Abstract
In the C4 plant maize (Zea mays L.), two ferredoxin isoproteins, Fd I and Fd II, are expressed specifically in mesophyll and bundle-sheath cells, respectively. cDNAs for these ferredoxins were introduced separately into the cyanobacterium Plectonema boryanum with a disrupted endogenous ferredoxin gene, yielding TM202 and KM2-9 strains expressing Fd I and Fd II. The growth of TM202 was retarded under high light (130 µmol/m2/s), whereas KM2-9 grew at a normal rate but exhibited a nitrogen-deficient phenotype. Measurement of photosynthetic O2 evolution revealed that the reducing power was not efficiently partitioned into nitrogen assimilation in KM2-9. After starvation of the cells in darkness, the P700 oxidation level under far-red illumination increased significantly in TM202. However, it remained low in KM2-9, indicating an active cyclic electron flow. In accordance with this, the cellular ratio of ATP/ADP increased and that of NADPH/NADP+ decreased in KM2-9 as compared with TM202. These results demonstrated that the two cell type-specific ferredoxins differentially modulate electron flow around photosystem I.
Keywords: C4 plant/cyclic electron flow/ferredoxin isoproteins
Introduction
The energy requirement for photosynthesis differs between C3 and C4 plants. In the overall reaction of photosynthetic CO2 fixation, three ATP and two NADPH molecules are required for each molecule of CO2 fixed in C3 plants. C4 plants, on the other hand, require an additional two (or three) ATP molecules for CO2 fixation (Hatch, 1987, 1992). Therefore, C4 plants need a higher capacity for photo-phospholylation to meet the requirement for the cellular balance of ATP and NADPH. In C4 plants of the NADP-malic enzyme type, most of the NADPH is produced in mesophyll cell (MC) chloroplasts. Thus, the major function of bundle-sheath cell (BSC) chloroplasts is to produce ATP, required to drive the Calvin cycle through cyclic electron flow around photosystem I (PSI) (Edwards and Walker, 1983). In fact, photosystem II (PSII) activity is low or undetectable in BSC chloroplasts in this C4 subtype (Woo et al., 1970; Meierhoff and Westhoff, 1993).
In the course of our study of ferredoxin (Fd) in maize, we found two distinctive Fd isoproteins distributed differentially in these two types of photosynthetic cells: MC-specific Fd I and BSC-specific Fd II (Kimata and Hase, 1989). Although they share a high sequence identity (∼90%), a critical amino acid substitution was found at position 65 (Asp for Fd I and Asn for Fd II). The unique Asn65 of Fd II was responsible for its weaker ability to interact with Fd-NADP+ reductase (FNR) when compared with Fd I and other Fds from C3 plants (Matsumura et al., 1999). We presumed that the structural and functional differences of Fd I and Fd II could be related to MC- and BSC-specific metabolic processes conducted by several redox enzymes requiring Fd as an electron donor. The nitrate assimilatory pathway (Moore and Black, 1979) and the enzymes for antioxidant re-reduction (Doulis et al., 1997) are localized exclusively in MC, while BSC is highly active in the cyclic electron flow, as described above.
In studies of the pathway of the cyclic electron flow, the involvement of NAD(P)H dehydrogenase (Ndh) and Fd-quinone reductase (FQR) has been reported. The genes for subunits of the Ndh complex have been found on the genomes of higher plant chloroplasts (Ohyama et al., 1986) and cyanobacteria (Ellersiek and Steinmüller, 1992) as homologs of the mitochondrial NADH: ubiquinone oxidoreductase (complex I). Analyses of mutants deficient in one or more genes for the subunits of the Ndh complex in Synechocystis sp. PCC 6803 (Ogawa, 1991; Mi et al., 1992) and tobacco (Burrows et al., 1998; Kofer et al., 1998; Shikanai et al., 1998) suggested the participation of this enzyme in cyclic electron flow. Ndh was localized at the thylakoid membrane (Nixon et al., 1989; Berger et al., 1993), and its subunit complex was isolated from pea (Sazanov et al., 1998) and Synechocystis (Matsuo et al., 1998). Differential expression of plastid-encoded Ndh genes was reported between MC and BSC chloroplasts of a C4 plant, Sorghum bicolor, and the enzyme was proposed to be involved mainly in cyclic electron flow within BSC (Kubicki et al., 1996). Another pathway mediated by putative FQR is not yet well characterized. This enzyme was presented as the binding site of antimycin A, which specifically inhibits cyclic electron flow (Bendall and Manasse, 1995). The molecular identity of this enzyme remains to be elucidated, but involvement of a novel cytochrome b has been shown (Miyake et al., 1995).
In this study we examined whether the MC- and BSC-specific Fds from maize play distinct roles in the photosynthetic electron transport systems to meet the differential requirements of ATP and NADPH in the two types of photosynthetic cells of C4 plants. We have developed a procedure in which the essential Fd gene in the cyanobacterium Plectonema boryanum is disrupted after the cDNAs for the two maize Fd isoproteins have been introduced into the cyanobacterial cells. The resulting transformants expressing maize Fd I and Fd II in place of the endogenous cyanobacterial Fd showed significant differences in their ability to partition electrons from PSI into descending pathways, such as cyclic electron flow and nitrogen assimilation, thus causing marked changes in the balance of cellular levels of ATP and NADPH. This is the first experimental evidence, to our knowledge, that different Fd isoproteins can contribute differentially as a determinant for the electron flow around PSI.
Results
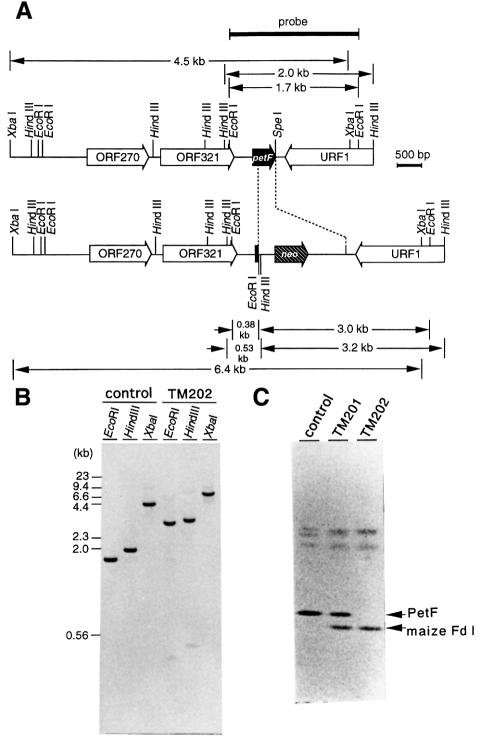
Cloning of a 4.8 kb genomic DNA fragment containing the petF gene
Using a pair of degenerated oligonucleotides corresponding to the highly conserved amino acid sequences among cyanobacterial Fds, a 176 bp DNA fragment was amplified from the genomic DNA of P.boryanum as described in Materials and methods. This fragment contained a reading frame corresponding to the expected amino acid stretch (from Cys at position 41 to the C-terminal Tyr) and was used as a probe for screening a genomic library of P.boryanum. One positive clone with a 11 kb insert was obtained. A 2 kb HindIII fragment and a 4.5 kb XbaI fragment of the insert, both of which hybridized with the probe, were subcloned and sequenced. The two fragments overlapped, and the complete nucleotide sequence of 4762 bp was elucidated. There was an open reading frame encoding a Fd polypeptide consisting of 99 amino acid residues. The N-terminal sequence of the purified Fd from the cyanobacterial cells, grown photomixotrophically, was determined to be Pro-Ser-Phe-Lys-Val-. This agreed with the sequence deduced from the nucleotide sequence except for the first Met. We confirmed that this gene was identical to the petF gene from P.boryanum PCC 73110 registered by Cassing et al. (1995).
