Telomeric recombination has been observed in telomerase-negative alternative lengthening of telomeres in human cancer cells and following telomerase inhibition or gene deletion. This study shows that telomeric recombination mechanisms can also be activated by dysfunctional telomeres without telomerase inhibition in telomerase-positive cells.
Abstract
Telomere maintenance is essential for cellular immortality, and most cancer cells maintain their telomeres through the enzyme telomerase. Telomeres and telomerase represent promising anticancer targets. However, 15% of cancer cells maintain their telomeres through alternative recombination-based mechanisms, and previous analyses showed that recombination-based telomere maintenance can be activated after telomerase inhibition. We determined whether telomeric recombination can also be promoted by telomere dysfunction. We report for the first time that telomeric recombination can be induced in human telomerase-positive cancer cells with dysfunctional telomeres.
INTRODUCTION
Telomere maintenance is essential for cellular immortality, and cancer cells must acquire a mechanism to counteract progressive telomere attrition that occurs as a result of successive cellular division and nuclease degradation (Watson, 1972; Olovnikov, 1973; Makarov et al., 1997; Palm and de Lange, 2008). In normal cells lacking a compensatory mechanism, telomere shortening eventually leads to replicative senescence triggered by a p53-dependent DNA damage response (Smogorzewska and de Lange, 2002). Mutation in checkpoint components or viral transformation can allow cells to bypass this senescence checkpoint, but further telomere shortening will lead to genomic instability, chromosomal fusion, breakage–fusion–bridge cycles, and eventually cell death. However, rare survivors are able to escape cell death by acquiring a mechanism to replenish their telomere length and become immortalized.
More than 85% of cancer cells maintain telomere length by the ribonucleoprotein telomerase that adds repetitive telomeric TTAGGG sequences to the 3′ end of chromosomes (Shay and Bacchetti, 1997). The catalytic core of the enzyme is composed of a telomerase reverse transcriptase (TERT) subunit and an internal telomerase RNA template subunit to catalyze de novo addition of telomeric tracts (Autexier and Lue, 2006). Telomere length can also be maintained by a recombination-based alternative lengthening of telomeres (ALT) mechanism (Dunham et al., 2000). Human tumors or immortalized cells utilizing ALT are characterized by a lack of telomerase activity and by exceptionally long and heterogeneous telomeres, ranging from <2 kb to >50 kb. ALT-associated promyelocytic leukemia bodies (APBs) are found in ALT cells and contain a high number of repair and recombination proteins, telomeric DNA, and telomere-specific proteins in addition to the standard promyelocytic leukemia (PML) body components, the PML protein and Sp100 (Yeager et al., 1999; Draskovic et al., 2009). Finally, human ALT tumors feature homologous recombination (HR)–mediated events leading to remarkably high levels of telomeric sister-chromatid exchange (T-SCE) and extrachromosomal telomeric DNA, of both linear (Ogino et al., 1998; Tokutake et al., 1998) and circular (termed t-circles) forms (Cesare and Griffith, 2004; Wang et al., 2004).
There have been several reports of telomerase-negative cancer cells that do not have all the characteristics typically associated with ALT cells (Cerone et al., 2005; Fasching et al., 2005; Marciniak et al., 2005) and a number of glioblastoma multiforme tumors and some types of soft tissue sarcomas that have neither detectable telomerase nor the usual phenotypic characteristics of ALT (Hakin-Smith et al., 2003; Costa et al., 2006), highlighting the potential for complex and varied mechanisms of telomere maintenance. The occurrence of telomere maintenance by recombination via more than one mechanism was first described in yeast (Teng and Zakian, 1999; Chen et al., 2001). In the absence of telomerase, type I survivors emerge with short telomeres and amplified subtelomeric sequences, while type II survivors maintain elongated telomeres. Recently, telomere maintenance without significant telomere elongation was also observed in telomerase-negative primary and tumor mouse cells with short telomeres, telomerase-positive mouse cells with short telomeres, and lymphocytes from dyskeratosis congenita patients with short telomeres due to a mutation in hTERT (Morrish and Greider, 2009). Telomere maintenance in these cells was found to be effected by subtelomeric or telomeric recombination, providing further evidence that several recombination-based mechanisms can also contribute to telomere maintenance in mammalian cells.
Given the fundamental role of telomere length maintenance in cellular immortalization, antitelomerase and antitelomere strategies represent promising approaches for the development of anticancer therapies. A promising antitelomerase approach consists of inhibiting telomerase activity, which leads to telomere shortening and growth arrest or apoptosis. However, one limitation of telomerase inhibition is the lag phase associated with the time required for telomeres to shorten sufficiently to observe antiproliferative effects. A consequence of this lag phase is that telomerase inhibition results in apoptosis of human cells with short telomeres while cells with long telomeres have time to adapt (Hahn, 1999; Zhang et al., 1999). Hence, there is concern that telomerase inhibition in cancer cells could lead to telomere maintenance by alternative mechanisms including recombination. Alternative telomere maintenance mechanisms are observed after telomerase inhibition with a dominant-negative hTERT (Bechter et al., 2004) or genetic deletion of telomerase (Hande et al., 1999; Niida et al., 2000; Chang et al., 2003; Morrish and Greider, 2009).
