The protumor functions for autophagy are largely attributed to its ability to promote cancer cell survival in response to stress. This study demonstrates an unexpected connection between autophagy and glucose metabolism that facilitates adhesion-independent growth driven by a strong oncogenic insult—mutationally active Ras.
Abstract
The protumorigenic functions for autophagy are largely attributed to its ability to promote cancer cell survival in response to diverse stresses. Here we demonstrate an unexpected connection between autophagy and glucose metabolism that facilitates adhesion-independent transformation driven by a strong oncogenic insult—mutationally active Ras. In cells ectopically expressing oncogenic H-Ras as well as human cancer cell lines harboring endogenous K-Ras mutations, autophagy is induced following extracellular matrix detachment. Inhibiting autophagy due to the genetic deletion or RNA interference–mediated depletion of multiple autophagy regulators attenuates Ras-mediated adhesion-independent transformation and proliferation as well as reduces glycolytic capacity. Furthermore, in contrast to autophagy-competent cells, both proliferation and transformation in autophagy-deficient cells expressing oncogenic Ras are insensitive to reductions in glucose availability. Overall, increased glycolysis in autophagy-competent cells facilitates Ras-mediated adhesion-independent transformation, suggesting a unique mechanism by which autophagy may promote Ras-driven tumor growth in specific metabolic contexts.
INTRODUCTION
Macroautophagy (hereafter called autophagy), which serves critical functions in maintaining cellular homeostasis and as an adaptive response to cellular stress, has both antitumor and protumor functions (Chen and Debnath, 2010). The tumor suppressor functions for autophagy were originally revealed through genetic studies of Beclin/ATG6 (Liang et al., 1999; Qu et al., 2003; Yue et al., 2003). Subsequently, multiple mechanisms have been uncovered by which autophagy potentially prevents tumor formation; these include the mitigation of genotoxic damage, suppression of a protumorigenic inflammatory response secondary to decreased necrosis, and the induction of oncogene-induced senescence (Degenhardt et al., 2006; Karantza-Wadsworth et al., 2007; Mathew et al., 2007, 2009; Young et al., 2009). In contrast, the protumorigenic functions for autophagy are almost exclusively attributed to its cytoprotective functions in tumor cells as they encounter common stresses during cancer progression (Chen and Debnath, 2010). For example, increased autophagy is observed in cells centrally located within solid tumors that lack access to nutrients and oxygen; genetic inhibition of autophagy results in the increased death of these stressed tumor cells (Degenhardt et al., 2006).
Autophagy is induced following extracellular matrix (ECM) detachment, which protects cells from detachment-induced cell death (anoikis) (Frisch and Francis, 1994; Fung et al., 2008). Because the ability to overcome anoikis is viewed as a critical hurdle in tumor development, one can hypothesize that detachment-induced autophagy enables the viability and fitness of tumor cells deprived of contact with ECM. In addition to autophagy, oncogene-mediated activation of growth factor signaling pathways also protects cells from anoikis (Gilmore, 2005); among these oncogenes, one of the most potent suppressors of anoikis is Ras, a small GTPase commonly mutated in a large number of human epithelial cancers (Khwaja et al., 1997).
In addition to its ability to inhibit anoikis, oncogenic Ras promotes glucose metabolism, which is critical for its capacity to support growth and proliferation during oncogenic transformation (Chiaradonna et al., 2006). Indeed, recent studies have begun to highlight the importance of the metabolic switch to aerobic glycolysis (termed the “Warburg effect”) that takes place in cancer cells as a critical driver of tumorigenesis (Vander Heiden et al., 2009). Remarkably, two proteins that mitigate cell stress, Oct1 and Hsf1, have both been demonstrated to facilitate tumorigenesis through the enhancement of glucose metabolism (Dai et al., 2007; Shakya et al., 2009). These studies broach important interconnections between stress pathways and cancer cell metabolism.
Surprisingly, although autophagy is similarly viewed as a salvage mechanism that affords basic components to sustain core metabolic functions during starvation or stress, the relationship between autophagy and metabolism remains largely unclear. Along with our previous work demonstrating that detachment-induced autophagy promotes cell survival during anoikis, the aforementioned studies highlighting the importance of stress pathways in modulating glucose metabolism motivated us to delineate the biological contributions of autophagy to Ras-mediated adhesion-independent transformation. Our experiments point to an unexpected requirement for autophagy competence in facilitating glycolysis, which promotes adhesion-independent transformation driven by oncogenic Ras.
RESULTS
Oncogenic Ras does not suppress ECM detachment-induced autophagy
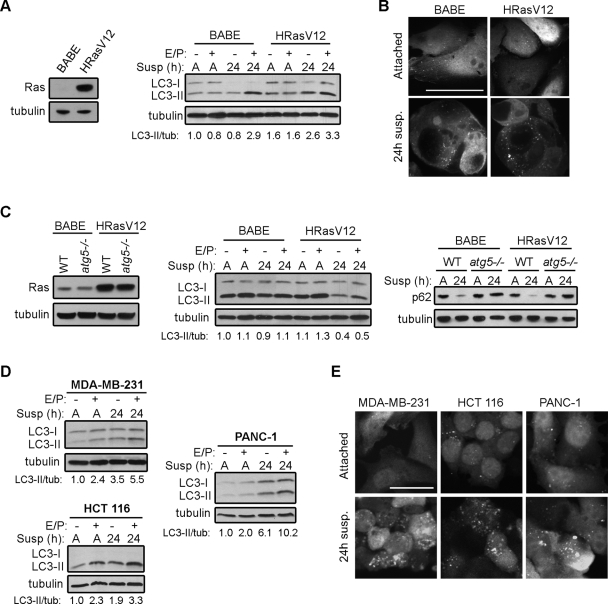
Several studies demonstrate that constitutive activation of Ras suppresses autophagy induction while others indicate that Ras/mitogen-activated protein kinase (MAPK) pathway activation can enhance autophagy during nutrient starvation (Pattingre et al., 2003; Furuta et al., 2004; Berry and Baehrecke, 2007). Given these paradoxical results, we first sought to clarify how oncogenic Ras modulates autophagy upon loss of cell–matrix contact, a cardinal stress during adhesion-independent transformation. We generated stable pools of MCF10A human mammary epithelial cells expressing oncogenic H-Ras (H-RasV12) as well as control cells expressing an empty vector (BABE) (Figure 1A, left). We then tested autophagy levels following substratum detachment by plating cells on poly-HEMA–coated plates to prevent cell–matrix adhesion. As previously reported, in BABE control cells, we observed an increase in the lysosomal turnover of phosphatidylethanolamine-lipidated LC3/ATG8 (LC3-II), commonly termed autophagic flux, following matrix detachment (Figure 1A, right) (Fung et al., 2008). Furthermore, MCF10A cells expressing H-RasV12 displayed an increase in LC3-II induction and lysosomal turnover following matrix detachment (Figure 1A, right). To further verify autophagosome induction in H-RasV12–transformed MCF10A cells, we stably expressed green fluorescent protein (GFP)–LC3 in H-RasV12 and vector control cells, and we assessed autophagosome formation (punctate GFP-LC3) using fluorescence microscopy. Following detachment, we observed robust induction of GFP-LC3 puncta in both empty vector and H-RasV12-expressing MCF10A cells in comparison to attached controls (Figure 1B). Because detached epithelial cells exhibit extensive clustering, we were unable to precisely enumerate puncta per cell; nonetheless, we consistently observed similar levels of punctate GFP-LC3 in H-RasV12–transformed MCF10A compared with BABE controls.