Targeted mutagenesis of the petF gene in the P.boryanum cells
To obtain a petF-disrupted mutant of P.boryanum, a kanamycin-resistant cassette with the petF gene flanking regions was introduced into the original strain (dg5) as described in Materials and methods. Resultant kanamycin-resistant cells were screened under photoautotrophic or mixotrophic conditions in the light or under heterotrophic conditions in darkness. However, no stable transformant was obtained after several trials under any growth conditions, indicative of the essential nature of the petF gene.
Next, we used another host strain, TM201, which had been transformed with a maize Fd I expression plasmid, pSVMmFD1 (see Figure 1 for plasmid construction), for targeted mutagenesis. The expression of maize Fd I was assessed by non-denaturing PAGE, followed by western blot analysis using the rabbit antibodies against maize Fd I (Figure 2C). As shown in the TM201 lane in Figure 2C, maize Fd I accumulated in addition to the endogenous petF gene product. We confirmed that this Fd I accumulation was dependent on the addition of isopropyl-β-d (–)-thiogalactopyranoside (IPTG) to the medium (data not shown). By targeted mutagenesis of the petF gene in TM201, kanamycin-resistant transformants were obtained at a frequency of 10–7–10–8 under mixotrophic conditions in the presence of 1 mM IPTG. One stable transformant, TM202, obtained after several rounds of segregation, was examined by Southern and western blot analyses (Figure 2B and C). Restriction patterns of EcoRI, HindIII and XbaI digests of the genomic DNA of TM202 (Figure 2B) matched completely those expected as the result of the mutagenesis (see Figure 2A for the restriction map). In western blot analysis (Figure 2C), the signal for the petF gene product disappeared in TM202. These data confirmed the complete disruption of the petF gene in TM202.
Fig. 1. Construction of a plasmid for the expression of maize Fd I in P.boryanum. A shuttle vector, pSVM30, between P.boryanum and E.coli, a derivative of pPBH201 with the lacIq gene, was used for the construction. A segment containing the trp promoter and the Fd coding region of pTMmFD1, which expresses the mature part of maize Fd I in E.coli (Matsumura et al., 1999), was excised at SspI sites and inserted into an end-filled PstI site of pSVM30 under the control of the lacIqgene, yielding pSVMmFD1.
Fig. 2. (A) Physical map of the 4.8 kb genomic DNA fragment containing the petF gene. Two open reading frames, ORF270 and ORF321, and one partial reading frame, URF1, other than that of the petF gene are contained in a 4762 bp DNA fragment cloned from the P.boryanum genome. These sequence data have been submitted to the DDBJ/EMBL/GenBank database under Accession No. AB017194. The position and direction of the neo gene introduced for the targeted mutagenesis are shown in the lower map. The position of a probe used for Southern analysis is indicated by a heavy bar. (B) Southern analysis of the genome of the petF-disrupted mutant expressing maize Fd I. Genomic DNAs (0.4 µg of each) from the original strain (control) and petF-disrupted mutant expressing maize Fd I (TM202) were digested with the restriction enzymes indicated, size fractionated and probed with a 1.7 kb EcoRI fragment as indicated in (A). Sizes of marker DNA fragments are shown on the left. (C) Western analysis of maize Fd I and the petF gene product in P.boryanum cells. Cells of the original strain (control), the transformant expressing maize Fd I (TM201) and the petF-disrupted mutant expressing maize Fd I (TM202) were grown photomixotrophically in the presence of 30 mM glucose and 200 µM IPTG. Total crude extracts of the cells (corresponding to 30 µg of protein) were loaded on the gel and immunolabeled with anti-maize Fd I antibodies that also reacted with the petF gene product.
Phenotypes of petF gene disruptants expressing maize Fd I and Fd II
Another petF gene disruptant, KM2-9, which expresses maize Fd II, was also obtained by the same procedure. Both TM202 and KM2-9 were dependent on the expression of maize Fd for growth. As shown in Figure 3, both strains hardly grew in the absence of IPTG, but significant growth was observed in the presence of IPTG at >10 µM. Thus, there seemed to be a critical cellular concentration of Fd for growth. The content of maize Fd in TM202 and KM2-9 grown in the presence of 200 µM IPTG was found to be comparable to that of the authentic Fd in the original strain (Figure 4).
Fig. 3. Effects of IPTG concentration on the growth of petF-disrupted P.boryanum cells expressing maize Fd I (TM202) and Fd II (KM2-9). Cells of TM202 and KM2-9 strains were grown photoautotrophically with different concentrations of IPTG under the light intensities and growth periods indicated.