An alternative to antitelomerase strategies is to target telomere integrity. Telomere integrity is maintained by the association of telomeres with a complex of six specific telomeric shelterin proteins (TRF1, TRF2, POT1, TIN2, TPP1, and Rap1), which cap telomere ends and prevent them from being recognized as double-strand breaks, repress DNA repair reactions, and regulate telomerase access to telomeres (Palm and de Lange, 2008). It has been suggested that alterations in telomere capping might facilitate the activation of ALT in human cells (Cesare and Reddel, 2008), most likely by allowing HR reactions at telomeres. Studies in S. cerevisiae and Kluyveromyces lactis revealed increased telomeric recombination after induction of telomere dysfunction through mutation or deletion of telomere-capping proteins (Teng et al., 2000; Grandin et al., 2001; Iyer et al., 2005). In addition, mutant telomerase RNA expression and the resulting incorporation of mutated telomeric repeats in K. lactis lead to increased recombination and elongated telomeres (Underwood et al., 2004). More recently, studies in yeast revealed that loss of the double-strand telomeric DNA-binding protein Rap1 or telomeric mutations that disrupt the binding of Rap1 can lead to recombinational telomere maintenance (Bechard et al., 2009; Sfeir et al., 2010). Finally, elevated levels of T-SCEs and formation of t-circles are observed in mice after POT1a deletion (Wu et al., 2006). However, it is not known whether telomere dysfunction can also increase telomere-specific HR in telomerase-positive human cells or whether this could eventually lead to telomere maintenance by mechanisms other than telomerase.
We previously reported that telomere destabilization induced by the expression of a mutant human telomerase RNA template (MuA-hTR) increases the sensitivity of human breast cancer cells such as YCC-B2 cells to chemotherapeutic drugs (Cerone et al., 2006). The MuA-hTR reconstitutes an active mutant telomerase enzyme that adds TTTGGG instead of wild-type TTAGGG repeats to the ends of chromosomes; thus the sensitization of the MuA-hTR–expressing cells to chemotherapy is most likely due to an improper recruitment of shelterin proteins and telomere uncapping (Guiducci et al., 2001a; Kim et al., 2001; Li et al., 2004; Cerone et al., 2006). In the current study, we further analyzed three MuA-hTR–expressing clones derived from the YCC-B2 cancer cell line (telomere length 11 kb) with telomere lengths of approximately 6 kb (clone 17), 15 kb (clone 23), and 8 kb (clone 27) as assessed by telomere terminal restriction fragment (TRF) analysis (Cerone et al., 2006). Quantitative fluorescence in situ hybridization (Q-FISH) analysis at the single-cell level showed an increase in telomere-length heterogeneity in all three clones expressing MuA-hTR compared with the control cells (Cerone et al., 2006). We demonstrate that telomere dysfunction increases HR at the telomeres of these human breast cancer cells as shown by increased levels of T-SCEs and variation of pq ratio. We also show that some of the MuA-hTR–expressing cells share some characteristics with ALT cells, including the presence of APBs and t-circles.
RESULTS
Increased telomere dysfunction–induced foci in MuA-hTR–expressing telomerase-positive breast cancer cells
Various tumor cell lines expressing different mutant telomerase RNAs induce a DNA damage response at telomeres and mild phenotypes such as growth inhibition to more severe phenotypes including apoptosis (Marusic et al., 1997; Guiducci et al., 2001b; Kim et al., 2001; Li et al., 2004; Cerone et al., 2006; Goldkorn and Blackburn, 2006; Stohr and Blackburn, 2008). However, in our earlier study, MuA-hTR expression had only a mild effect on cellular viability, probably because of the presence of endogenous hTR, which can likely partially restore normal telomere function due to the synthesis of wild-type telomeric repeats. We determined whether MuA-hTR expression could engage a primary DNA damage response at telomeres of YCC-B2 cells, which might account for its ability to exacerbate the antiproliferative effects of chemotherapeutic drugs (Cerone et al., 2006). The cytological manifestation of a DNA damage response at telomeres is called telomere dysfunction–induced foci (TIF) and can be observed by the colocalization of DNA damage protein foci with telomeric proteins (d’Adda di Fagagna et al., 2003; Takai et al., 2003).
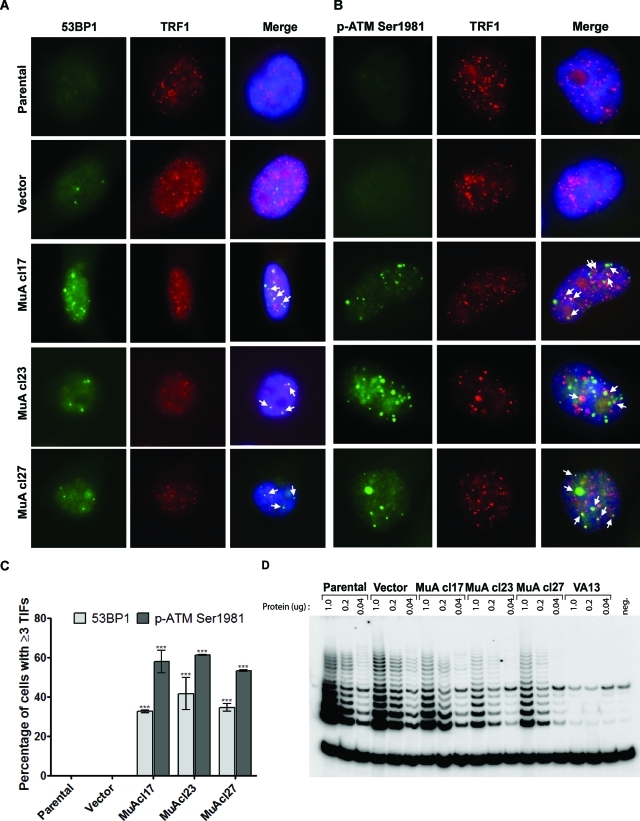
MuA-hTR expression resulted in the formation of foci containing the DNA damage protein 53BP1 and the telomeric protein TRF1 (Figure 1A). TIFs containing the phosphorylated (Ser1981) form of ataxia telangiectasia mutated (ATM) were also observed in cells expressing MuA-hTR, consistent with deprotected telomeres (Figure 1B). Quantification of TIF-positive cells is reported in Figure 1C. Hence, the incorporation of mutant telomere repeats in YCC-B2 cells can elicit a primary DNA damage response at telomeres, but this response is insufficient to lead to apoptosis or senescence (Cerone et al., 2006). Instead, MuA-hTR–expressing cells maintained growth rates similar to parental and vector-containing cells despite extensive, continuous passaging for 3 mo. Persistence of mutant telomerase activity and mutant telomeric repeats confirmed that the mutants’ proliferation was not due to loss of MuA-hTR expression.