FIGURE 1:
Oncogenic Ras does not suppress ECM detachment-induced autophagy. (A) Left: Ras expression in MCF10A cells expressing empty vector (BABE) or H-RasV12. Right: BABE and H-RasV12 MCF10A cells were grown attached (A) or suspended (susp) for the indicated times in the presence or absence of E64d and pepstatin A (E/P), lysed, and subjected to immunoblotting with antibodies against LC3 and tubulin. (B) GFP-LC3 puncta in MCF10A cells expressing empty vector (BABE) or H-RasV12 grown attached or suspended for 24 h. (C) Left: Ras expression in atg5+/+ (WT) and atg5−/− MEFs expressing empty vector or H-RasV12. Center: atg5+/+ (WT) MEFs expressing empty vector (BABE) and H-RasV12 were growth attached (A) or suspended (susp) for 24 h in the presence or absence of E64d and pepstatin A (E/P), lysed, and subjected to immunoblotting with antibodies against LC3 and tubulin. Right: atg5+/+ (WT) and atg5−/− MEFs expressing H-RasV12 or empty vector (BABE) were grown attached (A) or suspended (susp) for 24 h, lysed, and subjected to immunoblotting with antibodies against p62 and tubulin. (D) MDA-MB-231, HCT 116, and PANC-1 cells were grown attached (A) or suspended (susp) for 24 h in the presence or absence of E64d and pepstatin A (E/P) and subjected to immunoblotting with antibodies against LC3 and tubulin. (E) GFP-LC3 puncta in MDA-MB-231, HCT 116, and PANC-1 cells that were grown attached or detached for 24 h. Bar, 25 μm.
We next evaluated the effects of H-RasV12 on detachment-induced autophagy in immortalized mouse embryonic fibroblasts (MEFs) (Figure 1C). Remarkably, both H-RasV12 and control fibroblasts exhibited a high baseline level of LC3-II when grown in attached conditions; upon suspension, LC3-II levels decreased dramatically in the H-RasV12–expressing MEFs and, to a lesser extent, in vector controls during suspension. Upon addition of E/P, LC3-II levels increased following detachment in both cell types, indicating that both control and H-RasV12 fibroblasts exhibit LC3-II turnover during matrix detachment (Figure 1C, center). To more conclusively validate these results, we assessed the degradation of p62 (SQSTM1), a scaffold protein specifically degraded by autophagy, following detachment (Figure 1C, right). In both vector control and H-RasV12–transformed MEFs, we observed significantly reduced p62 levels following 24 h of suspension. In contrast, p62 levels remained elevated in both control and H-RasV12–transformed atg5−/− MEFs, supporting that the degradation of p62 during substratum detachment requires an intact autophagy pathway.
To extend these results, we evaluated detachment-induced autophagy in epithelial cancer cell lines that naturally harbor oncogenic Ras mutations. In three different carcinoma lines that possess activating K-Ras mutations—MDA-MB-231 breast carcinoma cells, HCT 116 colon carcinoma cells, and PANC-1 pancreatic carcinoma cells—both LC3-II induction and turnover increased upon substratum detachment (Figure 1D). In parallel, we examined autophagosome formation (GFP-LC3 puncta) following suspension. Similar to MCF10A cells, all three carcinoma cell lines displayed an increase in GFP-LC3 puncta following 24 h matrix detachment (Figure 1E). Altogether, our results support the robust induction of autophagy in both epithelial and fibroblast cells expressing H-RasV12 as well as in cancer cell lines harboring activating K-Ras mutations following matrix detachment; hence, Ras activation does not suppress autophagy during ECM detachment.
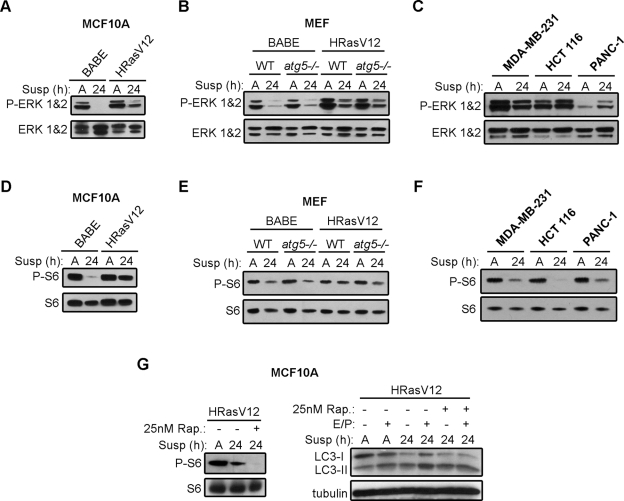
We next assessed whether constitutive Ras activation was sufficient to maintain activation of downstream signaling pathways following ECM detachment. We first tested whether oncogenic activation of Ras sustained activation of the MAPK pathway by examining levels of phosphorylated ERK. Both MCF10A cells and mouse fibroblasts (expressing empty vector) displayed a reduction in phosphorylated ERK1/2 levels following 24 h ECM detachment. In contrast, the phosphorylation of ERK1/2 remained elevated in both H-RasV12–transformed MCF10As and MEFs during ECM detachment (Figure 2, A and B). ERK1/2 phosphorylation was similarly maintained in MDA-MB-231 and HCT 116 cells; remarkably, in PANC-1 cells ERK1/2 phosphorylation was increased in matrix-detached cells when compared with attached controls (Figure 2C).
FIGURE 2:
Effects of ECM detachment on MAPK and mTORC1 signaling in Ras-transformed cells. (A–C) Empty vector (BABE) and H-RasV12–expressing MCF10A cells (A), atg5+/+ (WT) and atg5−/− MEFs (B), and K-Ras mutant carcinoma cell lines (C) were grown attached (A) or suspended (susp) for the indicated times and subjected to immunoblotting with antibodies against phosphorylated ERK1/2 and total ERK1/2 protein. (D–F) Empty vector (BABE) and H-RasV12–expressing MCF10A cells (D), atg5+/+ (WT) and atg5−/− MEFs (E), and K-Ras mutant carcinoma cell lines (F) were grown attached (A) or suspended (susp) for the indicated times and subjected to immunoblotting with antibodies against phosphorylated S6 and total ribosomal S6 protein. (G) H-RasV12 MCF10A cells were grown attached (A) or suspended (susp) for 24 h in the presence or absence of E64d and pepstatin A (E/P) and subjected to immunoblotting with antibodies against phosphorylated S6, S6, LC3, and tubulin. When indicated, cells were treated with 25 nM rapamycin for 5 h before harvest.
Sustained activation of mTORC1, the archetypal negative regulator of autophagy, has been proposed to mediate autophagy inhibition downstream of oncogenic Ras (Furuta et al., 2004; Maiuri et al., 2009). Thus we measured mTORC1 activation in H-RasV12–transformed cells following ECM detachment by assessing the phosphorylation status of ribosomal protein S6, a downstream mTOR target. Upon detachment, S6 phosphorylation decreased sharply in control MCF10A cells, supporting reduced activation of the mTORC1 pathway. Notably, S6 phosphorylation was partially decreased in H-RasV12–transformed cells following 24 h suspension (Figure 2D). In fibroblasts, both control and H-RasV12–transformed MCF10A cells demonstrated decreased levels of phosphorylated S6 during suspension (Figure 2E). Similarly, in K-Ras mutant cancer cells, S6 phosphorylation was reduced following ECM detachment (Figure 2F). Because we observed a partial decrease in S6 phosphorylation during ECM detachment, particularly in H-RasV12 MCF10A cells, we treated suspended cells with rapamycin to assess whether robust inhibition of mTORC1 was able to further enhance detachment-induced autophagy. Upon rapamycin treatment, we were unable to detect S6 phosphorylation in H-RasV12 MCF10A cells following 24 h suspension; however, we did not observe any further increase in LC3-II induction or turnover upon rapamycin treatment (Figure 2G). This result supports that autophagy can be potently induced in H-RasV12 MCF10A cells following ECM detachment without complete suppression of mTORC1 activity.