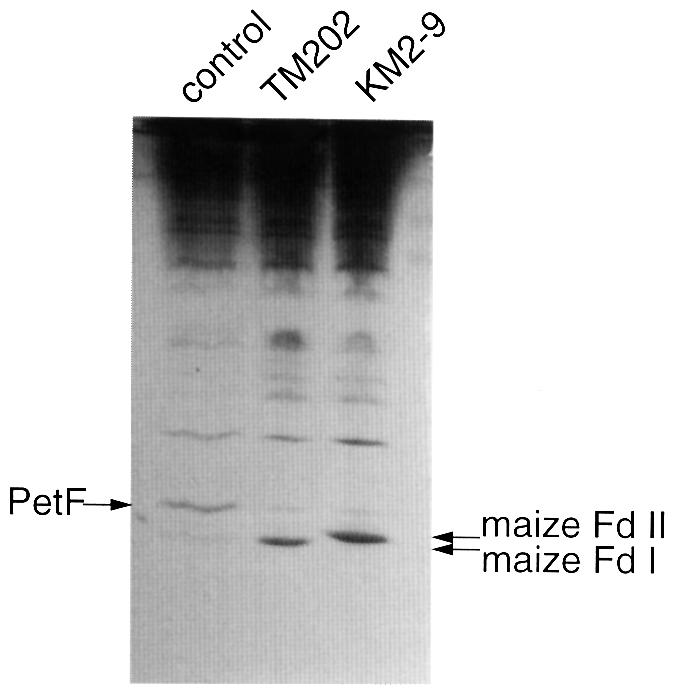
Fig. 4. Expression of maize Fd I and Fd II in petF-disrupted P.boryanum cells. Cells of the original strain (control) and the petF-disrupted mutants expressing maize Fd I (TM202) and Fd II (KM2-9) were grown photoautotrophically in the presence of 200 µM IPTG under a light intensity of 50 µmol/m2/s. Total crude extracts of the cells (corresponding to 10 µg of chlorophyll) were subjected to non-denaturing PAGE (Kimata and Hase, 1989) and stained with Coomassie Blue. The amount of protein loaded on the gel was 23, 23 and 35 µg for the control, TM202 and KM2-9, respectively. The faint bands detected at the same migration as the petF gene product in the TM202 and KM2-9 lanes are not the remaining petF gene product, but other proteins, as confirmed by immunodetection with anti-Fd antibodies (data not shown).
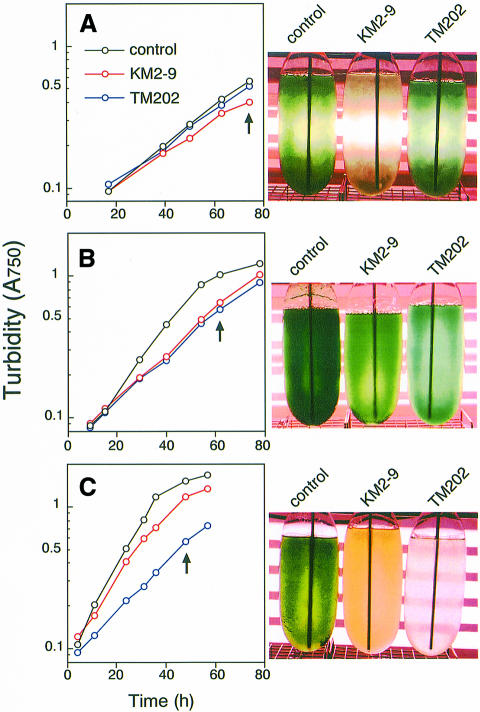
Growth of TM202 and KM2-9 was monitored under different light intensities (Figure 5). Both mutant strains showed significant growth under a wide range of light intensities, although their growth rates and tonalities varied depending on the light intensity. Under medium light (50 µmol/m2/s, Figure 5B), the two mutant strains grew at similar rates (doubling times of 11, 20 and 19 h for the control, TM202 and KM2-9, respectively). The color of the cells, which was considered to reflect the content of antenna pigments such as chlorophyll and phycocyanin, showed slight variation: the difference in the chlorophyll content was at most 2-fold. However, under high light (130 µmol/m2/s, Figure 5C), both mutant strains showed distinct phenotypes; TM202 grew more slowly (doubling time 16 h) than the control or KM2-9 (doubling times 9 and 12 h, respectively). The color of the TM202 cells was lighter than other strains, which probably reflected more extensive photobleaching. KM2-9 exhibited a rather yellowish color, which may reflect the lower content of certain photosynthetic pigments due to nitrogen deficiency, as described later. Under low light (25 µmol/m2/s, Figure 5A), the mutant strains also showed different growth rates, but with reverse order to those observed under high light; KM2-9 (doubling time 30 h) grew more slowly than the control (original strain) or TM202 (doubling times 17 and 20 h, respectively). We utilized the cells grown under medium light for the following experiments and transiently placed them under each experimental light condition, in order to minimize the changes in physiological status between the strains.
Fig. 5. Growth curves and tonalities of petF-disrupted P.boryanum cells expressing maize Fd I (TM202) and Fd II (KM2-9) and the original strain (control). (A) Cells were grown photoautotrophically in the presence of 200 µM IPTG under low light (25 µmol/m2/s), (B) under medium light (50 µmol/m2/s) and (C) under high light (130 µmol/m2/s). Growth was monitored by absorption at 750 nm. Culture bottles of the three strains were photographed at the growth stages indicated by an arrow in each panel.
Photosynthetic electron transfer activities of TM202 and KM2-9 in response to inorganic nitrogen
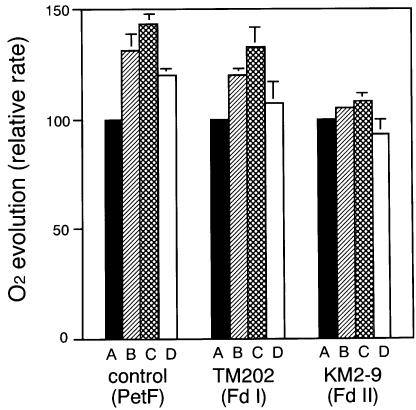
Nitrogen assimilation, as well as carbon assimilation, requires photosynthetic reducing power. Three Fd- dependent enzymes, nitrate reductase, nitrite reductase and Fd-dependent glutamate synthase, are involved in the reductive assimilation of nitrate into the amino group of glutamic acid in cyanobacterial cells. To evaluate the utilization of the reducing power for nitrogen and carbon assimilations, in vivo O2 evolution of the cyanobacterial cells was measured in a medium containing bicarbonate only or both bicarbonate and nitrate (Figure 6). Light-dependent O2 evolution on the basis of chlorophyll was similar in all three strains under all conditions, except TM202 under high light (>80 µmol/m2/s) as described below. In response to the addition of nitrate, O2 evolution increased significantly in the control and TM202 over the wide range of light intensities examined. In contrast, no such significant increase was observed in KM2-9, suggesting that the reducing power was not efficiently partitioned for nitrogen assimilation. This phenomenon, in combination with the observations of the yellowish cell color (Figure 5), suggests that KM2-9 was subject to nitrogen deficiency. We further examined whether O2 evolution was enhanced by two other inorganic nitrogen compounds, nitrite and ammonium, which are metabolic intermediates of nitrate assimilation and are themselves good nitrogen sources for cultures of this cyanobacterium (Figure 7). A significant increase in O2 evolution was observed with both compounds in the control and in TM202. The differences in the extent of the increase between the compounds were partly explicable based on the reductant stoichiometries in their assimilation into glutamate. The larger increase in O2 evolution after the addition of nitrite compared with that of nitrate suggests that reduction of nitrate by nitrate reductase may be a rate-limiting step in the nitrogen assimilation pathway of this cyanobacterium. In KM2-9, any increase in O2 evolution due to the addition of nitrogen compounds was negligible or of a lesser order than in the other two strains. These data suggested that the capacity for nitrogen assimilation in KM2-9 was lowered as a whole, rather than arrested at a specific step catalyzed by a certain Fd-dependent enzyme.