FIGURE 1:
Increased formation of 53BP1- or p-ATMSer1981–containing TIFs in the MuA-hTR–expressing clones. (A) YCC-B2 parental, vector-containing, and MuA-hTR–expressing cells were coimmunostained with an antibody recognizing 53BP1 (BP13, green) and an antibody recognizing TRF1 (#370, red). 53BP1 is colocalized with TRF1, indicating telomere dysfunction–induced foci (yellow). (B) YCC-B2 cells were also coimmunostained with an antibody against the activated and phosphorylated (Ser1981) form of ATM (10H11.E12, green) and anti-TRF1 (#370, red). Telomere dysfunction–induced foci are shown in yellow. (C) Quantification of TIF-positive cells. Cells with three or more foci colocalizing with TRF1 were counted as TIF positive. For each cell type, 100 cells were counted. Data are mean ± standard error of the mean (SEM) for three independent experiments. Asterisks indicate statistical significance from vector-containing cells (***P < 0.001). (D) Relative telomerase activity was measured by TRAP using serial fivefold dilutions of protein extracts.
Previous studies have shown that hTR levels are limiting in telomerase-positive human cell lines and that increasing hTR levels results in increased telomerase activity and telomere lengthening (Cristofari and Lingner, 2006; Pickett et al., 2009). To address the possibility that increased telomere length of MuA-hTR–expressing YCC-B2 clone 23 and increased telomere-length heterogeneity of all three clones expressing mutant hTR could result from increased endogenous wild-type telomerase activity, we assessed the telomerase activity of the three YCC-B2 clones expressing the mutant hTR compared with the parental YCC-B2 cells. We found that telomerase activity was not increased (Figure 1D) in the cells expressing the mutant hTR, consistent with previous studies reporting the retention of endogenous wild-type telomerase activity levels upon expression of mutant hTR (Kim et al., 2001).
Telomeric recombination between sister chromatids (T-SCEs) after MuA-hTR expression
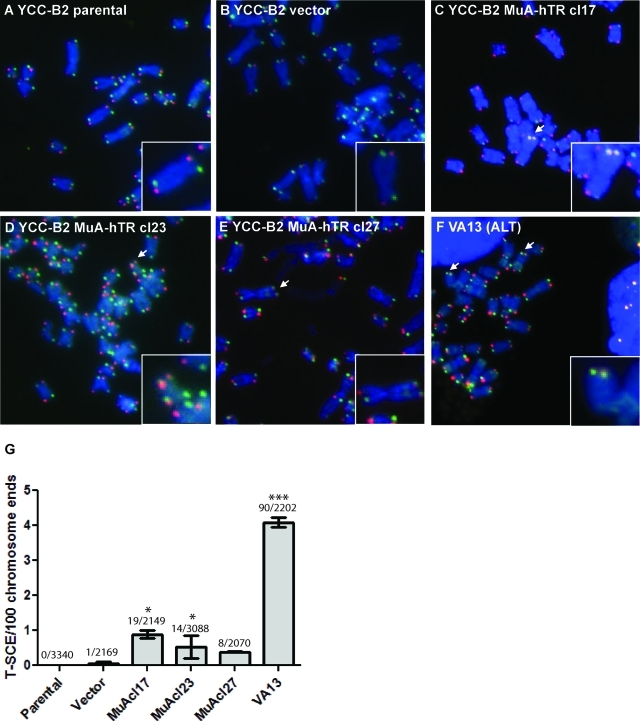
Telomere protection is essential in repressing HR of sister telomeres (T-SCEs). Elevated levels of T-SCEs are observed in cells utilizing the HR-based ALT pathway (Bechter et al., 2004; Londono-Vallejo et al., 2004). We verified whether the potential telomere deprotection in YCC-B2 cells could favor T-SCE. T-SCEs can be detected by the chromosome-orientation FISH (CO-FISH) technique using a fluorescein isothiocyanate (FITC)–conjugated probe against the C-rich strand and a Cy3-conjugated probe against the G-rich strand, and exchanges between two sister chromatids can be visualized by a double signal at one telomere. Consistent with recombination occurring at telomeres of MuA-hTR–expressing cells, T-SCE events increased in the MuA-hTR–expressing clones when compared with vector-containing cells (Figure 2, A–E). The increased T-SCE events were statistically significant for MuA-hTR–expressing clones 17 and 23, which showed an average of 0.89% and 0.53% T-SCE events, respectively, compared with 4.1% for the control VA13 cell line (Figure 2, C–F). Quantification of T-SCE events is reported in Figure 2G.
FIGURE 2:
Telomeric recombination between sister chromatids (T-SCE) after MuA-hTR expression. (A–F) Representative CO-FISH images of metaphase spreads of YCC-B2 cells. Slides were hybridized with an FITC-conjugated probe (FITC-[TTAGGG]3) against the C-rich strand (green) and a Cy3-conjugated probe(Cy3-[CCCTAA]3) against the G-rich strand (red). Chromosomes were stained with DAPI (blue). Chromosomes from (A) parental cells and (B) vector-containing cells exhibit two telomeric signals of each color per chromosome. No significant increase in T-SCEs was found in these cells. (C–E) The three MuA-hTR–expressing clones contain low levels of T-SCEs represented by chromosomes with double signals at both sister telomeres. (F) The VA13 control cell line also shows T-SCEs. Examples of T-SCEs are indicated by arrows and enlarged in inset. (G) Quantification of the T-SCEs for each cell type. The total number of T-SCE out of the total number of chromosomes analyzed is indicated over bars. Error bars give standard deviations for two independent experiments. Asterisks indicate significant differences from vector-containing cells (***P < 0.001 and *P < 0.05).