Reduced H-RasV12–driven soft agar transformation in autophagy-deficient MEFs
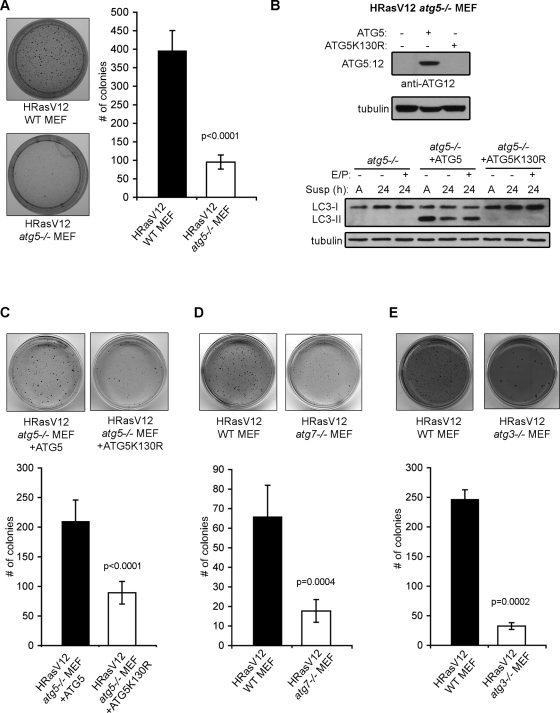
Because autophagy was robustly induced in Ras-transformed cells upon loss of cell–matrix contact, we next interrogated the functional contribution of autophagy to H-RasV12–driven anchorage-independent growth. For these experiments, we initially tested how the genetic deletion of three critical autophagy regulators, atg5, atg7, and atg3, individually influence anchorage-independent transformation by oncogenic Ras. All three proteins are essential components of the ubiquitin-like conjugation pathways that control the early step of autophagosome formation; thus the genetic deletion of any of these ATGs is sufficient to completely inhibit autophagy (Ohsumi, 2001; Kuma et al., 2004; Komatsu et al., 2005; Sou et al., 2008). We first compared the ability of atg5+/+ and atg5−/− MEFs transformed with H-RasV12 to form colonies in soft agar. H-RasV12–transformed atg5−/− MEFs displayed an approximate fourfold decrease in colony formation compared with wild-type autophagy-competent controls (Figure 3A); importantly, both atg5+/+ and atg5−/− cells expressed equivalent levels of H-RasV12 (Figure 1C, left).
FIGURE 3:
Decreased anchorage-independent growth in autophagy-deficient MEFs expressing H-RasV12. (A) Soft agar colony formation in H-RasV12 expressing atg5+/+ (WT) and atg5−/− MEFs. (B) atg5−/− MEFs reconstituted with wild-type murine ATG5 or ATG5 K130R were subjected to immunoblotting with antibodies against ATG12 (to detect the ATG12-ATG5 complex) and tubulin. As indicated, cells were grown attached (A) or suspended (susp) for 24 h in the presence or absence of E64d and pepstatin A (E/P) and subjected to immunoblotting with antibodies against LC3 and tubulin as a loading control. (C) Soft agar colony formation in H-RasV12–expressing atg5−/− MEFs expressing ATG5 or ATG5K130R. (D and E) Soft agar colony formation in H-RasV12 expressing wild-type (WT), atg7−/−, and atg3−/− MEFs. The above results represent the mean ± SEM from three or more independent experiments. P value was calculated using Student's t test.
To verify that these differences resulted directly from autophagy inhibition upon ATG5 deletion, we constituted atg5−/− cells with either wild-type mouse ATG5 or ATG5 K130R, a lysine mutant unable to conjugate to ATG12 and therefore unable to induce autophagy. Rescue of H-RasV12 atg5−/− MEFs with wild-type ATG5 restored ATG5–ATG12 complex levels, whereas expression of ATG5 K130R did not (Figure 3B). This rescue of H-RasV12 atg5−/− MEFs with wild-type ATG5 restored autophagy induction, indicated by the production of LC3-II in attached conditions and following suspension. In contrast, both H-RasV12 atg5−/− MEFs, as well as those expressing ATG5 K130R, were unable to induce autophagy during suspension (Figure 3B). Furthermore, the rescue of H-RasV12–transformed atg5−/− MEFs with wild-type ATG5, but not ATG5 K130R, was able to restore soft agar colony formation (Figure 3C), further supporting that autophagy competence functionally contributes to Ras-driven transformation. Similarly, soft agar transformation mediated by H-RasV12 was also abrogated in atg7−/− and atg3−/− cells. Colony formation was reduced almost fourfold in H-RasV12 atg7−/− MEFs compared with wild-type controls (Figure 3D), and H-RasV12 atg3−/− MEFs displayed the most profound defect in soft agar colony formation, almost eightfold, compared with wild-type controls (Figure 3E). These results support that the elimination of autophagy in mouse fibroblasts, achieved via the genetic deletion of multiple ATGs, potently inhibits the transformation potential of H-RasV12.
Reduced soft agar transformation upon ATG knockdown in Ras-transformed epithelial cells
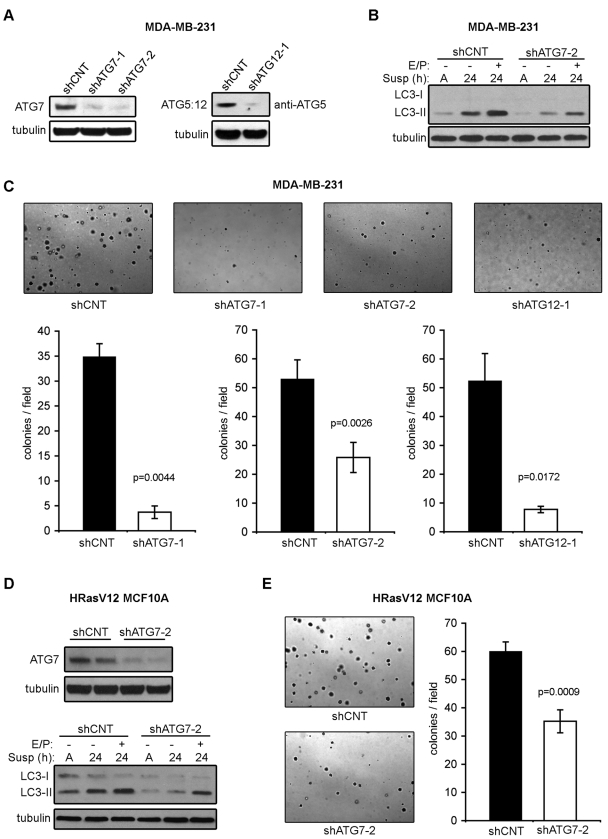
We next determined whether the acute reduction of autophagy in the context of preexisting oncogenic Ras activation was similarly able to inhibit adhesion-independent transformation. First, we stably expressed two independent short-hairpin RNAs (shRNAs) against ATG7 (shATG7-1 and shATG7-2) as well as a hairpin directed against ATG12 (shATG12-1) in MDA-MB-231 cells. Analysis of target protein levels by Western blot revealed high-level knockdown of ATG7 with both shATG7-1 and 2 and reduction of the ATG5–ATG12 complex in shATG12-1–expressing MDA-MB-231 cells (Figure 4A). Of these three hairpins, shATG7-2 gave the most robust reduction in autophagy, based on immunoblotting for LC3-II (unpublished data). MDA-MB-231 cells expressing this shRNA exhibited an approximately 50% decrease in both basal and detachment-induced autophagy (Figure 4B). Furthermore, the expression of all three of these shATGs in MDA-MB-231 cells resulted in a significant decrease in soft agar colony formation, ranging from approximately 50–90%, depending on the shRNA used (Figure 4C).
FIGURE 4:
Effects of ATG knockdown on adhesion-independent transformation in MDA-MB-231 cells and H-RasV12 MCF10A cells. (A) MDA-MB-231 cells transduced with lentiviral vectors encoding shRNAs against the indicated ATGs (shATGs) were subjected to immunoblotting with antibodies against ATG7, ATG5 (to detect ATG12–ATG5 complex), and tubulin. (B) MDA-MB-231 cells expressing shATG7-2 or shCNT were grown attached (A) or suspended (susp) for the indicated times in the presence or absence of E64d and pepstatin A (E/P) and subjected to immunoblotting with antibodies against LC3 and tubulin. (C) Representative images and quantification of soft agar colony formation in MDA-MB-231 cells expressing the indicated shATGs. (D) H-RasV12 MCF10A cells expressing shCNT or shATG7-2 were grown attached (A) or suspended (susp) for the indicated times in the presence or absence of E64d and pepstatin A (E/P) and subjected to immunoblotting with antibodies against ATG7, LC3, and tubulin. (E) Representative images and quantification of soft agar colony formation in H-RasV12 MCF10A cells expressing shCNT or shATG7-2. The above results represent the mean ± SEM from three or more independent experiments. P value was calculated using Student's t test.