Fig. 6. The rate of O2 evolution dependent on CO2 only, and on both CO2 and NO3– as a function of light intensity in the original strain (control) and petF-disrupted P.boryanum cells expressing maize Fd I (TM202) and Fd II (KM2-9). Cells were grown photoautotrophically in the presence of 200 µM IPTG under medium light (50 µmol/m2/s), suspended in nitrate-free BG11 medium, and after addition of 10 mM NaHCO3 only (closed symbols) or both 10 mM NaHCO3 and 1 mM NaNO3 (open symbols) to the medium, O2 evolution was measured under increasing light intensities from 20 to 120 µmol/m2/s.
Fig. 7. Relative rate of O2 evolution dependent on CO2 only, and on both CO2 and an inorganic nitrogen compound (NO3–, NO2– or NH4+) in the original strain (control) and petF-disrupted P.boryanum cells expressing maize Fd I (TM202) and Fd II (KM2-9). Cells were grown photoautotrophically in the presence of 200 µM IPTG under medium light (50 µmol/m2/s), suspended in nitrate-free BG11 medium and, after addition of 10 mM NaHCO3, O2 evolution at 120 µmol/m2/s light intensity was measured in the presence of no additional reagents (A), 1 mM NaNO3 (B), 1 mM NaNO2 (C) or 1 mM NH4Cl (D). Relative rates, compared with that of CO2-dependent O2 evolution in each strain (normalized as 100), are expressed as the mean ± SD of three independent determinations.
TM202 showed a unique feature in the light dependency of its photosynthetic rate (Figure 6): O2 evolution reached saturation level under a light intensity lower than that of the control or KM2-9, indicating that photoinhibition may have occurred. Such a light response seemed to be consistent with its slower growth rate and lighter color under higher illumination (Figure 5).
The oxidation level of P700 under far-red light in TM202 and KM2-9
Because cyclic electron flow is known to be enhanced in BSC of C4 plants, we examined whether the cell type-specific Fds show differential activity in electron flow around PSI in the cyanobacterial cells. To assess this possibility, the oxidation level of P700 under far-red light was measured in the control, TM202 and KM2-9 (Figure 8). In cyanobacteria, respiratory electron flow shares electron carriers with the intersystem chain of photosynthetic electron flow (Scherer, 1990). Thus, P700 can be reduced by donation of electrons from both the cytosolic respiratory donors and photoreductant generated by PSI under far-red light (>710 nm). As shown in Figure 8A, the cells of all three strains grown photoautotrophically showed marginal oxidation of P700 by far-red light at an intensity of 9 W/m2. When these cells were transferred to the dark for >24 h, the P700 oxidation level, observed by the same far-red light illumination, increased gradually in the control and TM202 cells and reached close to the maximum after 72 h (Figure 8B, left). The increased signal was diminished upon the addition of glucose at 30 mM (Figure 8B, right). Also, when these strains were grown heterotrophically with 30 mM glucose in the dark, they did not show such a high oxidation level of P700 (data not shown). Therefore, the increased oxidation of P700 in the control and TM202 cells after the dark treatment was shown to be caused by a shortage of respiratory substrate. On the other hand, in KM2-9, P700 was kept at a lower oxidation level even after the same dark treatment (Figure 8B, left). This was not due to a lower population of P700 in KM2-9, because the cells showed a higher oxidation level when illuminated with a stronger far-red light (at 11 W/m2, Figure 8B, bottom). Respiratory activity (denoted in parentheses under each measurement in Figure 8) of all three strains was decreased to similar levels by the dark treatment and increased by the addition of glucose; the observed higher respiratory activity of KM2-9 cells is probably due to its lower chlorophyll content, which is used as a basis for measurement. This indicates that, after the dark treatment, electron donation from respiratory substrate decreased in KM2-9 cells as well as in the control and TM202 cells. Therefore, these results lead us to suggest that cyclic electron flow from the photoreductant generated by PSI to an electron donor site of P700 was considerably enhanced in KM2-9, compared with the other two strains, as the most probable explanation. Another possible explanation is described in the Discussion.
Fig. 8. Oxidation and reduction kinetics of P700 induced by FR light (>710 nm) and 50 ms saturating MT light in the original strain (control) and petF-disrupted P.boryanum cells expressing maize Fd I (TM202) and Fd II (KM2-9). (A) Cells were grown photo autotrophically in the presence of 200 µM IPTG under medium light (50 µmol/m2/s). (B) Cells were grown photoautotrophically in the presence of 200 µM IPTG under medium light and then transferred to the dark for 72 h. All measurements were performed using the cells that were suspended in BG11 at 100 µg Chl/ml. The intensity of the FR light irradiated was 9 W/m2 except for the measurement at the bottom of (B), which was performed at 11 W/m2. Where indicated, glucose at 30 mM was added to the suspensions of cells in the dark, 2 min before the measurements. The respiratory activity of the cells is denoted in parentheses under each measurement, which was expressed as pmoles of O2 consumed per milligram of chlorophyll per minute.
Adenine and pyridine nucleotide pools in TM202 and KM2-9
Higher activity of cyclic electron flow was expected to cause an enhanced level of ATP and a lower level of NADPH. Therefore, the cellular balance of ATP to NADPH would differ between the control, TM202 and KM2-9. To examine this possibility, steady-state levels of ATP, ADP, NADP+ and NADPH were determined on the basis of chlorophyll in all the strains grown under two different light intensities (Table I). Although the cellular content of chlorophyll may vary depending on physiological conditions, the ratios of ATP to ADP and of NADPH to NADP+ were applicable as a parameter to evaluate their cellular status. The proportion of ATP in KM2-9 cells increased to about twice the levels in the other two strains under both light intensities. On the other hand, the proportion of NADPH in KM2-9 cells decreased, especially under high light. This result was consistent with the dominant cyclic electron flow observed in KM2-9.
