assays were incubated for 10 min at 37°C, then stopped with 400 μL 0.5 mol L−1 Na2CO3. The absorbance of p-nitrophenol was measured at 400 nm (ε400 = 18.300 M−1 cm−1 at pH 10.2) in a Beckman DU 800 spectrometer (Paolo Alto, CA, USA). All assays were performed in duplicate.
For assays with intact biomass (cell-bound glucosidase activities) the cell suspensions were standardized to an OD600 of 1, the assays were incubated for 1 h at 37°C. Prior to measurement, the cells were removed by centrifugation (6000 g, 5 min).
One unit of glucosidase activity corresponds to 1 μmol of p-nitrophenol released per min at 37°C. Cell-bound activities are expressed as units per grams of dry cell mass.
RESULTS AND DISCUSSION
Identification of a β-glucosidase (bgl) gene from Oenococcus oeni
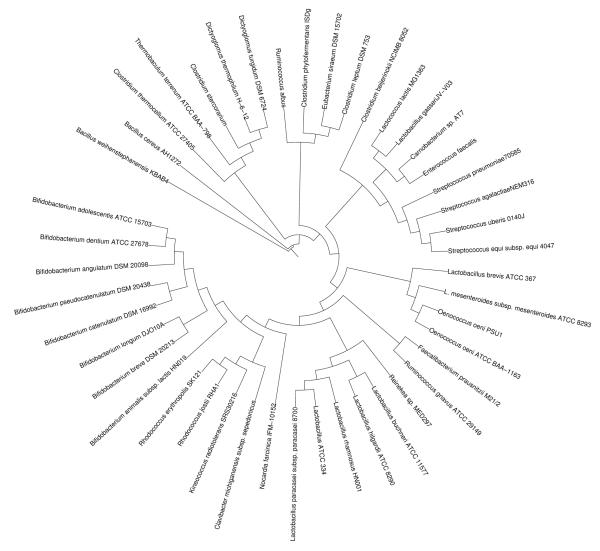
Peptide sequences from the β-glucosidase of L. brevis (Michlmayr et al. 2009) matched a sequence of L. brevis ATCC 367 annotated as “beta glucosidase related glycosidase” (GenBank accession no. YP_796051). Fig.1 shows a phylogenetic tree constructed from related protein sequences obtained by a BLASTp search in the NCBI database. Noticeable is the high occurrence of the gene in intestinal bacteria including members of the genera Enterococcus, Bifidobacterium, Clostridium and Lactobacillus. No similar sequences could be found in the genomes of L. plantarum and Pediococcus spp.. Interestingly, the closest similarities were found in the genomes of L. mesenteroides subsp. mesenteroides (accession no. YP_818356) and O. oeni (ZP_01543735, YP_811088).
Fig. 1.
Distance-matrix tree of β-glucosidases. Protein sequences were aligned with T-coffee (Notredame et al. 2000). The neighbour-joining tree was created with Phylip (Felsenstein 1993).
According to the predicted protein sequence, the β-glucosidase (bgl) of O. oeni belongs to the glycoside hydrolase family 3. Members of this family show a broad specificity toward glycosylated plant metabolites. Based on topology prediction software (http://www.expasy.ch/tools/#proteome) the protein can be considered to be cytoplasmic, since no transmembrane regions or target sequences are predicted.
The bgl gene of O. oeni is initiated with the rare start codon TTG. While there is no possible start codon (ATG) upstream of this position, expression of the gene initiated with the next ATG codon in frame downstream resulted in inactive protein. As shown in figure 2, a ribosomal binding site (AAGGAG) is located upstream of the TTG codon. Expression in E. coli was successful using both ATG or TTG as start codon when pET-21a was used as expression vector (constructs pHM2ATG and pHM2TTG, see table 1).