In parallel, we generated stable pools of H-RasV12 MCF10A cells expressing shRNA against ATG7 (shATG7-2). These cells demonstrated potent ATG7 knockdown and decreased LC3-II in both attached and detached conditions (Figure 4D). Colony formation in H-RasV12 MCF10A cultures expressing shATG7-2 was reduced by approximately 50% when compared with cells expressing control shRNA (shCNT) (Figure 4E), suggesting ATG7 knockdown was sufficient to partially suppress H-RasV12–induced soft agar growth in MCF10A cells. Hence, consistent with our data in ATG-deficient fibroblasts, epithelial cells with oncogenic Ras displayed a reduction in soft agar colony formation following RNA interference (RNAi)–mediated knockdown of ATGs. Furthermore, it is important to note that although we were able to achieve high levels of ATG knockdown, such perturbations produced partial reductions in autophagic capacity, up to 50% of control. Nonetheless, such levels of autophagy reduction resulted in robust decreases in soft agar colony formation, pointing to a critical role for autophagy in Ras-mediated anchorage-independent transformation.
Effects of autophagy inhibition on detachment-induced apoptosis (anoikis) in Ras-transformed cells
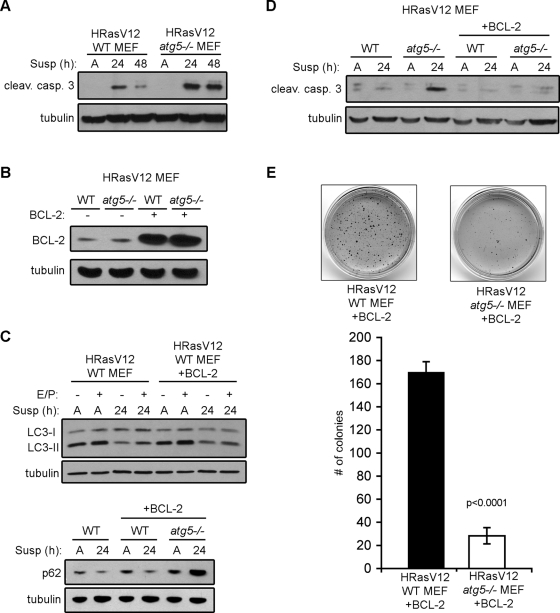
Because our previous work indicates that detachment-induced autophagy protects nontransformed MCF10A cells from anoikis, we hypothesized that autophagy may similarly promote the survival of Ras-transformed cells deprived of cell–matrix contact (Fung et al., 2008). To test this prediction, H-RasV12 atg5+/+ and atg5−/− MEFs were grown either attached or suspended for 24–48 h and protein lysates were immunoblotted for cleaved caspase-3. Although previous work indicates that fibroblasts do not undergo anoikis, we found that cleaved caspase-3 did indeed increase in H-RasV12 autophagy-competent fibroblasts upon ECM detachment. Furthermore, compared with wild-type controls, the levels of cleaved caspase-3 in H-RasV12 atg5−/− MEFs following suspension were higher. This corroborates that autophagy deficiency leads to increased detachment-induced apoptosis in H-RasV12–transformed cells (Figure 5A).
FIGURE 5:
Bcl-2 inhibition of apoptosis is not sufficient to restore anchorage-independent growth in autophagy-deficient cells. (A) H-RasV12 expressing atg5+/+ (WT) and atg5−/− MEFs were grown attached (A) or suspended (susp) for the indicated times and subjected to immunoblotting with antibodies against cleaved capase-3 and tubulin. (B) Bcl-2 expression levels in H-RasV12 atg5+/+ (WT) and atg5−/− MEFs in the presence or absence of stable ectopic expression of Bcl-2. (C) The indicated cell types were grown attached (A) or suspended (susp) for 24 h with or without E64d and pepstatin A (E/P) and subjected to immunoblotting with antibodies against LC3, p62, and tubulin. (D) The indicated cell types, all expressing H-RasV12, were grown attached (A) or suspended for 24 h and subjected to immunoblotting with antibodies against cleaved caspase-3 and tubulin. (E) Soft agar colony formation of H-RasV12 atg5+/+ (WT) and atg5−/− MEFs stably expressing BCL-2. Results represent the mean ± SEM from three independent experiments. P value was calculated using Student's t test.
On the basis of these results, we interrogated whether the ectopic expression of the anti-apoptotic molecule Bcl-2 was sufficient to promote adhesion-independent growth and survival in H-RasV12 atg5−/− MEFs. To test this hypothesis, we generated H-RasV12 atg5+/+ and atg5−/− MEFs stably expressing Bcl-2 (Figure 5B). As Bcl-2 has previously been shown to suppress autophagy in certain cell types via its interaction with Beclin 1, we determined the effects of Bcl-2 expression on detachment-induced autophagy in H-RasV12 MEFs but did not identify any significant effects on LC3-II induction or turnover or on p62 degradation during ECM detachment (Figure 5C) (Pattingre et al., 2005). In contrast, Bcl-2 potently reduced apoptosis in H-RasV12–transformed atg5+/+ and atg5−/− cells following matrix detachment, as indicated by immunoblotting for cleaved caspase-3 (Figure 5D).
We next evaluated whether Bcl-2 expression was sufficient to restore adhesion-independent transformation in H-RasV12 atg5−/− MEFs. However, we continued to detect reduced levels of H-RasV12–driven soft agar growth in Bcl-2–expressing autophagy-deficient cells when compared with wild-type counterparts (Figure 5E). These results indicate that autophagy inhibition in H-RasV12–transformed cells can promote anoikis; however, protecting autophagy-deficient cells from apoptosis is not sufficient to restore adhesion-independent transformation, raising the possibility that autophagy facilitates Ras transformation via other mechanisms.
Autophagy inhibition results in decreased proliferation of Ras-transformed cells
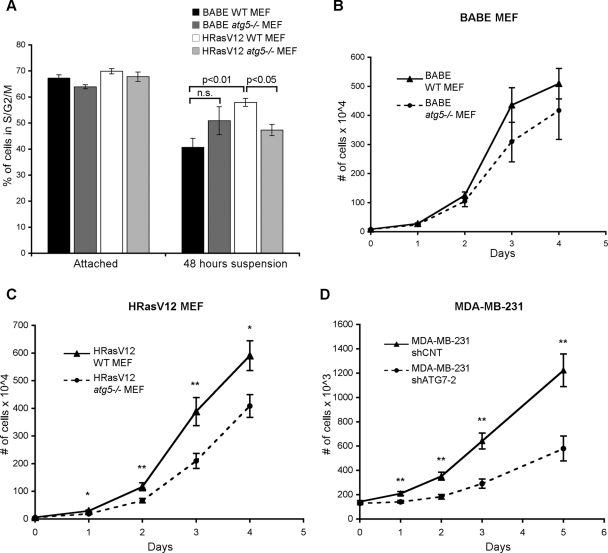
The aforementioned results motivated us to test the functional contributions of autophagy to the proliferation of H-RasV12–transformed cells. First, we tested the effects of ECM detachment on the proliferation capacity of autophagy-competent and autophagy-deficient MEFs expressing either vector control (BABE) or H-RasV12. Cells grown attached or in suspension for 48 h were subjected to flow cytometric analysis for DNA content corresponding to the S and G2/M phases of the cell cycle (Figure 6A). In vector control (BABE) wild-type MEFs, we observed a decrease in the percentage of cycling cells (S + G2/M), from 67.3% ± 1.3% in attached conditions to 40.7% ± 3.5% after 48 h of suspension (Figure 6A, black bar). In contrast, 57.9% ± 1.5% of H-RasV12–transformed wild-type (atg5+/+) cells remained in S + G2/M following 48 h of suspension (Figure 6A, white bar). Thus H-RasV12–transformed cells continue to proliferate upon loss of cell–matrix contact. However, in H-RasV12 atg5−/− MEFs, incapable of autophagy, the ability of H-RasV12 to promote proliferation in the absence of cell–matrix contact was attenuated, with only 47.3% ± 2.1% of cells remaining in cycle following 48 h of suspension (Figure 6A, light gray bar). Interestingly, we noted that control (BABE) atg5−/− MEFs (dark gray bars) proliferated slightly better than atg5+/+ cells during detachment; such results are consistent with previous studies demonstrating that reduced autophagy due to Beclin/ATG6 haploinsufficiency or genetic deletion of Ambra1 can promote cell proliferation (Qu et al., 2003; Fimia et al., 2007). Nevertheless, in the context of H-RasV12 expression, autophagy inhibition curtailed rather than enhanced proliferation during ECM detachment.