Mutant telomerase RNA expression is associated with changes in telomere pq ratios
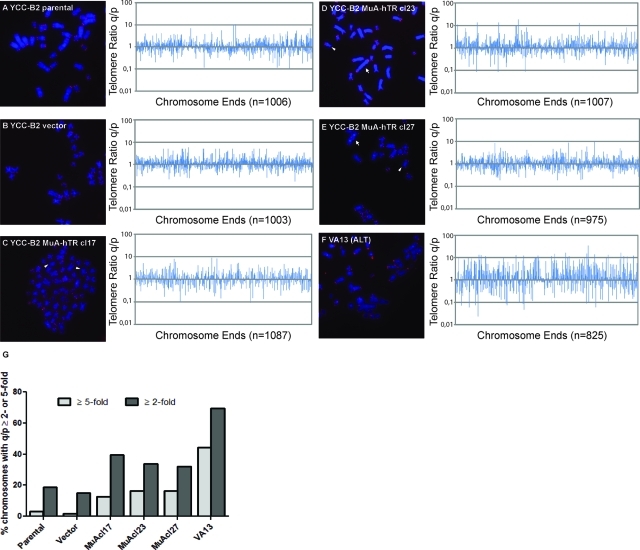
To determine whether the observed telomere-length heterogeneity in MuA-hTR–expressing cells could be due to telomeric recombination, we measured the pq ratio of the telomere signals. In telomerase-positive cells, telomere lengths of the p and q arms of a chromosome should conserve a constant ratio. However, telomere recombination results in either increased or decreased telomere length at one chromosome arm. Thus the pq ratio of the telomeres of a chromosome that has undergone recombination will vary from the pq ratio of the other copies of that chromosome in a cell population (Perrem et al., 2001). Originally, pq ratio analysis was performed on a specific chromosome in a growing population of cells; however, the analysis can also be expanded to all chromosomes of a metaphase spread, as described in Morrish and Greider (2009). Telomere lengths were assessed by Q-FISH and TFL-Telo analysis, and pq ratios were measured and plotted (Figure 3, A–E). Significant increases in pq ratios were observed in the three MuA-hTR–expressing clones when compared with parental and vector-containing cells. The pq ratios for each metaphase were plotted along the x-axis of the graph from left to right. The distribution of the changes along the x-axis of each graph demonstrates that changes in pq ratios were observed in all metaphases examined and cannot be attributed to changes in only a subset of metaphases. Chromosomes with ends lacking detectable telomeric signals (signal-free ends, or SFEs) or telomere fusions were given a pq ratio value of 5 and were not plotted for better clarity. The VA13 cell line was used as a control for elevated pq ratios (Figure 3F). To quantify the changes in pq ratios, the percentage of chromosomes showing a variation greater than twofold or greater than fivefold was measured (Figure 3G). All three clones showed a higher proportion of chromosomes with a qp ratio ≥ twofold and a qp ratio ≥ fivefold compared with parental and vector-containing cells. We also observed telomere fusions with detectable telomere signals as well as SFEs in the MuA-hTR–expressing cells (10.1%, 13.5%, and 15.5% for the MuA-hTR–expressing clones 17, 23, and 27, respectively, versus 1.9% and 1.2% for the parental and vector-containing cells and 30.8% for the control VA13 cells), although we did not detect them previously (Cerone et al., 2006). We propose that extensive passaging might have allowed the emergence of fusions and SFEs.
FIGURE 3:
Pq ratio variation is increased in cells expressing MuA-hTR. (A–F, left panel) Representative FISH images of metaphase spreads of YCC-B2 parental, vector-containing, MuA-hTR–expressing, and VA13 control cells. Telomeres were hybridized with a Cy3-[CCCTAA]3 telomere probe (red), and chromosomes were stained with DAPI (blue). Cy3 signal intensity correlated with telomere length. Arrows show examples of telomere fusions with detectable telomere signal at the site of fusions. Arrowheads indicate telomere ends without detectable signal (signal-free ends [SFEs]). (A–F, right panel) The ratios (q/p) of telomere signals for the q arm and the p arm for different chromosomes are plotted along the x-axis of the graph from left to right. The distribution of the changes along the x-axis of each graph demonstrates that changes in pq ratios were observed in all metaphases examined and cannot be attributed to changes in only a subset of metaphases. Telomere ratios (q/p) are represented on a log scale on the y-axis. SFEs and telomere fusions were given a pq ratio value of 5 but were excluded from these graphs for better clarity. The total number of chromosome ends analyzed is indicated below graphs. (G) The percentage of chromosomes with a telomere pq ratio greater than twofold (q/p ≥ 2 or q/p ≤ 0.5) or telomere pq ratio greater than fivefold (q/p ≥ 5 or q/p ≤ 0.2) is represented.
MuA-hTR expression results in the accumulation of circular extrachromosomal telomeric DNA
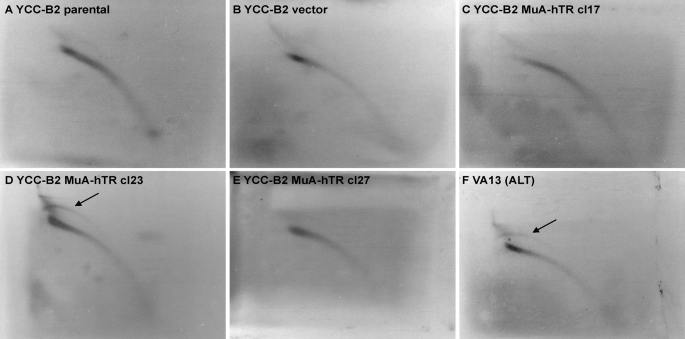
ALT cells contain abundant t-circles or extrachromosomal telomeric repeat (ECTR) DNA in a circular form. T-circles are generated by resolution of the t-loop junction, which creates a truncated telomere in addition to a t-circle. We analyzed the MuA-hTR–expressing cells for t-circles by performing two-dimensional pulse-field gel electrophoresis (2D-PFGE) followed by hybridization with a telomeric-specific probe (Cesare and Griffith, 2004). No arcs were detected in the YCC-B2 parental and vector-containing cells or MuA-hTR–expressing clone 17 and clone 27 (Figure 4, A–C and E). We detected an arc of double-stranded circular DNA in both the control VA13 cells and the YCC-B2 MuA-hTR clone with the longest telomeres (cl23) (Figure 4, D and F), suggesting that the expression of a mutant telomerase RNA can lead to the formation of t-circles. The generation of t-circles in the MuA-hTR clone harboring the longest telomeres is in accordance with a recent study suggesting that the generation of t-circles is a consequence of a telomere-length control mechanism, by which telomeric repeats would be trimmed when the telomeres reach a certain threshold length (Pickett et al., 2009).