Heterologous expression and characterization of the β-glucosidase from O. oeni
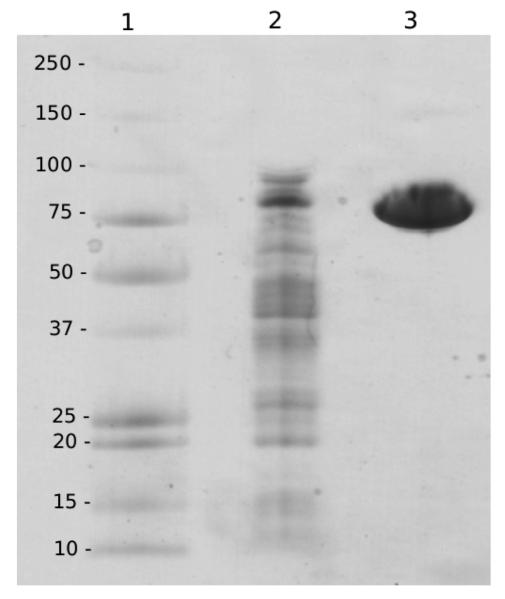
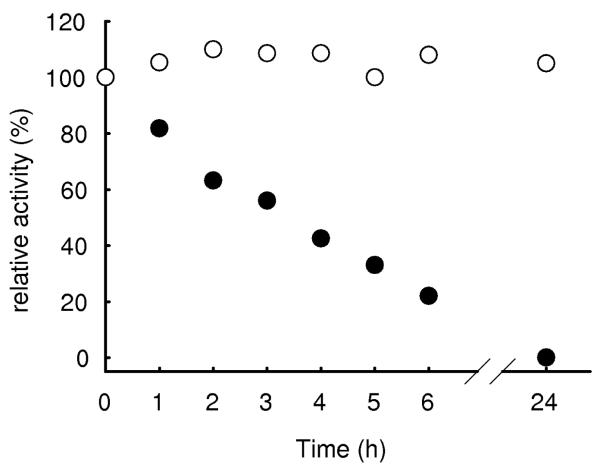
The gene from O. oeni ATCC BAA-1163 (ZP_01543735) was cloned into an expression vector (pET-16b, Novagen) and expressed in E. coli. Lactose induced expression of a 250 mL culture yielded 4,500 U of β-glucosidase with a specific activity of 9.7 U mg−1. After purification the enzyme was electrophoretically pure and showed a molecular weight of ca. 80 kDa per subunit on the SDS gel (fig. 3). A molecular weight of 140 kDa was estimated by gel filtration, indicating that the enzyme is a homo-dimer. Although no activity loss was recorded within one month of storage of a stock solution (0.12 mg protein ml−1), the enzyme was instable when diluted to measuring concentrations (half life ca. 3 h, figure 4). This is in contrast to the observation that the β-glucosidase from L. brevis is tetrameric and displayed good stability (Michlmayr et al. 2009). A possible explanation could be improper protein folding due to the N-terminal His-tag of pET-16b, nevertheless, when pET-21a (His-tag C-terminal) was used, the same results were achieved. Furthermore, when the β-glucosidase from L. brevis was expressed with pET-21a, the resulting enzyme was tetrameric (310 kDa) and displayed the same stability as the native enzyme (data not shown). Also expression at lower temperatures (16°C) didn't result in better stability. For further characterization, the enzyme could be stabilized by the addition of bovine serum albumin (BSA, 0.1 mg/ml) to the dilution buffer. Fig. 4 shows stability in buffer with and without BSA.
Fig. 3.
SDS PAGE with coomassie blue staining. Lane 1: High Precision Dual Colour molecular weight marker (kDa). Lane 2: crude extract after expression of the β-glucosidase from O. oeni in E. coli (4 μg). Lane 3: recombinant β-glucosidase from O. oeni after IMAC purification (4 μg).
Fig. 4.
Stabilizing effect of bovine serum albumine (BSA) onthe recombinant β-glucosidase from O. oeni. The enzyme (~ 0.6 μg ml−1) was stored in 20 mM citrate-phosphate buffer pH 7.0 with 0.1 mg ml−1 BSA (○) and in buffer without BSA (●).