FIGURE 6:
Reduced proliferation upon autophagy inhibition in H-RasV12 expressing MEFs and MDA-MB-231 cells. (A) The indicated cell types were grown attached or subjected to ECM detachment for 48 h and analyzed by flow cytometry to quantify the percentage of cells with DNA content corresponding to the S and G2/M (S + G2/M) phases of the cell cycle. Results are the mean ± SEM from three or more independent experiments. Statistical significance was calculated using ANOVA. (B) Proliferation curves of empty vector (BABE) atg5+/+ (WT) and atg5−/− MEFs cultured in attached, nutrient-rich conditions. (C) Proliferation curves of H-RasV12 expressing atg5+/+ (WT) and atg5−/− MEFs in attached, nutrient-rich conditions. (D) Proliferation curves of MDA-MB-231 cells expressing shCNT or shATG7-2 in attached, nutrient-rich conditions. For (B–D), p value was calculated at each time point using Student's t test, with statistical significance indicated as follows: *p < 0.05; **p < 0.01.
To extend these results, we then measured whether H-RasV12–transformed atg5−/− cells displayed similar defects in proliferation in the absence of the stresses imposed by substratum detachment. Thus we grew the various cell types in nutrient replete, attached conditions in which only basal levels of autophagy were present. Upon enumerating cell numbers from cultures, we found that nontransformed wild-type and atg5−/− MEFs exhibited minimal differences in proliferation (Figure 6B). In contrast, upon transformation with H-RasV12, autophagy-deficient cells failed to proliferate as well as controls (Figure 6C). Similarly, acute ATG7 knockdown in MDA-MB-231 cells led to a profound decrease in proliferation compared with controls (Figure 6D). Overall, these results indicate that autophagy induction is necessary for optimal cell proliferation in H-RasV12–expressing cells following ECM detachment and that oncogenic Ras activation engenders an increased reliance on basal autophagy for cell expansion in attached conditions.
Increased glucose metabolism in autophagy-competent cells
Owing to the decreased proliferation observed in Ras-transformed cells upon autophagy inhibition, we hypothesized that the difference in adhesion-independent transformation we observed between Ras-transformed autophagy-competent and deficient cells may arise from changes in protein synthesis or in cellular metabolism, two processes that directly impact the capacity for cell growth and proliferation. Both nitrogen-starved, autophagy-deficient yeast and early ATG5-deficient embryos display a decrease in de novo protein translation compared with wild-type controls (Onodera and Ohsumi, 2005; Tsukamoto et al., 2008). Therefore we speculated that H-RasV12 atg5−/− MEFs would exhibit diminished rates of protein synthesis compared with H-RasV12 wild-type MEFs in the absence of ECM contact. Although we observed decreased de novo protein synthesis in empty vector (BABE)–expressing atg5−/− MEFs compared with wild-type following 24 h detachment, only minor differences were present when we compared H-RasV12–expressing wild-type and atg5−/− MEFs (Supplemental Figure S1).
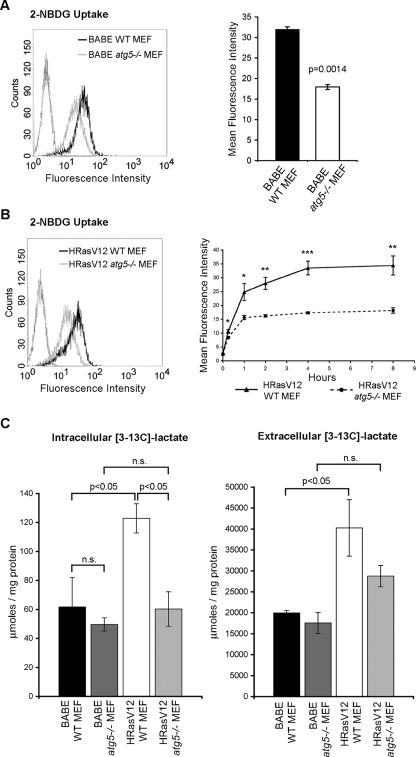
Like many oncogenes, H-RasV12 enhances glycolysis, which is associated with increased glucose uptake and lactate production; importantly, increased aerobic glycolysis is required for Ras-driven tumors to maintain energy production and enhance biosynthetic pathways. Remarkably, we found that glucose uptake, determined by uptake of 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-d-glucose (2-NBDG), was significantly reduced in empty vector atg5−/− MEFs compared with atg5+/+ controls (Figure 7A). As expected, H-RasV12 expression resulted in increased glucose uptake (Supplemental Figure S2A). When we compared glucose uptake between H-RasV12 atg5+/+ and H-RasV12 atg5−/− MEFs over an 8-h time course, we found reduced glucose uptake in H-RasV12 atg5−/− MEFs compared with H-RasV12 atg5+/+ cells at all time points examined (Figure 7B). We also observed a decreased level of glucose uptake in H-RasV12 atg5−/− MEFs following ECM detachment (Supplemental Figure S2B). Enhanced glucose uptake is often associated with an increase in glycolytic flux, resulting in the enhanced production of lactate. Thus, to determine the glycolytic status of autophagy-proficient and deficient cells, we used 13C-NMR spectroscopy to assess metabolic fluxes. Cells were labeled with [1-13C]glucose, and de novo lactate production was monitored. Despite reduced glucose uptake in atg5−/− MEFs compared with atg5+/+ controls, there was no concomitant decrease in lactate production in atg5−/− MEFs (Figure 7C). In fact, we observed equivalent levels of both intracellular and extracellular lactate production by wild-type and atg5−/− MEFs. However, upon H-RasV12 transformation, both intracellular and extracellular [3-13C]lactate production was decreased in H-RasV12–transformed atg5−/− cells in comparison to atg5+/+ controls (Figure 7C). Furthermore, compared with atg5+/+ controls, H-RasV12 atg5−/− MEFs exhibited decreased levels of [3-13C]alanine, which is produced via the transamination of the glycolytic end product pyruvate (Supplemental Figure S2C).
FIGURE 7:
Reduced glucose metabolism in autophagy-deficient MEFs. (A) Levels of glucose uptake (2-NBDG uptake, mean fluorescence intensity) in empty vector (BABE) atg5+/+ (WT) and atg5−/− MEFs following 2.5 h incubation. Statistical significance was calculated using Student's t test. (B) 2-NBDG uptake (mean fluorescence intensity) after 1 h (left histogram) and over an 8-h time course (right graph) in H-RasV12 expressing atg5+/+ (WT) and atg5−/− MEFs. P value was calculated at each time point using Student's t test, with statistical significance indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001. (C) Levels of 13C-labeled intracellular lactate and extracellular lactate detected by NMR following 24 h of labeling with [1-13C]glucose. Results represent the mean ± SEM from three independent experiments. Statistical significance was calculated using ANOVA.
Next, we evaluated whether defects in glucose metabolism were present in MDA-MB-231 cells upon ATG7 knockdown. Although glucose uptake was not significantly reduced in ATG7-depleted cells (unpublished data), they did exhibit a significant decrease in the enzymatic activity of lactate dehydrogenase (LDH), which is required for the conversion of pyruvate to lactate. Furthermore, LDH-A protein levels were also decreased in ATG7-depleted MDA-MB-231 cells (Supplemental Figure S2D). LDH-A levels were not altered in empty vector atg5−/− or H-RasV12 atg5−/− MEFs compared with wild-type controls (Supplemental Figure S2E). Altogether, these results implicate that reduced autophagy results in a concomitant decrease in glycolytic capacity.