Table I. Steady-state levels of ATP, ADP, NADPH and NADP+ (all in nmol/mg Chl) in petF-disrupted P.boryanum cells expressing maize Fd I (TM202) and Fd II (KM2-9) and the original strain (control) grown under different light intensities.
| Strain | ATP | ADP | ATP/ADP | NADPH | NADP+ | NADPH/NADP+ |
|---|---|---|---|---|---|---|
| Low light (25 µmol/m2/s) | ||||||
| TM202 (maize Fd I) | 130 ± 14 | 68 ± 7 | 1.9 ± 0.3 | 16 ± 1 | 35 ± 9 | 0.50 ± 0.13 |
| KM2-9 (maize Fd II) | 392 ± 36 | 81 ± 2 | 4.8 ± 0.4 | 24 ± 6 | 61 ± 17 | 0.39 ± 0.01 |
| control | 123 ± 5 | 62 ± 4 | 2.0 ± 0.2 | 10 ± 4 | 25 ± 1 | 0.40 ± 0.17 |
| High light (130 µmol/m2/s) | ||||||
| TM202 (maize Fd I) | 75 ± 9 | 66 ± 9 | 1.2 ± 0.3 | 12 ± 2 | 11 ± 4 | 1.2 ± 0.3 |
| KM2-9 (maize Fd II) | 467 ± 18 | 187 ± 6 | 2.4 ± 0.1 | 12 ± 1 | 64 ± 19 | 0.18 ± 0.06 |
| control | 158 ± 12 | 123 ± 13 | 1.3 ± 0.1 | 18 ± 8 | 18 ± 14 | 1.3 ± 0.8 |
Extraction and measurement of the nucleotides from the cells were performed as described in Materials and methods. The ratios of ATP/ADP and NADPH/NADP+ are also presented for comparisons between different strains. The values are means of three independent determinations, with the error shown as the SD.
Discussion
Maize has two Fd isoproteins, Fd I and Fd II, localized specifically in MC and BSC, respectively. In overall structure, the two Fds share high homology, but they have a critical amino acid substitution, which confers different ability to interact with FNR (Matsumura et al., 1999). In this study, their functions were examined in vivo using the cyanobacterium P.boryanum, whose endogenous Fd gene (petF) was inactivated. Disruption of petF with a kanamycin-resistant gene cassette was not successful with the original strain. On the next trial, the complete segregation of petF-disrupted cells was achieved when the mutagenesis was conducted using cells already carrying plasmid-borne Fd I and Fd II cDNAs. Two resulting petF-deficient strains, TM202 and KM2-9, were found to express Fd I and Fd II, respectively, as the sole Fd molecule with a cellular concentration comparable to the authentic Fd (PetF) in the original strain (Figure 4). It was demonstrated that the expression of these maize Fds over a certain, minimum level was essential to render the host cells viable (Figure 3). Interestingly, growth profiles of TM202 and KM2-9 in response to different light intensities varied significantly (Figure 5). The slower growth and the apparent photobleaching of TM202 under high light could be due to photoinhibition caused by the lower activity of cyclic electron flow, as discussed below. The reason for the slower growth of KM2-9 under low light is not clear at present.
By further analyses of the photosynthetic capacities of the two mutant strains, we found that they have distinct characteristics in their electron donation towards nitrogen assimilation and the electron flow around PSI.
In KM2-9, the partitioning of photosynthetic energy to the nitrogen assimilation pathway seemed to be suppressed as a whole, because no significant increase in photosynthetic rate was observed upon the addition of the assimilable inorganic nitrogen compounds, nitrate, nitrite and ammonia (Figures 6 and 7). Nitrate reductase, nitrite reductase and glutamate synthase are Fd-dependent enzymes involved in this pathway, and electron transfer from photoreduced Fd to these enzymes is ongoing simultaneously with other Fd-dependent pathways. Therefore, the capacity for electron donation from Fd to these nitrogen assimilatory enzymes is considered to be lowered when PetF is substituted for maize Fd II.
An enhanced capacity for cyclic electron flow around PSI in KM2-9 was indicated by two different experiments. Measurement of the P700 oxidation level indicated that electron flow from photo-excited PSI back to P700 occurred more actively in KM2-9 than in the control or in TM202 (Figure 8). Cellular ratios of ATP/ADP and NADPH/NADP+ suggested that utilization of photoreducing power for ATP synthesis was dominant over that for the generation of NADPH in KM2-9, as compared with the control or with TM202 (Table I). Because the redox level of P700 is determined by the balance of the rate of electron flow into and out of PSI, it could be argued that the lower P700 oxidation level of KM2-9 (Figure 8B) is due to a lower rate of oxidation of PSI by maize Fd II. However, we presume that this possibility is unlikely because maize Fd II is able to replace the function of PetF over a wide range of light intensity, and because Figure 6 shows that the rate of photosynthetic electron transfer of KM2-9 is similar to that of the control, which indicates that electron flow from PSI to Fd II is normal in KM2-9.
The levels of ATP and NADPH need to be coordinately regulated to meet their requirements for assimilation of carbon and nitrogen, two major metabolic pathways consuming photosynthetic energy (Noctor and Foyer, 1998). Nitrate assimilation into glutamic acid requires 2.5 times as many electrons as the reduction of CO2 to triose, but only one-third as much ATP (Noctor and Foyer, 1998). In KM2-9 cells, electron donation from the photosynthetic electron transport system to the reduction of NADP+ appeared to be suppressed at the expense of enhanced formation of ATP by cyclic electron flow. Thus we suppose that the resulting lower NADPH/NADP+ ratio and/or higher ATP/NADPH ratio in KM2-9 are/is the main reason(s) for the decrease in electron donation toward the nitrogen assimilation pathway. Under higher light conditions, the ATP/NADPH ratio of KM2-9 (∼39, calculated from Table I) deviated further from that of the control strain (∼9). This is consistent with the fact that KM2-9 shows a more prominent phenotype of nitrogen deficiency under such light conditions (Figure 5).
In contrast to KM2-9, TM202 possessed an ATP/NADPH ratio (∼8 and 6 under low and high light, respectively) lower than that of the control strain (∼12 and 9), which may be due to its lower capacity for cyclic electron flow. Cyclic electron flow has been proposed to contribute to protection from photoinhibition (Herber and Walker, 1992) and other stresses, such as heat stress and salt stress (Canaani, 1990). The slow growth rate of TM202 under high light (Figure 5) and the early saturation of photosynthetic rate with increasing light intensity (Figure 6) suggested that this strain may suffer photoinhibition even at light intensities that are suitable for rapid growth of the control and KM2-9 strains.
These combined results strongly suggest that the MC-specific Fd I and BSC-specific Fd II are also functionally distinct in maize. Therefore, the function of these Fds appears to be differentiated to meet the demands for NADPH and ATP supply in the cells in which they reside: both the generation and consumption of NADPH are mostly restricted to MC, and the main function of BSC chloroplasts is to produce ATP, required to drive the Calvin cycle through cyclic electron flow.