FIGURE 4:
The incorporation of mutant telomeric repeats results in accumulation of t-circles. 2D-PFGE of TRFs from YCC-B2 (A) parental, (B) vector-containing, (C–E) MuA-hTR–expressing, and (F) VA13 cells, digested with RsaI and HinfI. Hybridization was performed in gel with a γ-32P–labeled C-rich probe under denaturing conditions. The black arrows indicate circular telomeric DNA.
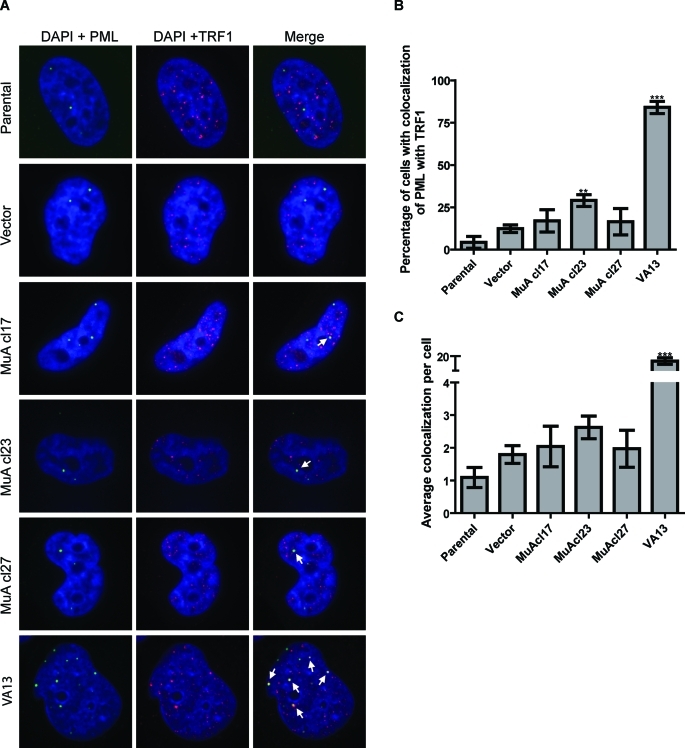
Increased formation of PML bodies associated with telomeric DNA in telomerase-positive cells
ALT cells contain specific nuclear structures called APBs (Yeager et al., 1999). We analyzed MuA-hTR–expressing cells for the presence of APBs by immunostaining with antibodies against the PML and TRF1 proteins (Figure 5A). We found an increased proportion of cells with APBs in the three MuA-hTR–expressing clones compared with the parental and vector-containing cells (Figure 5, B and C), although the increase compared with vector-containing cells was statistically significant only in YCC-B2 MuA-hTR clone 23. The VA13 cell line was used as a positive control for APBs.
FIGURE 5:
MuA-hTR–expressing cells contain APBs. (A) Immunostaining with an antibody against the PML protein (N-19, green) and an antibody against the telomeric-binding protein TRF1 (#370, red). APBs were detected by the colocalization of the fluorescent signals in merged images (yellow). Images represent only one slice in the Z-stack. Arrows indicate colocalization. (B) Quantification of APBs in the YCC-B2 MuA-hTR–expressing cells compared with parental, vector-containing, and VA13 control cells. The percentage of cells showing four or more colocalization events per nucleus is shown. A minimum of 50 cells were scored for each population. Three-dimensional images were obtained using a Leica DMI600B confocal laser scanning microscope (63×), and colocalization was analyzed with Volocity 5.4.1 software. (C) The average number of APBs per cell is represented. Data are mean ± SEM for three independent experiments. Asterisks indicate significant differences from vector-containing cells (***P < 0.001 and **P < 0.01).
DISCUSSION
There is concern that targeting the enzyme telomerase as an anticancer therapy might lead to resistance mechanisms, including the activation of the recombination-based pathways of telomere maintenance in the cancer cells treated. Telomeric recombination after telomerase inhibition in conjunction with an MSH6 defect has been reported (Bechter et al., 2004). Additionally, genetic deletion of telomerase can also lead to activation of alternative telomere maintenance mechanisms (Hande et al., 1999; Niida et al., 2000; Chang et al., 2003; Morrish and Greider, 2009). The induction of telomere dysfunction is currently investigated as an alternative anticancer strategy that would be independent of telomere length and mechanism of telomere maintenance (telomerase or recombination based) (Marusic et al., 1997; Guiducci et al., 2001a; Kim et al., 2001; Li et al., 2004; Cerone et al., 2006; Goldkorn and Blackburn, 2006; Stohr and Blackburn, 2008). Nonetheless, it is also important to consider the possibility that such approaches could lead to the emergence of alternative mechanisms of telomere maintenance. Here we report for the first time that telomeric recombination can be induced by dysfunctional telomeres without telomerase inhibition. Although telomerase and ALT can coexist when hTERT is introduced experimentally in ALT cells (Cerone et al., 2001; Grobelny et al., 2001; Perrem et al., 2001), it was not known whether endogenous telomere maintenance pathways can coexist spontaneously in tumor cells. In this study, we report that the expression of a mutant telomerase RNA template can favor telomeric recombination in telomerase-positive cells, as shown by the increased level of T-SCEs and variation of pq ratios. Some MuA-hTR–expressing cells also exhibited additional characteristics of ALT cells, including APBs and t-circles, although they conserved telomere lengths that were less heterogeneous than ALT cells (Cerone et al., 2006).