Autophagy-competent cells exhibit increased sensitivity to diminished glucose availability
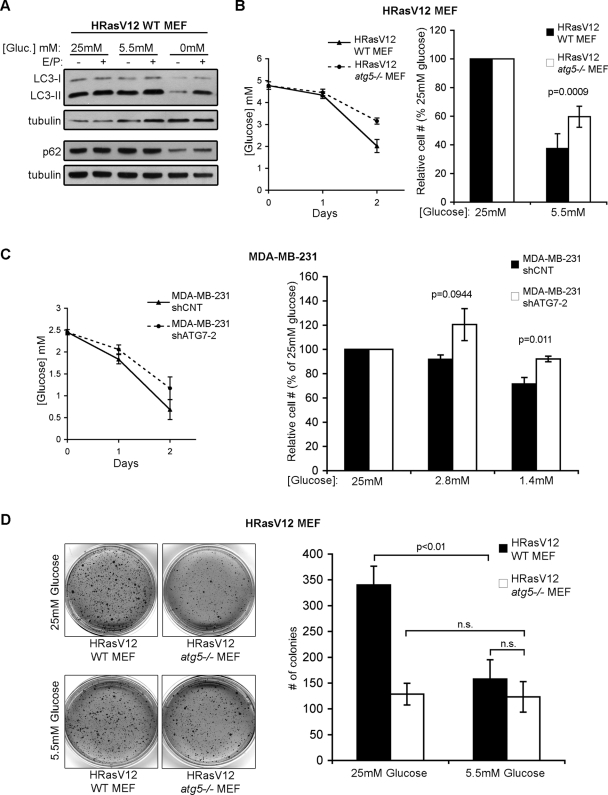
Because glycolysis was decreased in Ras-transformed, autophagy-deficient cells, we next sought to determine the effects of varying glucose concentrations on autophagy-competent versus autophagy-deficient cells. We first assessed whether autophagy is stimulated in response to decreasing media glucose concentrations because previous studies support that autophagy is induced upon glucose starvation or treatment with 2-deoxy-glucose (Aki et al., 2003; DiPaola et al., 2008). Although we observed a robust induction of autophagy following 9 h of complete glucose withdrawal, a similar increase in autophagy was not detected in H-RasV12–expressing MEFs upon lowering glucose concentrations from the standard 25 mM to 5.5 mM for up to 48 h (Figure 8A).
FIGURE 8:
The proliferation and transformation of autophagy-competent cells are more sensitive to diminished glucose availability than autophagy-deficient cells. (A) H-RasV12 expressing WT MEFs were cultured in media containing 25 mM or 5.5 mM glucose for 48 h or in the complete absence of glucose (0 mM) for 9 h. E64d and pepstatin A (E/P) were added when indicated to measure autophagic flux. Cells were lysed and subjected to immunoblotting with antibodies against LC3, p62, and tubulin as a loading control. (B) Left: Media glucose levels from cultures of H-RasV12 atg5+/+ (WT) and atg5−/− MEFs grown in 5.5 mM glucose over 2 d. Right: Relative percentage of viable cells in 5.5 mM glucose compared with 25 mM glucose following 4 d of culture. (C) Left: Media glucose levels from cultures of MDA-MB-231 cells expressing shCNT or shATG7-2 grown in 2.8 mM glucose over 2 d. Right: Relative percentage of viable cells grown in 2.8 mM and 1.4 mM glucose media compared with 25 mM glucose following 3 d of growth. (D) Soft agar colony formation of H-RasV12 expressing atg5+/+ (WT) and atg5−/− MEFs in 25 and 5.5 mM glucose conditions. Results represent the mean ± SEM from four independent experiments. Statistical significance was calculated using ANOVA.
Glycolytic cells typically display exquisite sensitivity to diminishing concentrations of glucose; accordingly, we assessed how autophagy competence versus deficiency impacted glucose consumption and proliferation in H-RasV12–expressing cells. First, we measured the consumption of glucose in H-RasV12 atg5+/+ and atg5−/− cells grown over 2 d in 5.5 mM glucose; in accordance with the above results, media glucose concentrations declined more precipitously in H-RasV12 wild-type MEF cultures compared with H-RasV12 atg5−/− cultures (Figure 8B). Furthermore, following 4 d of culture in 5.5 mM glucose, cell numbers in H-RasV12 wild-type cultures were reduced by 62.5% in comparison to those grown in 25 mM. In contrast, the expansion of H-RasV12 atg5−/− cells was not as profoundly attenuated by similar reductions in glucose concentration; these cells exhibited only a 40.4% reduction in cell number when cultured in 5.5 mM glucose compared with 25 mM glucose. This increased sensitivity of H-RasV12 atg5+/+ MEFs to lower media glucose levels is in accordance with the increases in glycolytic capacity and glucose uptake we observed in H-RasV12 atg5+/+ MEFs (Figure 8B). To corroborate these results, we performed parallel experiments in MDA-MB-231 cells following acute ATG7 depletion. When grown in 2.8 mM glucose, cells expressing shATG7-2 consumed glucose at a lower rate than shCNT cells (Figure 8C). In addition, the expansion of shATG7-2 cells was not as sensitive to lower glucose concentrations as shCNT cells (Figure 8C).
On the basis of these results, we hypothesized that declining glucose concentrations would attenuate the rate of adhesion-independent transformation of H-RasV12–transformed autophagy-competent cells but have little effect on autophagy-deficient counterparts. Accordingly, we observed a significant reduction in soft agar colony formation in H-RasV12 atg5+/+ MEFs grown in 5.5 mM glucose compared with those grown in 25 mM glucose. In contrast, adhesion-independent transformation in H-RasV12 atg5−/− MEFs was not affected by declining glucose concentrations. Remarkably, at the lower glucose concentration (5.5 mM), we observed comparable levels of soft agar colony formation between H-RasV12–transformed autophagy-competent and deficient cells (Figure 8D). These results support that the ability of autophagy to promote adhesion-independent transformation is highly dependent on glucose levels and point to a previously unrecognized role for autophagy competence in facilitating glycolysis and proliferation during oncogenic Ras-mediated transformation.
DISCUSSION
Overall, our studies demonstrate that in the context of a potent oncogene, mutationally active Ras, autophagy both promotes adhesion-independent transformation and facilitates glycolysis. The genetic deletion or RNAi-mediated knockdown of autophagy regulators (ATGs) causes a potent decrease in anchorage-independent growth in soft agar, indicating that an intact autophagy pathway is required for robust adhesion-independent transformation by oncogenic Ras. Furthermore, autophagy inhibition during Ras transformation results in reduced proliferation and decreased glucose metabolism. The decreased rate of glycolysis found in Ras-transformed autophagy-deficient cells correlates with decreased sensitivity to declining glucose concentrations in comparison with autophagy-competent counterparts, both in terms of proliferation and adhesion-independent transformation.
In addition, our results indicate that oncogenic Ras does not suppress autophagy during ECM detachment. These data differ from previous reports in both Ras-expressing cells in vitro as well as Drosophila development in vivo (Furuta et al., 2004; Berry and Baehrecke, 2007), both of which demonstrate that Ras activation suppresses autophagy. Ras-mediated suppression of autophagy is proposed to arise secondary to the constitutive activation of the PI3K/mTOR pathway, a negative regulator of autophagy induction. However, upon ECM detachment of several K-Ras mutant cancer lines, ribosomal protein S6 phosphorylation, an established readout of mTORC1 activity, is rapidly decreased, indicating that Ras is unable to sustain mTORC1 activation in cells deprived of matrix contact. Furthermore, detached MEFs expressing H-RasV12 exhibit decreased S6 phosphorylation comparable to nontransformed controls. On the other hand, in H-RasV12–transformed MCF10A cells, S6 phosphorylation is only slightly reduced following matrix detachment compared with nontransformed counterparts. Interestingly, although we were able to completely inhibit mTORC1 activity by treating H-RasV12 MCF10A cells with rapamycin during suspension, this did not further augment autophagy. This result indicates that the enhanced level of mTORC1 activity that persists in H-RasV12 MCF10A cells following detachment is not sufficient to suppress autophagy induction. From these results, we speculate that a partial reduction in mTORC1 activity in H-RasV12–expressing MCF10A cells may be sufficient to promote detachment-induced autophagy. Alternatively, other mTORC1-independent pathways may promote autophagy in detached cells expressing oncogenic Ras. Importantly, our results support that oncogenic Ras activation does not inhibit detachment-induced autophagy in mammalian cells.