Two major pathways involving the Ndh complex and FQR have been presented to mediate the cyclic electron flow. Ndh catalyzes the reduction of plastoquinone using a stromal reductant such as NADH or NADPH. Recent analysis of the isolated Ndh complex from pea indicates that the immediate reductant is NADH (Sazanov et al., 1998). It is also proposed that the Ndh could have a dual function in cyclic electron flow and a putative chlororespiratory pathway. In cyanobacteria, the purified Ndh complex from Synechocyctis PCC 6803 was shown to use NADPH as its preferred electron donor rather than NADH (Matsuo et al., 1998). The possibility of Fd as an electron donor for Ndh has also been presented for cyanobacteria (Mi et al., 1995) and higher plants (Endo et al., 1998). FQR is a putative enzyme that reduces plastoquinone using Fd as a direct electron donor and has been characterized as a binding site for antimycin A, a specific inhibitor of cyclic electron flow. Miyake et al. (1995) showed a Fd-dependent, antimycin A-sensitive cyclic electron flow in maize thylakoid, which was mediated by a novel cytochrome b. The localization of the antimycin A binding site indicates its association with PSI, and involvement of the PsaE subunit of PSI in the cyclic electron flow activity of FQR is suggested (Bendall and Manasse, 1995). An antimycin-sensitive FQR-mediated pathway has not been found in cyanobacteria.
Because maize Fd II conferred the greater activity of cyclic electron flow, it is assumed that Fd II has the greater electron transfer ability towards a certain component of the pathway in the cyanobacteria. This assumption is expected to be true also in the case of higher plants. Because cyclic electron flow is known to be dependent on Fd in maize (Miyake et al., 1995), maize Fd II may have greater electron transfer ability towards FQR and/or Ndh compared with Fd I. Moreover, because the expression of Ndh was shown to be enhanced in BSC of a C4 plant S.bicolor (Kubicki et al., 1996), it is worthwhile to examine whether Fd II is a preferred, physiological electron donor for Ndh. We found that the affinity of maize leaf FNR for Fd I is higher than that for Fd II (Matsumura et al., 1999). Therefore, FQR and/or Ndh in maize may also discriminate between Fd I and Fd II, but in an opposite fashion to that of maize FNR. If that is the case, it would also be interesting to investigate the structure enabling these enzymes to distinguish between Fd I and Fd II.
Finally, in addition to the in vivo experiments performed in this study, an in vitro experiment using isolated thylakoid membranes would be desirable in order to clarify the direct effect of the two Fds on the photosynthetic electron transfer pathways. We are currently constructing such an in vitro system.
Materials and methods
Cultivation of the cyanobacteria
Plectonema boryanum IAM-M101 strain dg5 (Fujita et al., 1996) and its derivatives TM201, TM202 and KM2-9 obtained in this study were cultured at 30°C in BG11 (Ripkka et al., 1979) supplemented with 20 mM HEPES–NaOH pH 7.5 and bubbled with 2% (v/v) CO2 in air under continuous illumination provided by fluorescent lamps (4000 lux). For photomixotrophic growth in the light and heterotrophic growth in the dark, 30 mM glucose was added to the medium. For TM201, the medium was supplemented with chloramphenicol (25 µg/ml), and for TM202 and KM2-9, chloramphenicol, kanamycin (15 µg/ml) and IPTG (200 µM) were added. When necessary, P.boryanum cells were harvested by centrifugation (3000 g for 10 min) and stored at –80°C. For plate culture, 1% (w/v) agar was added to the medium.
PCR and cloning of the 4.8 kb region of genomic DNA containing the petF gene
A pair of degenerated primers, 5′-TG(TC)CG(AGTC)GC(AGTC)GGT GCTTGCT-3′ and 5′-TAGTAGAG(GT)TCTTC(TC)TCTTTGT-3′, were used for the amplification of a part of the Fd gene in P.boryanum. These sequences were derived from two conserved regions of known cyanobacterial Fds. PCR was carried out under the following conditions of thermal cycling using total genomic DNA from P.boryanum as a template: first one cycle (95°C, 2 min; 55°C, 1 min; 72°C, 2 min) and 24 successive cycles (95°C, 1 min; 55°C, 1 min; 72°C, 2 min). The amplified DNA fragment (176 bp) was subcloned into a T-vector (Novagen) and used as a probe to screen a genomic library of P.boryanum constructed with λDASH II (Stratagene) as described previously (Fujita et al., 1998). The probe was labeled with a random-prime kit (Pharmacia) in the presence of [α-32P]dCTP. A 2 kb HindIII fragment and a 4.5 kb XbaI fragment from a positive clone were subcloned into pUC19, and a set of nested deletions was generated by exonuclease III treatment. DNA sequences were determined by a dye-terminator method with a DNA sequencer model 370A (Perkin Elmer Applied Biosystems).
Purification of Fd from P.boryanum and analysis of the N-terminal sequence
Plectonema boryanum cells (∼20 g) grown mixotrophically were suspended in 50 ml of 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and 10 mM β-mercapto ethanol and disrupted by extensive sonication. The crude extract was centrifuged at 10 000 g for 10 min at 4°C, and the supernatant was fractionated by the addition of ammonium sulfate to 70% saturation. The resulting precipitated materials were removed by centrifugation as above. The supernatant was applied directly to a column of butyl-Toyopearl (20 ml; Tosoh, Japan) and eluted with a linear gradient of ammonium sulfate from 70 to 0% in 50 mM Tris–HCl pH 7.5. Fd fractions with reddish color were combined, dialyzed against 50 mM Tris–HCl pH 7.5 and 200 mM NaCl, and further purified on a MonoQ HR5/5 column (Pharmacia) developed with a linear gradient of NaCl from 200 to 500 mM in Tris–HCl pH 7.5. The purified Fd was denatured by treatment with 5% (w/v) trichloroacetic acid and sequenced by manual Edman degradation, essentially according to the method described previously (Hase et al., 1976).
Expression of maize Fd cDNAs in P.boryanum
To express maize Fd I cDNA (Hase et al., 1991) in P.boryanum, an expression plasmid was constructed from a shuttle vector pPBH201 between P.boryanum and Escherichia coli (Walton et al., 1993). Details of the gene manipulation are shown in Figure 1. The construction of pTMmFD1, which expresses the mature part of maize Fd I under the trc promoter in E.coli, was described previously (Matsumura et al., 1999). A segment containing the promoter and the Fd coding region of pTMmFD1 was excised and introduced into shuttle vector pSVM30, a derivative of pPBH201 with a lacIq gene. The resulting pSVMmFD1 consisted of the maize Fd I cDNA under the trc promoter, the lacIq gene for controlling the trc promoter, a cyanobacterial cryptic plasmid pGL3 carrying a replication origin in P.boryanum and the cat (chloramphenicol acetyltransferase) gene. For the expression of maize Fd II cDNA, the expression plasmid pSVMmFD2 was constructed from pTMmFD2 (Matsumura et al., 1999) and pSVM30 by the procedure described above. These plasmid DNAs were each introduced into the cyanobacterial cells by electroporation (Fujita et al., 1992), and transformed cells were selected on a BG11 agar plate containing glucose (30 mM) and chloramphenicol (25 µg/ml).