In previous reports using HT1080, MCF-7, LNCaP, HCT116, and UM-UC-3 tumor cell lines, the introduction of mutant template telomerase RNA decreased cell proliferation and viability and increased senescence or apoptosis (Marusic et al., 1997; Guiducci et al., 2001b; Kim et al., 2001; Li et al., 2004; Goldkorn and Blackburn, 2006; Stohr and Blackburn, 2008). However, the expression of a mutant telomerase RNA in YCC-B2 breast cancer cells had only a mild effect on cell viability and proliferative ability in the absence of chemotherapeutic drugs (Cerone et al., 2006). The expression of a mutant telomerase RNA template leads to the synthesis of mutant telomeric sequences that are predicted to interfere with the binding of shelterin proteins and proper capping of the telomeres (Marusic et al., 1997; Guiducci et al., 2001b; Kim et al., 2001; Cerone et al., 2006). In our study, improper capping of YCC-B2 cells led to recombination at telomeres without affecting cell proliferation and viability. What favored recombination rather than growth defects and arrest is unclear. Differences in the levels of MuA-hTR expression due to the promoter used to drive its expression or cell-specific particularities could have contributed to a milder dysfunction that was not sufficient to induce cell death but rather favored the activation of HR at telomeres. Information about the exact number of mutant repeats at each telomere and their organization and intercalation between wild-type repeats could be useful to understand the regulation of the balance between decreased proliferation and increased recombination. Loss of tumor suppression function might also be permissive for telomeric recombination. More than 95% of ALT cell lines are impaired in the p53 pathway (Bryan et al., 1995; Henson and Reddel, 2010), and in a more recent study in glioblastoma multiforme patients, 78% of all ALT-positive tumors were found to be defective for p53 while only 21% of telomerase-positive tumors were defective for p53 (Chen et al., 2006). Together, the results suggest that p53 deficiency could favor activation of HR events. Despite one study reporting rapid growth inhibition of HCT116 p53–null cells after MuA-hTR expression (Li et al., 2004), we suggest that the nonfunctional p53 status of YCC-B2 cells (Cerone et al., 2006) has contributed to the development of recombination in these cells.
Increased recombination and ALT activation in mammalian cells have been linked to disruption of telomere capping. Deletion of POT1a in mouse results in aberrant HR at the telomeres as shown by the formation of t-circles and elevated level of T-SCEs (Wu et al., 2006). Mutation in POT1b also increases levels of T-SCEs as well as telomere end-to-end fusions (He et al., 2006; Wu et al., 2006). Simultaneous deletion of TRF2 and Ku in mouse embryonic fibroblasts in a ligase 4–null background also creates levels of T-SCEs similar to levels observed in ALT cells (Celli et al., 2006). Finally, deletion of the basic domain of TRF2 increases formation of t-circles in human cells (Wang et al., 2004). Human POT1 and TRF2, respectively, bind to single- and double-strand TTAGGG repeats with extremely high specificity (Bianchi et al., 1999; Loayza et al., 2004). Thus it is not surprising that the effects we observe after expression of a mutant hTR that dictates the synthesis of TTTGGG sequences resemble the effects created by POT1 and TRF2 deletions.
Different mechanisms could explain how telomere uncapping can lead to telomere recombination (reviewed in Cesare and Reddel, 2008). T-loop formation is a mechanism remarkably similar to the HR process, and many recombination proteins including RAD51, RAD52, XRCC3, and NBS1 and telomeric proteins TRF1, TRF2, and TIN2 possess t-loop formation activity in vitro (Verdun and Karlseder, 2006). Thus HR may contribute to telomere capping by participating in the formation of the t-loop. However, recombination events at telomeres must be tightly regulated in order to prevent T-SCEs, t-loop resolution, and ultimately telomere maintenance by recombination. In this study we show that the introduction of a mutated template telomerase RNA in telomerase-positive breast cancer cells impairs the tight regulation of HR at telomeres to allow inappropriate recombination of telomeres. Aberrant control of telomeric recombination in YCC-B2 MuA-hTR–expressing cells likely results from disruption of TRF1, TRF2, and POT1 binding on mutated telomeres, a defective p53 pathway and subsequent disturbance of normal telomere damage responses, and possibly yet unknown factors.
MATERIALS AND METHODS
Cell culture
YCC-B2 breast cancer cells (Park et al., 1998) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). VA13 ALT cells were cultured in α-DMEM supplemented with 10% FBS.
CO-FISH
Cells were incubated in fresh medium containing 5′-BrdU/5′–BrdC (3:1 ratio; Sigma, St. Louis, MO) for 16 h, and Colcemid (Life Technologies, Rockville, MD) was added to the medium for the last 3 h of incubation to accumulate mitotic cells. Chromosome preparations were then obtained according to standard cytogenetic methods. Slides were treated with 0.5 mg/ml RNase A (Fermentas, Hanover, MD) in phosphate-buffered saline (PBS) for 10 min at 37°C and then stained with Hoechst 33258 (Invitrogen, Carlsbad, CA) in 2× NaCl–sodium citrate for 15 min at room temperature. Cells were exposed to 254 nm UV light (GS Gene linker; Bio-Rad, Hercules, CA) at a dose of 180 mJ, and the BrdU/C-substituted DNA strands were digested with 800 U of Exonuclease III (Promega, Madison, WI) in 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 5 mM dithiothreitol for 10 min at room temperature. Dehydrated slides were hybridized with an FITC-conjugated (T2AG3)3 peptide nucleic acid (PNA) probe (Cambridge Research Biochemicals, Cleveland, UK) at a final concentration of 0.5 μg/ml in 70% formamide, 0.5% blocking reagent (Roche), and 10 mM Tris-HCl (pH 7.2) for 2 h at room temperature; rinsed with 70% formamide, 0.1% bovine serum albumin (BSA), and 10 mM Tris-HCl (pH 7.2) for 5 s; and hybridized with a Cy3-conjugated (C3TA2)3 PNA probe (Cambridge Research Biochemicals) at a final concentration of 0.5 μg/ml in 70% formamide, 0.5% blocking reagent (Roche, Indianapolis, IN), and 10 mM Tris-HCl (pH 7.2) for 2 h at room temperature. After hybridization, cells were washed two times in 70% formamide, 0.1% BSA, and 10 mM Tris-HCl (pH 7.2) for 30 min and three times in 0.1 M Tris-HCl (pH 7.2), 0.15 M NaCl, and 0.08% Tween-20 for 5 min. The slides were then dehydrated through an ethanol series (70%, 95%, 100%), air-dried, and mounted in a 4′,6-diamino-2-phenylindole (DAPI)/antifade solution (Chemicon, Temecula, CA). Images were captured using a Zeiss M1 fluorescence microscope (63× and 100×). Only T-SCE events observed with both leading- and lagging-strand probes simultaneously were scored as positive.