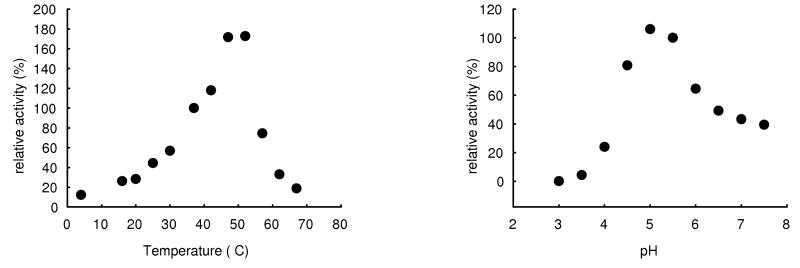
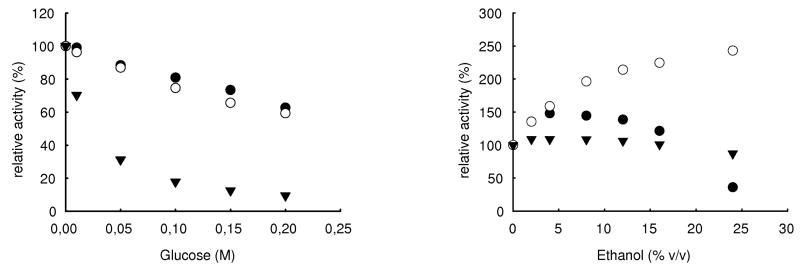
Specific activities of the β-glucosidase expressed with pET-16b were 81 U mg−1 for pNPβGlc, 46 U mg−1 for pNPβXyl and 5.7 U mg−1 for pNPαAra, rhamnosidase activity was not detected. A pH optimum between 5.0 and 5.5 (fig. 5a) and a Km of 0.17 mmol L−1 were determined for pNPβGlc. Maximum activity was observed at ca. 50°C. Figure 6 shows the influence of ethanol and glucose on the β-glucosidases from O. oeni, L. brevis and A. niger (crude preparation, Sigma). While the glucosidase from A. niger is not influenced by ethanol, the bacterial enzymes seem to be activated. Unlike the bgl of L. brevis, the glucosidase from O. oeni is denaturated in higher ethanol concentrations, probably because of improper folding as mentioned above. Fig. 6 also shows that the glucosidase from A. niger is strongly inhibited by glucose, while the bacterial enzymes still retain 60% of initial activity in 0.2 mol L−1 (3.6% w/v) glucose.
Fig. 5.
pH (a) and temperature dependance (b) of the recombinant β-glucosidase from O. oeni.
Fig. 6.
Influence of ethanol (a) and glucose (b) on the β-glucosidases from Oenococcus oeni (●), Lactobacillus brevis (○) and Aspergillus niger (▼) (crude preparation, Sigma). 100 % of relative activity refers to the specific activities of the pure enzymes.
In tests with a glycoside extract from wine (Muskat), all enzymes (1 U mL−1 in the assay) were able to release 50% of the total bound glucose (Table 2) after 30 minutes of reaction. Under the same conditions, the enzymes from O. oeni and L. brevis showed only low activity towards cellobiose.
Table 2.
Release of glucose from different substrates by the β-glucosidases from Lactobacillus brevis (native), Oenococcus oeni (recombinant) and Aspergillus niger (crude preparation) after 30 min reaction with 1 U mL−1 in the assay
| Substrate | Substrate concentration (mmol L−1) |
L. brevisa (mmol L−1) |
O. oenia (mmol L−1) |
A. nigera (mmol L−1) |
|---|---|---|---|---|
| n-Octyl-β-D-glucoside | 10 | 10.4 | 9.9 | 6.9 |
| Cellobioseb | 10 | 3.6 (1.8) | 0.13 (0.063) | 19.3 (9.63) |
| Wine extract | 1.7 | 0.88 | 0.92 | 0.95 |
Amount of glucose detected by HPLC analysis
The values in parentheses refer to the amount of hydrolysed cellobiose
Glucosidase activities of LAB
Several species with possible interest for winmaking including Oenococcus kitaharae (Endo and Okada 2006) were assayed for cell-bound and intracellular β-glucosidase activities after growth in different media (table 3). According to the current data (GenBank), among the tested strains only L. brevis, L. mesenteroides subsp. mesenteroides and O. oeni possess glucosidase genes as described above. It is therefore interesting that only these species exhibited detectable intracellular activities after growth in medium with glucose or apple juice. Unfortunately there is no genomic information available on O. kitaharae, which displayed no glucosidase activity at all when grown on glucose or apple juice.
Table 3.