Isolation of a petF-disrupted mutant
To disrupt the petF gene, a plasmid harboring a kanamycin-resistant (neo) gene, whose ends were connected with the 5′- and 3′-flanking regions of the petF gene, was constructed as follows. One of the nested deletion clones (3.1 kb fragment) used for DNA sequencing of the 4.5 kb XbaI fragment, in which the total 3′-region and coding region from the C-terminus up to Glu25 of the petF gene had been deleted, was recloned into EcoRI (end-filled) and SacI sites of pUC19 to obtain pUC802. The 1 kb SpeI–XbaI fragment, which contained the entire 3′-non-coding region of the petF gene, was ligated into the XbaI site of pUC802 to give pUC802/1. A SacI fragment (1.9 kb) containing the neo gene from pMC19 (Fujita et al., 1992) was inserted into a unique SacI site of pUC802/1, locating the junction region of the 3.1 and 1 kb fragments in the same orientation with the petF gene. The resulting plasmid, pUC802/2, was linearized by digestion with XbaI and introduced into P.boryanum cells by electroporation. Transformants were selected on kanamycin-containing plates and further tested for sensitivity to ampicillin to distinguish double recombinants from single ones, as described previously (Fujita et al., 1996).
Southern blot analysis
Total genomic DNAs were prepared from the original strain (dg5) and a mutant strain (TM202) according to the method of Wilson (1988) and digested with appropriate restriction endonucleases. The digests (0.4 µg each) were electrophoresed on 0.8% agarose gel, transferred to nylon membrane (Hybond-N+, Amersham), and proved with a 1.7 kb EcoRI fragment containing the petF gene (see Figure 2A) labeled with [α-32P]dCTP as described above.
Western blot analysis
Total crude extracts of cyanobacterial cells were prepared by sonication as described above and analyzed by PAGE on a non-denaturing gel with a linear gradient of 15–25% acrylamide (Kimata and Hase, 1989). Proteins on the gels were electrotransferred to a PVDF membrane (Immobilon, Millipore) for immunolabeling with anti-maize Fd I antibodies (Hase et al., 1991).
Measurement of evolution and consumption of oxygen
Evolution and consumption of dioxygen were followed with a Clark-type oxygen electrode equipped with an oxygen monitoring system (Hansatech, Norfolk, UK). For the analysis of photosynthetic activity, cells growing in the log phase were harvested and suspended in BG11 without NaNO3. After NaHCO3 was added to the cell suspension to a final concentration of 10 mM, the evolution of O2 by cells (10–20 µg Chl/ml) was monitored at 30°C under 660 nm light at intensities from 20 to 120 µmol/m2/s from the light emitting diodes; NaNO3, NaNO2 or NH4Cl at 1 mM was added when necessary. For the analysis of respiratory activity, the consumption of O2 by cells (100 µg Chl/ml in BG11) was monitored in darkness at 30°C.
Measurement of the redox state of P700
The redox state of P700 was monitored in terms of the change in the absorbance difference between 810 and 860 nm in a pulse-amplitude-modulation chlorophyll fluorometer with the dual-wavelength emitter–detector unit ED-P700 DW (Walz, Germany). The cell suspension (100 µg Chl/ml in BG11) was pipetted into a cuvette (KS101, Walz), and multi-branched fiberoptics connecting to the emitter–detector unit and to far-red light (FR; >710 nm) and saturating multiple-turnover light (MT; 50 ms length, 1500 W/m2) were attached to the cuvette. MT was applied with a xenon discharge lamp (XMT103; Walz).
Determination of adenine nucleotides
The cells were extracted using either cold perchloric acid (Schmid et al., 1989) or hot ethanol and KOH (Chapman et al., 1971), in both cases from 4 ml of cell culture. The two methods gave essentially identical results with respect to the adenine nucleotide levels. The resulting extracts were stored at –20°C. For ATP determinations, 200 µl of the cell extract were added to 50 µl of 75 mM potassium phosphate buffer pH 7.3, 15 mM MgCl2. For ATP plus ADP determinations, 200 µl of the cell extract were added to 50 µl of the same solution as above, except that it contained 0.5 mM phosphoenolpyruvate and 20 µg of pyruvate kinase. These mixtures were incubated at 30°C for 15 min and then held at 0°C until assayed. The resulting ATP was determined by the luciferase reaction using ATP bioluminescence assay kit CLSII (Boehringer Mannheim, Germany) according to the manufacturer’s instructions with a fluorescence spectrometer F-2500 (Hitachi, Tokyo, Japan). Inhibition of the luciferase reaction by perchloric acid and ethanol was corrected for the calculation.
Determination of pyridine nucleotides
The NADP+ and NADPH were extracted using cold perchloric acid and hot ethanol plus KOH as described above, respectively. For storage of the extract for NADPH, NADPH was converted enzymically to NADP+ by the addition of 5 µl of 1 M α-oxoglutarate in 1 M NH4Cl pH 7.0 and 1 µl of glutamate dehydrogenase (2400 U/ml) to 2 ml of neutralized extract. After a 15 min incubation at room temperature, the reaction was stopped by the addition of 10 µl of 5 N HCl, and the enzyme was inactivated by heating for 10 min at 50°C. The extract was then cooled and neutralized by the addition of 9 µl of 5 N NaOH. NADP+ samples at neutral pH (not above pH 7.0) could be stored frozen for several weeks. Resulting NADP+ was determined by an enzymic cycling method using 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide as a terminal electron acceptor (Matsumura and Miyachi, 1980). No significant inhibition of the reaction by either perchloric acid or ethanol was observed.
Acknowledgments
Acknowledgements
We thank Dr Douglas K.Walton (College of St Scholastica, Minnesota, USA) for providing the shuttle vector pPBH201. This work was supported in part by Grants-in-Aid for Research on Priority Areas (Nos 09274101 and 09274103 to F.S. and T.H.) from the Ministry of Education, Science and Culture of Japan.
References
- Bendall D.S. and Manasse,R.S. (1995) Cyclic photophosphorylation and electron transport. Biochim. Biophys. Acta, 1229, 23–38. [Google Scholar]
- Berger S., Ellersiek,U., Westhoff,P. and Steinmüller,K. (1993) Studies on the expression of NDH-H, a subunit of the HAD(P)H-plastoquinone-oxidoreductase of higher-plant chloroplasts. Planta, 190, 25–31. [Google Scholar]
- Burrows P.A., Sazanov,L.A., Svab,Z., Maliga,P. and Nixon,P.J. (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J., 17, 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani O. (1990) The role of cyclic electron flow around photosystem I and excitation energy distribution between the photosystems upon acclimation to high ionic strength in Dunaliella.Photochem. Photobiol., 52, 591–599. [Google Scholar]
- Cassing A., Boehme,H. and Schrautemeier,B. (1995) Nucleotide sequence, promoter structure, and expression of the petF1 gene (accession No. U33848) encoding the [2Fe–2S] Fd I from the nitrogen-fixing nonheterocystous cyanobacterium Plectonema boryanum PCC 73110. Plant Physiol., 109, 1499. [Google Scholar]
- Chapman A.G., Fall,L. and Atkinson,D.E. (1971) Adenylate energy charge in Escherichia coli during growth and starvation. J. Bacteriol., 103, 1072–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulis A.G., Debian,N., Kingston-Smith,A.H. and Foyer,C.H. (1997) Differential localization of antioxidants in maize leaves. Plant Physiol., 114, 1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G. and Walker,D. (1983) C3, C4: Mechanisms, and Cellular and Environmental Regulation, of Photosynthesis. Blackwell Scientific, Oxford, UK.