Immunofluorescence
For detection of APBs, cells were grown on coverslips for 24 h, fixed in 4% formaldehyde for 10 min at room temperature, and permeabilized in 0.5% NP-40 (Sigma) for 10 min at room temperature. Slides were incubated with 1:200 goat anti-PML (N-19, Santa Cruz Biotechnology, Santa Cruz, CA) and 1:4000 rabbit anti-TRF1 (kindly provided by Titia de Lange) in phosphate-buffered gelatin (PBG; 0.2% fish gelatin [Sigma], 0.2% BSA in PBS) at 37°C for 2 h. Slides were washed in PBG and incubated with 1:250 Cy3-conjugated donkey anti-rabbit (Jackson ImmunoResearch, West Grove, PA) and 1:250 FITC-conjugated donkey anti-goat (Jackson ImmunoResearch) at 37°C for 1 h. Slides were washed in PBG and mounted as described above. Three-dimensional images were captured using a Leica DMI600B confocal laser scanning microscope (63×) equipped with a Hamamatsu EM-CCD camera. At least 50 cells per condition were captured using Volocity 5.4.1 (Improvision, Lexington, MA), and the experiment was repeated three times. Images were obtained at 0.2-μm vertical intervals for each cell, and colocalization was analyzed using Volocity. For TRF1 foci, a threshold of 15 SD above the mean of the total fluorescence intensity was used, and foci below 0.05 μm3 were excluded. For PML foci, a threshold of 9 SD was used, and foci below 0.05 μm3 were excluded. Erosion was applied to every PML foci. The protocol calculated the number of PML foci touching the TRF1 foci.
TIFs were visualized in interphase cells, fixed, and permeabilized as described above. Slides were incubated with 1:100 mouse anti-53BP1 (BP13; Upstate, Temecula, CA) or 1:500 goat anti–ATM Ser1981 (10H11.E12; Cell Signaling, Danvers, MA) and 1:4000 rabbit anti-TRF1 (kindly provided by Titia de Lange) in PBG (0.2% fish gelatin [Sigma], 0.2% BSA in PBS) at 37°C for 2 h. Slides were washed in PBG and incubated with 1:250 Cy3-conjugated donkey anti-rabbit (Jackson ImmunoResearch) and 1:250 FITC-conjugated donkey anti-goat (Jackson ImmunoResearch) at 37°C for 1 h. Slides were washed in PBG and mounted as described above. Images were captured using a Zeiss M1 fluorescence microscope (63× and 100×).
Q-FISH and pq ratios
Metaphase spreads from Colcemid-arrested cells were prepared according to standard cytogenetic methods. Cells were fixed in 4% formaldehyde in PBS for 2 min, then washed three times in 1× PBS for 5 min. Slides were treated in a preheated solution of 1 mg/ml pepsin in 10 mM glycine (pH 2.0) for 10 min at 37°C, washed twice in 1× PBS for 2 min, and then subjected to a second round of formaldehyde fixation and 1× PBS washes. After dehydration through an ethanol series, cells were hybridized with a Cy3-conjugated (C3TA2)3 PNA probe, washed, and mounted as described above for CO-FISH. Telomere lengths were measured with the TFL-Telo v2.0 software kindly provided by Peter Lansdorp (British Columbia Cancer Center, Vancouver, Canada), and values were used to measure telomere qp ratios. A total of 15–30 metaphases were examined for each clone. Chromosomes with single-free ends or end-to-end fusions were given a q/p score of 5 (Morrish and Greider, 2009). Twofold ratios values (q/p ≥ 2 or q/p ≤ 0.5) and fivefold ratio values (q/p ≥ 5 or q/p ≤ 0.2) were plotted and normalized according to the total number of chromosomes examined.
Telomeric repeat amplification protocol (TRAP) and TRF analysis
Protein extracts were prepared using 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate lysis buffer, and TRAP assays were performed as previously described (Marie-Egyptienne et al., 2009). To compare telomerase activity between extracts, serial dilutions of telomerase extension products were assayed in the PCR reaction. Genomic DNA was digested with RsaI and HinfI, and the fragments were separated by 2D-PFGE (Cerone et al., 2005). After denaturation and neutralization, agarose gels were dried for 2 h at 60°C. In-gel hybridization was performed using γ-32P-ATP–labeled (C3TA2)4 probe and exposed to x-ray film.
Acknowledgments
We thank J. Arturo Londoño-Vallejo for critical discussion and comments on the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research to C.A. (MOP-81215) and the Canadian Cancer Society (grant #700428). M.E.B. was a Cole Foundation Fellow and is supported by a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ). C.A. is a Chercheur National of the FRSQ.