Cell-bound and intracellular activities of LAB after growth on different media. Activities are displayed as U per gram of dry cell mass. Undetectable activity is indicated by “-”
| Glucose | Apple juice | Cellobiose | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| cells | extract | cells | extract | cells | extract | |
| Lactobacillus brevis SK3 | 3.3 | 9.7 | 27 | 45 | 11 | - |
| L. brevis DSM 20054 | 6.9 | 54 | 15 | 66 | 4.6 | - |
| Leuconostoc mesenteroides SK1/2 a | 25 | 15 | 99 | 72 | 15 | - |
| L. mesenteroides DSM 20193 b | - | - | 2.6 | - | 120 | - |
| Oenococcus oeni ATCC BAA-1163 | 13 | 13 | 14 | 14 | 18 | 16 |
| Oenococcus kitaharae DSM 17330 | - | - | - | - | 174 | - |
|
Lactobacillus plantarum DSM 20174 |
6.8 | - | 8.2 | - | 20 | - |
| Pediococcus pentosaceus 784a | 1.3 | - | 12 | - | 43 | - |
| Pediococcus acidilactici SK1/1 | 2.2 | - | 2.1 | - | 16 | - |
L. mesenteroides subsp. mesenteroides
L. pseudomesenteroides
On the other hand, all strains grown on cellobiose (except O. oeni ATCC BAA-1163) showed high cell bound, but undetectable intracellular activities. This indicates a distinction between intracellular and parietal (possibly membrane bound) mechanisms which could serve different metabolic functions. Considering the currently available genetic information, it seems reasonable to propose that glycosidic activities of LAB could also be caused by the membrane associated sugar phosphotransferase system (PTS) which plays a key role in procaryotic carbohydrate metabolism (Barabote and Saier 2005). Phosphoglucosidases (EC 3.2.1.86) of LAB are thought to be part of a PTS specific for cellobiose or lactose (De Vos and Vaughan 1994). Nagaoka et al. (2008) reported the expression of 5 distinct phosphoglycosidases in Lactobacillus gasseri after growth on lactose. The genome of O. oeni PSU-1 contains four genes encoding for phosphoglucosidases (YP_809876, YP_809979, YP_809980, YP_810767, and several transporter proteins specific for cellobiose and β-glucosides (PTS system IIABC components; TransportDB, http://www.membranetransport.org). Nevertheless, O. oeni ATCC BAA-1163 was hardly able to grow on cellobiose (OD600 = 0.4 after 7 days), although this strain has at least one operon for a cellobiose specific PTS (accession NZ_AAUV01000047, loci OENOO_52043 - OENOO_52046). In contrast to L. brevis and L. mesenteroides this strain also displayed high intracellular β-glucosidase activity after growth on cellobiose.
CONCLUSIONS
Considering the data presented in this study, there are two major implications:
First, the β-glucosidase of O. oeni is capable to act on grape derived glycosides. It remains to be shown whether this enzyme or its possible homologues from L. brevis and L. mesenteroides can be used as biocatalysts for the aroma release in wine or fruit juices. Although the tolerance to ethanol and glucose prove useful, immobilization techniques may be required to reach stability in wine conditions. It is also worth considering the possibility to use food-grade expression hosts like Lactobacillus plantarum for over-expression of such enzymes.
Secondly, it's not quite clear where the metabolic function of these enzymes lies. Intracellular activities in O. oeni, L. mesenteroides and L. brevis which could be ascribed to the presented enzyme family were high after growth in rich media (glucose, apple juice), but only detectable in O. oeni ATCC BAA-1163 when the cells were grown on cellobiose as carbon source. Furthermore, the recombinant enzyme from O. oeni showed only low activity on cellobiose. A possible function of such glycosidases might be the hydrolysis of plant glycosides detrimental for the growth of LAB, rather than the supply of glucose in depleted media. It would be interesting to monitor the expression levels of both the β-glucosidase and the phosphoglucosidases of O. oeni in glucose rich and glucose depleted media as well to achieve a better understanding of their metabolic role.
ACKNOWLEDGEMENTS
The authors appreciate the support given to KD Kulbe by the Austrian Science Fund (FWF project 20246-B11) and the Research Centre Applied Biocatalysis, Graz. We thank Prof. G. Spano for his invaluable scientific advice. Many thanks to Viktoria Hell who did the HPLC work and to Johannes Stadlmann from the Department of Chemistry (University of Natural Resources and Applied Life Sciences Vienna, Austria) for doing the LC-ESI-QTOF-MS/MS analysis.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boido E, Lloret A, Medina K, Carrau F, Dellacassa E. Effect of β-glycosidase activity of Oenococcus oeni on the glycosylated flavor precursors of Tannat wine during malolactic fermentation. J Agric Food Chem. 2002;50:2344–2349. doi: 10.1021/jf0109367. [DOI] [PubMed] [Google Scholar]