The mechanisms through which autophagy modulates oncogenic transformation are context dependent (Chen and Debnath, 2010). Here we demonstrate that autophagy is required for robust Ras-driven transformation; cells deleted or depleted of multiple independent ATGs all exhibit decreased anchorage-independent transformation in soft agar. Recently, we discovered that detachment-induced autophagy protects cells from anoikis, which we proposed to facilitate oncogenic transformation (Fung et al., 2008). However, because the ectopic overexpression of the antiapoptotic protein Bcl-2 does not enhance soft agar growth in Ras-transformed autophagy-deficient cells, the ability of autophagy to facilitate Ras transformation cannot be completely explained by its ability to protect cells from apoptosis during ECM detachment.
These studies point to a previously unrecognized tumor-promoting function for autophagy that manifests during oncogenic Ras transformation. For example, upon matrix detachment, increased numbers of atg5−/− cells continue to proliferate compared with atg5+/+ controls. In fact, the enhanced proliferation of autophagy-deficient cells has been proposed as a potential mechanism by which autophagy might exert tumor suppressive effects (Qu et al., 2003; Fimia et al., 2007). However, unlike nontransformed autophagy-deficient cells, atg5 genetic deletion impedes, rather than enhances, the ability of H-RasV12–transformed MEFs to proliferate during ECM detachment. Similarly, when cultured in attached nutrient-rich conditions, Ras-transformed atg5−/− cells exhibit a marked decrease in proliferation compared with their autophagy-competent counterparts. In addition, we have also found that MCF10A cells expressing H-RasV12 occasionally undergo growth arrest or cell death following lentiviral-driven introduction of shRNAs against ATGs; in contrast, nontransformed cells consistently remain viable and continue to proliferate upon ATG knockdown (unpublished data). These results support that autophagy competence is required for cells to proliferate and expand during oncogenic Ras transformation.
Increasing evidence indicates that stress response pathways play diverse, multifaceted roles necessary for oncogenic transformation. For example, heat shock protein 1 (HSP-1), an important mediator of the heat shock response, has been implicated as an important facilitator of Ras transformation, which correlates with its ability to modulate both proliferative capacity and glucose metabolism (Dai et al., 2007). Here we demonstrate that autophagy similarly supports increased glucose metabolism, suggesting a previously unrecognized mechanism by which autophagy may contribute to tumorigenesis. H-RasV12–transformed, autophagy-competent MEFs display enhanced glucose uptake compared with their autophagy-deficient counterparts. In addition, using 13C-NMR analysis of glucose metabolism, we observe augmented glycolytic flux in H-RasV12–expressing autophagy-competent cells as evidenced by increased production of lactate and alanine from glucose. Notably, we have also observed reduced glucose uptake in nontransformed, autophagy-deficient cells, but unlike H-RasV12–transformed cells, these reductions do not correlate with significant changes in lactate production or in monolayer proliferation. Increased glycolysis in tumors, first observed by Otto Warburg, is crucial to support both the increased energy and synthetic demands required for high rates of proliferation. This metabolic shift in tumor cells is coordinated by up-regulating critical components of glycolysis, resulting in enhanced glucose uptake and lactate production even in the presence of ample oxygen (Vander Heiden et al., 2009). It is currently unclear whether reduced autophagy specifically elicits changes in glucose metabolism or causes more global metabolic shifts during Ras transformation. We are presently evaluating whether and how other metabolic pathways are affected by the loss or reduction of autophagy.
Although glucose withdrawal and energy depletion have been shown to be potent activators of autophagy as a survival response, we have unexpectedly found that the reduction or elimination of autophagy competence can actually reduce glycolytic capacity in a Ras-transformed cell. Hence, we speculate that autophagy may promote oncogenic Ras-driven tumor growth in specific metabolic microenvironments. In support, decreasing glucose concentrations inhibits soft agar colony formation in H-RasV12–expressing wild-type cells to levels approaching that of H-RasV12 autophagy-deficient cells. In contrast, both the proliferation and adhesion-independent transformation of autophagy-deficient cells are relatively insensitive to reductions in glucose availability. These alterations in glucose metabolism in autophagy-deficient cells may similarly impact transformation by other oncogenes, such as Myc and PI3K, which orchestrate global metabolic changes that contribute to the transformed phenotype, similar to activating mutations in Ras. Thus we are presently examining the impact of autophagy inhibition on glucose metabolism and transformation driven by other oncogenes.
MATERIALS AND METHODS
Cell culture
Noburu Mizushima (Tokyo Medical and Dental University) generously provided atg5+/+ and atg5−/− MEFs (simian virus 40 T-antigen immortalized). Masaaki Komatsu (Tokyo Metropolitan Institute) generously provided atg7+/+ and atg7−/−, atg3+/+, and atg3−/− MEFs (simian virus 40 T-antigen immortalized). All mouse fibroblasts were cultured in DMEM containing 25 mM glucose (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. MCF10A cells were cultured as described previously (Debnath et al., 2003), and MDA-MB-231 cells were grown in DMEM containing 25 mM glucose supplemented with 10% FBS, penicillin, and streptomycin. When indicated, MEFs and MDA-MB-231 cells were grown in DMEM containing 25 mM, 5.5 mM, 2.8 mM, or 1.4 mM glucose supplemented with 10% dialyzed FBS, penicillin, and streptomycin.
Antibodies and chemicals
A peptide corresponding to the N-terminus common to human, mouse, and rat MAP1LC3 was used to create α-LC3 rabbit polyclonal antibody (Fung et al., 2008). Other antibodies used included the following: α-p62 (Progen Biotechnik, Heidelberg, Germany), α-phospho-ERK1/2 (Invitrogen, Carlsbad, CA), α-ERK1/2 (Invitrogen), α-phopho-ribosomal protein S6 (Ser240/244, Cell Signaling Technology, Danvers, MA), α-ribosomal protein S6 (Cell Signaling Technology), α-ATG12 (Cell Signaling Technology), α-ATG7 (Santa Cruz Biotechnology, Santa Cruz, CA), α-ATG5 (Cell Signaling Technology), α-cleaved caspase-3 (Cell Signaling Technology), α-BCL-2 (BD Biosciences, Franklin Lakes, NJ), α-LDH-A (Cell Signaling Technology), and α-α-tubulin (Sigma-Aldrich, St. Louis, MO). Chemicals utilized included poly(2-hydroxyethyl methacrylate) (poly-HEMA), E64d, and pepstatin A (all from Sigma-Aldrich).
Generation of stable lines
The following retroviral vectors for stable gene expression have been described previously: pBABEpuro-H-RasV12, pBABEneo-H-RasV12, pBABEneo-Bcl-2, and pBABEpuroGFP-LC3 (Debnath et al., 2002). For retroviral transduction, vesicular stomatitis virus G–pseudotyped retroviruses were generated, and cells were infected and selected as previously described (Debnath et al., 2003).
RNA interference
pLKO.1 lentiviral expression plasmids containing shRNAs against ATG7 and ATG12 were purchased from Sigma-Aldrich (Mission shRNA), and viral particles were produced using a third-generation lentiviral packaging system in HEK293T cells. Following infection and drug selection, early passage stable pools (maximum of 3–5 passages) were utilized because the extended propagation of cells results in the loss of RNAi-mediated ATG silencing and autophagy inhibition. The target sequences for hairpins directed against ATG7 (NM_006395) are shATG7-1 (TRCN0000007584): GCCTGCTGAGGAGCTCTCCA and shATG7-2 (TRCN0000007587): CCCAGCTATTGGAACACTGTA, and the target sequence directed against ATG12 (NM_004707) is shATG12-1 (TRCN0000007393): TGTTGCAGCTTCCTACTTCAA.