Abbreviations used:
- 2D-PFGE
two-dimensional pulse-field gel electrophoresis
- ALT
alternative lengthening of telomeres
- APB
ALT-associated PML body
- ATM
ataxia telangiectasia mutated
- BSA
bovine serum albumin
- CO-FISH
chromosome-orientation fluorescence in situ hybridization
- DAPI
4′,6-diamino-2-phenylindole
- ECTR
extrachromosomal telomeric repeat
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- HR
homologous recombination
- hTERT
human telomerase reverse transcriptase
- hTR
human telomerase RNA
- MuA-hTR
mutant human telomerase RNA template
- PBG
phosphate-buffered gelatin
- PBS
phosphate-buffered saline
- PML
promyelocytic leukemia
- PNA
peptide nucleic acid
- Q-FISH
quantitative FISH
- SEM
standard error of the mean
- SFE
signal-free end
- TIF
telomere dysfunction-induced foci
- TRAP
telomeric repeat amplification protocol
- TRF
terminal restriction fragment
- T-SCE
telomeric sister-chromatid exchanges
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0173) on November 30, 2010.
REFERENCES
- Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Bechard LH, Butuner BD, Peterson GJ, McRae W, Topcu Z, McEachern MJ. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol Cell Biol. 2009;29:626–639. doi: 10.1128/MCB.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter OE, Zou Y, Walker W, Wright WE, Shay JW. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 2004;64:3444–3451. doi: 10.1158/0008-5472.CAN-04-0323. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene. 2005;24:7893–7901. doi: 10.1038/sj.onc.1208934. [DOI] [PubMed] [Google Scholar]
- Cerone MA, Londono-Vallejo JA, Autexier C. Mutated telomeres sensitize tumor cells to anticancer drugs independently of telomere shortening and mechanisms of telomere maintenance. Oncogene. 2006;25:7411–7420. doi: 10.1038/sj.onc.1209727. [DOI] [PubMed] [Google Scholar]
- Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, et al. Association of mutant TP53 with alternative lengthening of telomeres and favorable prognosis in glioma. Cancer Res. 2006;66:6473–6476. doi: 10.1158/0008-5472.CAN-06-0910. [DOI] [PubMed] [Google Scholar]
- Costa A, et al. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. 2006;66:8918–8924. doi: 10.1158/0008-5472.CAN-06-0273. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londono-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci USA. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Fasching CL, Bower K, Reddel RR. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 2005;65:2722–2729. doi: 10.1158/0008-5472.CAN-04-2881. [DOI] [PubMed] [Google Scholar]
- Goldkorn A, Blackburn EH. Assembly of mutant-template telomerase RNA into catalytically active telomerase ribonucleoprotein that can act on telomeres is required for apoptosis and cell cycle arrest in human cancer cells. Cancer Res. 2006;66:5763–5771. doi: 10.1158/0008-5472.CAN-05-3782. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 2001;20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobelny JV, Kulp-McEliece M, Broccoli D. Effects of reconstitution of telomerase activity on telomere maintenance by the alternative lengthening of telomeres (ALT) pathway. Hum Mol Genet. 2001;10:1953–1961. doi: 10.1093/hmg/10.18.1953. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Anglana M, Wang A, Bacchetti S. Transient expression of wild-type or biologically inactive telomerase allows the formation of artificial telomeres in mortal human cells. Exp Cell Res. 2001a;265:304–311. doi: 10.1006/excr.2001.5189. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001b;20:714–725. doi: 10.1038/sj.onc.1204145. [DOI] [PubMed] [Google Scholar]
- Hahn WC. Inhibition of telomerase limits the growth of human cancer cells. Nature Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, McKay MJ, Reddel RR, Royds JA. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- Hande MP, Samper E, Lansdorp P, Blasco MA. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Reddel RR. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Iyer S, Chadha AD, McEachern MJ. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol. 2005;25:8064–8073. doi: 10.1128/MCB.25.18.8064-8073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc Natl Acad Sci USA. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5‘-TAGGGTTAG-3‘ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Marciniak RA, Cavazos D, Montellano R, Chen Q, Guarente L, Johnson FB. A novel telomere structure in a human alternative lengthening of telomeres cell line. Cancer Res. 2005;65:2730–2737. doi: 10.1158/0008-5472.CAN-04-2888. [DOI] [PubMed] [Google Scholar]
- Marie-Egyptienne DT, Brault ME, Nimmo GA, Londono-Vallejo JA, Autexier C. Growth defects in mouse telomerase RNA-deficient cells expressing a template-mutated mouse telomerase RNA. Cancer Lett. 2009;275:266–276. doi: 10.1016/j.canlet.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish TA, Greider CW. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 2009;5:e1000357. doi: 10.1371/journal.pgen.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Shinkai Y, Hande MP, Matsumoto T, Takehara S, Tachibana M, Oshimura M, Lansdorp PM, Furuichi Y. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA. Mol Cell Biol. 2000;20:4115–4127. doi: 10.1128/mcb.20.11.4115-4127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Nakabayashi K, Suzuki M, Takahashi E, Fujii M, Suzuki T, Ayusawa D. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem Biophys Res Commun. 1998;248:223–227. doi: 10.1006/bbrc.1998.8875. [DOI] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Park KH, et al. Telomerase activity and telomere lengths in various cell lines: changes of telomerase activity can be another method for chemosensitivity evaluation. Int J Oncol. 1998;13:489–495. doi: 10.3892/ijo.13.3.489. [DOI] [PubMed] [Google Scholar]
- Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol. 2001;21:3862–3875. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr BA, Blackburn EH. ATM mediates cytotoxicity of a mutant telomerase RNA in human cancer cells. Cancer Res. 2008;68:5309–5317. doi: 10.1158/0008-5472.CAN-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutake Y, Matsumoto T, Watanabe T, Maeda S, Tahara H, Sakamoto S, Niida H, Sugimoto M, Ide T, Furuichi Y. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem Biophys Res Commun. 1998;247:765–772. doi: 10.1006/bbrc.1998.8876. [DOI] [PubMed] [Google Scholar]
- Underwood DH, Carroll C, McEachern MJ. Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot Cell. 2004;3:369–384. doi: 10.1128/EC.3.2.369-384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Watson JD. Origin of concatameric T4 DNA. Nature New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhang X, Mar V, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]