- Coulon S, Chemardin P, Gueguen Y, Arnaud A, Galzy P. Purification and characterization of an intracellular β-glucosidase from Lactobacillus casei ATCC 393. Appl Biochem Biotechnol A. 1998;74:105–114. [Google Scholar]
- De Vos WM, Vaughan EE. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- D'Incecco N, Bartowsky E, Kassara S, Lante A, Spettoli P, Henschke P. Release of glycosidically bound flavour compounds of Chardonnay by Oenococcus oeni during malolactic fermentation. Food Microbiol. 2004;21:257–265. [Google Scholar]
- Endo A, Okada S. Oenococcus kitaharae sp. nov., a non-acidophilic and non-malolactic-fermenting oenococcus isolated from a composting distilled shochu residue. Int J Syst Evol Microbiol. 2006;56:2345–2348. doi: 10.1099/ijs.0.64288-0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington; Seattle: 1993. [Google Scholar]
- Grimaldi A, Bartowsky E, Jiranek V. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J Appl Microbiol. 2005;99:1061–1069. doi: 10.1111/j.1365-2672.2005.02707.x. [DOI] [PubMed] [Google Scholar]
- Grimaldi A, McLean H, Jiranek V. Identification and partial characterization of glycosidic activities of commercial strains of the lactic acid bacterium, Oenococcus oeni. Am J Enol Vitic. 2000;51:362–369. [Google Scholar]
- Gueguen Y, Chemardin P, Labrot P, Arnaud A, Galzy P. Purification and characterization of an intracellular β-glucosidase from a new strain of Leuconostoc mesenteroides isolated from cassava. J Appl Microbiol. 1997;82:469–476. [Google Scholar]
- Günata Z, Blondeel C, Vallier MJ, Lepoutre JP, Sapis JC, Watanabe N. An Endoglycosidase from Grape Berry Skin of Cv. M. Alexandria Hydrolyzing Potentially Aromatic Disaccharide Glycosides. J Agric Food Chem. 1998;46:2748–2753. [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Maicas S, Mateo JJ. Hydrolysis of terpenyl glycosides in grape juice and other fruit juices: A review. Appl Microbiol Biotechnol. 2005;67:322–335. doi: 10.1007/s00253-004-1806-0. [DOI] [PubMed] [Google Scholar]
- Mateo JJ, Jiménez M. Monoterpenes in grape juice and wines. J Chromatogr A. 2000;881:557–567. doi: 10.1016/s0021-9673(99)01342-4. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Graf A, Stadlmann J, Dragosits M, Redl A, Maurer M, Kleinheinz M, Sauer M, Altmann F, Gasser B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb Cell Fact. 2009;8:29. doi: 10.1186/1475-2859-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlmayr H, Schuemann C, Barreira Braz da Silva N, Kulbe KD, del Hierro AM. Isolation and basic characterization of a β-glucosidase from a strain of Lactobacillus brevis isolated from a malolactic starter culture. J Appl Microbiol. 2009 doi: 10.1111/j.1365-2672.2009.04461.x. (article in press, doi: 10.1111/j.1365-2672.2009.04461.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S, Honda H, Ohshima S, Kawai Y, Kitazawa H, Tateno Y, Yamazaki Y, Saito T. Identification of five phospho-β-glycosidases from Lactobacillus gasseri ATCC33323T cultured in lactose medium. Biosci Biotechnol Biochem. 2008;72:1954–1957. doi: 10.1271/bbb.80089. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucl Acids Res. 2007;35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribéreau-Gayon P, Glories Y, Maujean A, Dubourdieu D. Handbook of Enology. 2nd edn. Vol. 2. John Wiley & Sons Ltd; Chichester, England: 2006. The chemistry of wine stabilization and treatments; pp. 205–227. [Google Scholar]
- Sestelo ABF, Poza M, Villa TG. β-Glucosidase activity in a Lactobacillus plantarum wine strain. World J Microbiol Biotechnol. 2004;20:633–637. [Google Scholar]
- Ugliano M. Enzymes in winemaking. In: Moreno-Arribas MV, Polo MC, editors. Wine Chemistry and Biochemistry. 1st edn. Springer Science+Business Media LLC; New York, USA: 2009. pp. 103–126. [Google Scholar]
- Ugliano M, Moio L. The influence of malolactic fermentation and Oenococcus oeni strain on glycosidic aroma precursors and related volatile compounds of red wine. J Sci Food Agric. 2006;86:2468–2476. [Google Scholar]
- Ugliano M, Genovese A, Moio L. Hydrolysis of wine aroma precursors during malolactic fermentation with four commercial starter cultures of Oenococcus oeni. J Agric Food Chem. 2003;51:5073–5078. doi: 10.1021/jf0342019. [DOI] [PubMed] [Google Scholar]
- Williams PJ, Cynkar W, Francis IL, Gray JD, Hand PG, Coombe BG. Quantification of glycosides in grapes, juices, and wines through a determination of glycosyl glucose. J Agric Food Chem. 1995;43:121–128. [Google Scholar]