Substratum detachment assays
Tissue culture plates coated with 6 mg/ml poly-HEMA in 95% ethanol were incubated at 37°C until dry. Cells were plated on poly-HEMA–coated plates at a density of 500,000–750,000 cells/well in six-well plates in their appropriate complete growth medium. The lysosomal inhibitors, E64d and pepstatin A, were added directly to the culture media at 10 μg/ml at 4–6 h before lysis to evaluate autophagic flux.
Immunoblotting
Attached or suspended cells were lysed in RIPA lysis buffer plus 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO3, 10 nM calyculin A, and protease inhibitors. Lysates were clarified by centrifugation for 15 min at 4°C, and protein concentrations were assessed using a bicinchoninic acid (BCA) protein assay (Thermo, Waltham, MA). Samples containing equal amounts of protein were boiled in SDS sample buffer (15–50 μg total protein per lane), resolved using SDS–PAGE, and transferred to polyvinylidene diflouride membrane. Membranes were blocked in phosphate-buffered saline (PBS) + 0.1% Tween 20 with 5% nonfat dry milk, incubated with the indicated primary antibodies overnight at 4°C, washed, incubated with horseradish peroxidase-conjugated secondary antibodies, and analyzed by enhanced chemiluminescence.
Soft agar colony formation assay
To evaluate anchorage-independent growth, 1 × 104 H-RasV12 MEF cells, 1 × 104 H-RasV12 MCF10A cells, or 2 × 104 MDA-MB-231 cells were resuspended in 0.35% agarose in growth media. Cells were plated on a solidified bed of 0.5% agarose in growth media in 3.5-cm plates. Plates containing H-RasV12 MEF and H-RasV12 MCF10A cells were fixed and stained with 0.005% crystal violet after 14 d, and plates containing MDA-MB-231 cells were fixed following 21 d of growth. Bright-field images of H-RasV12 MCF10A and MDA-MB-231 colonies were taken using a 4× objective. Plates with H-RasV12 MEFs were scanned. The number of colonies per field (for MCF10A and MDA-MB-231 cells) and number of colonies per plate (for MEFs) were counted using MetaMorph (version 6.0).
Glucose uptake, LDH activity, and media glucose concentration analysis
To measure glucose uptake, cells were incubated with growth media containing 100 μM 2-NBDG (Invitrogen) for the indicated time points up to 8 h, washed three times with PBS, and detached for FACS analysis. Mean fluorescence intensity of cells was obtained using a Becton Dickinson FACSCalibur flow cytometer, and data were analyzed using CellQuest Pro v5.1.1 software. LDH activity was measured using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega, Madison, WI). To measure glucose concentrations, conditioned media were collected from cells on the days indicated and assayed using the Amplex Red Glucose/Glucose Oxidase Assay Kit (Invitrogen).
Cell cycle analysis and growth assays
For cell cycle analysis following ECM detachment, cells were either suspended on poly-HEMA–coated plates (suspension) or grown attached on tissue culture plastic for 48 h. Cells in suspension were washed and incubated in 0.25% trypsin for 10 min at 37°C to break up cell clumps and generate single-cell suspensions. Both suspended and attached cells were collected and fixed in 70% ice-cold EtOH. Cells were stained with 20 μg/ml PI in 0.1% Triton X-100 in PBS with 200 µg/ml RNase A and incubated at 37°C for 15 min. Cell cycle profiles were collected using a Becton Dickinson FACSCalibur flow cytometer, and data were analyzed using FlowJo v. 8.8.6 cell cycle analysis software. For cell growth assays, 4 × 104 MEFs or 1.1 × 105 MDA-MB-231 cells were plated on 6-cm plates, and cells were counted using a hemocytometer every 24 h for up to 5 d.
De novo protein translation analysis
The indicated cell types were grown either attached or suspended for 24 h. Cells were then incubated with methionine-free DMEM (Invitrogen) for 1 h, followed by 100 μM Click-iT AHA (Invitrogen), a methionine analog, in methionine-free DMEM to pulse label newly synthesized proteins for 3 h. Cells were lysed, and incorporated Click-iT AHA was detected using the TAMRA Click-iT Protein Analysis Detection Kit (Invitrogen) following the manufacturer's instructions. Total protein levels were measured and equilibrated using BCA assay (Thermo), and 10 μg total protein was loaded per well. Following detection of newly synthesized proteins, gels were stained with SYPRO Ruby gel stain (Invitrogen) to detect total protein levels. TAMRA fluorescence (newly synthesized proteins) and SYPRO Ruby fluorescence (total protein) were detected using a FLA-5100 imager (FujiFilm, Tokyo, Japan) and analyzed using Multi Gauge software v3.X. Newly synthesized protein levels were normalized to total protein levels.
NMR acquisition and analysis
Cells were grown in medium containing equal concentrations (12.5 mmol/L) of [1-13C]glucose and unlabeled glucose for 24 h, after which medium was collected and cell extracts were prepared using the dual-phase extraction method (Tyagi et al., 1996). Briefly, cells were rinsed with ice-cold saline, fixed in 10 ml ice-cold methanol and scraped from the culture flask surface, and vigorously vortexed, upon which 10 ml ice-cold chloroform was added, followed by 10 ml ice-cold deionized water, to the fixed cells. After phase separation, the aqueous phase was lyophilized, and the protein pellet was air dried. Protein concentration was determined using the BCA assay. To acquire 13C spectra, the aqueous fraction was reconstituted in 500 μl deuterium oxide, and NMR spectra were acquired on a 600-MHz INOVA spectrometer (Varian; Palo Alto, CA) using a 30° flip angle, 3.5-s repetition time, and broadband proton decoupling. Analysis of the NMR spectra was performed using ACD/Spec Manager software version 9.15 (Advanced Chemistry Development, Toronto, Canada). The concentrations of metabolites were determined with respect to an external reference, 2,2,3,3-tetradeutero-3-(trimethylsilyl)propionic acid (TSP), with a known concentration.
Statistics
Experimental groups were compared using Student's t test for pairwise comparisons or analysis of variance (ANOVA), followed by Tukey's honestly significant difference test.
Supplementary Material
Acknowledgments
We thank Davide Ruggero for critically reading the manuscript and Noboru Mizushima and Masaaki Komatsu for providing reagents. Grant support includes the National Institutes of Health (RO1CA126792 to JD and RO1 CA130819 to SR); a University of California, San Francisco, Program for Breakthrough Biomedical Research Integrative Award (to JD and SR); a Culpeper Medical Scholar Award (Partnership for Cures) to JD; and the California Tobacco-Related Diseases Research Program (18XT-0106) to JD. RL is a Department of Defense Breast Cancer Research Program Predoctoral Scholar (W81XWH-08-1-0759).
Abbreviations used:
- ATG
autophagy gene
- ECM
extracellular matrix
- E/P
E64d and pepstatin A
- HSP-1
heat shock protein 1
- LDH
lactate dehydrogenase
- shRNA
short-hairpin RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0500) on November 30, 2010.
REFERENCES
- Aki T, Yamaguchi K, Fujimiya T, Mizukami Y. Phosphoinositide 3-kinase accelerates autophagic cell death during glucose deprivation in the rat cardiomyocyte-derived cell line H9c2. Oncogene. 2003;22:8529–8535. doi: 10.1038/sj.onc.1207197. [DOI] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2010;584:1427–1435. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaradonna F, Sacco E, Manzoni R, Giorgio M, Vanoni M, Alberghina L. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25:5391–5404. doi: 10.1038/sj.onc.1209528. [DOI] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPaola RS, et al. Therapeutic starvation and autophagy in prostate cancer: a new paradigm for targeting metabolism in cancer therapy. Prostate. 2008;68:1743–1752. doi: 10.1002/pros.20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- Gilmore AP. Anoikis. Cell Death Differ. 2005;12(suppl 2):1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya A, Cooksey R, Cox JE, Wang V, McClain DA, Tantin D. Oct1 loss of function induces a coordinate metabolic shift that opposes tumorigenicity. Nat Cell Biol. 2009;11:320–327. doi: 10.1038/ncb1840. [DOI] [PubMed] [Google Scholar]
- Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Tyagi RK, Azrad A, Degani H, Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magn Reson Med. 1996;35:194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.