- Ellersiek U. and Steinmüller,K. (1992) Cloning and transcription analysis of the ndh(A-I-G-E) gene cluster and the ndhD gene in the cyanobacterium Synechocystis sp. PCC6803. Plant Mol. Biol., 20, 1097–1110. [DOI] [PubMed] [Google Scholar]
- Endo T., Shikanai,T., Sato,F. and Asada,K. (1998) NAD(P)H dehydrogenase-dependent, antimycin A-sensitive electron donation to plastoquinone in tobacco chloroplasts. Plant Cell Physiol., 39, 1226–1231. [Google Scholar]
- Fujita Y., Takahashi,Y., Chuganji,M. and Matsubara,H. (1992) The nifH-like (frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol., 33, 81–92. [PubMed] [Google Scholar]
- Fujita Y., Takagi,H. and Hase,T. (1996) Identification of the chlB gene and the gene product essential for light-independent chlorophyll biosynthesis in the cyanobacterium Plectonema boryanum. Plant Cell Physiol., 37, 313–323. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Takagi,H. and Hase,T. (1998) Cloning of the gene encoding a protochlorophyllide reductase: the physiological significance of the co-existence of light-dependent and -independent protochlorophyllide reduction systems in the cyanobacterium Plectonema boryanum. Plant Cell Physiol., 39, 177–185. [DOI] [PubMed] [Google Scholar]
- Hase T., Wada,K. and Matsubara,H. (1976) Amino acid sequence of the major component of Aphanothece sacrum ferredoxin. J. Biochem., 79, 328–343. [DOI] [PubMed] [Google Scholar]
- Hase T., Kimata,Y., Yonekura,K., Matsumura,T. and Sakakibara,H. (1991) Molecular cloning and differential expression of the maize ferredoxin gene family. Plant Physiol., 96, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D. (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta, 895, 81–106. [Google Scholar]
- Hatch M.D. (1992) C4 photosynthesis: an unlikely process full of surprises. Plant Cell Physiol., 33, 333–342. [Google Scholar]
- Herber U. and Walker,D.A. (1992) Concerning a dual function of coupled cyclic electron transport in leaves. Plant Physiol., 100, 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y. and Hase,T. (1989) Localization of ferredoxin isoproteins in mesophyll and bundle sheath cell in maize leaf. Plant Physiol., 89, 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer W., Koop,H.-U., Wanner,G. and Steinmüller,K. (1998) Mutagenesis of the genes encoding subunits A, C, H, I, J, and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Gen. Genet., 258, 166–173. [DOI] [PubMed] [Google Scholar]
- Kubicki A., Funk,E., Westhoff,P. and Steinmüller,K. (1996) Differential expression of plastome-encoded ndh genes in mesophyll and bundle sheath chloroplasts of the C4 plant Sorghum bicolor indicates that the complex I-homologous NAD(P)H-plastoquinone oxidoreductase is involved in cyclic electron transport. Planta, 199, 276–281. [Google Scholar]
- Matsumura H. and Miyachi,S. (1980) Cycling assay for nicotinamide adenine dinucleotides. Methods Enzymol., 69, 465–470. [Google Scholar]
- Matsumura T., Kimata-Ariga,Y., Sakakibara,H., Sugiyama,T., Murata,H., Takao,T., Shimonishi,Y. and Hase,T. (1999) Complementary DNA cloning and characterization of ferredoxin localized in bundle-sheath cells of maize leaves. Plant Physiol., 119, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Endo,T. and Asada,K. (1998) Properties of the respiratory NAD(P)H dehydrogenase isolated from the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol., 39, 263–267. [DOI] [PubMed] [Google Scholar]
- Meierhoff K. and Westhoff,P. (1993) Differential biogenesis of photosystem II in mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plant: the non-stoichiometric abundance of the subunits of photosystem II in the bundle-sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta, 191, 23–33. [Google Scholar]
- Mi H., Endo,T., Schreiber,U., Ogawa,T. and Asada,K. (1992) Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol., 33, 1233–1237. [Google Scholar]
- Mi H., Endo,T., Ogawa,T. and Asada,K. (1995) Thylakoid membrane-bound pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol., 36, 661–668. [Google Scholar]
- Miyake C., Schreiber,U. and Asada,K. (1995) Ferredoxin-dependent and antimycin A-sensitive reduction of cytochrome b-559 by far red light in maize thylakoids: participation of a menadiol-reducible cytochrome b-559 in cyclic electron flow. Plant Cell Physiol., 36, 743–748. [Google Scholar]
- Moore R.C. and Black,C.C. (1979) Nitrogen assimilation pathways in leaf mesophyll and bundle sheath cells of C4 photosynthesis plants formulated from comparative studies with Digitaria sanguinalis (L.) Scoop. Plant Physiol., 64, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.J., Gounaris,K., Coomber,S.A., Hunter,C.N., Dyer,T.A. and Barber,J. (1989) psbG is not a photosystem two gene but may be an ndh gene. J. Biol. Chem., 264, 14129–14135. [PubMed] [Google Scholar]
- Noctor G. and Foyer,C.H. (1998) A re-evaluation of the ATP:NADPH budget during C3 photosynthesis: a contribution from nitrate assimilation and its associated respiratory activity? J. Exp. Bot., 49, 1895–1908. [Google Scholar]
- Ogawa T. (1991) A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC 6803. Proc. Natl Acad. Sci. USA, 88, 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K. et al. (1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature, 322, 572–574. [Google Scholar]
- Ripkka R., Deruelles,J., Waterbury,J.B., Herdman,M. and Stanier,R.Y. (1979) Genetic assignments, strain histories, and properties of pure cultures of cyanobacteria. J. Gen. Microbiol., 111, 1–61. [Google Scholar]
- Sazanov L.A., Burrows,P.A. and Nixon,P.J. (1998) The plastid ndh genes code for an NADH-specific dehydrogenase: isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl Acad. Sci. USA, 95, 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S. (1990) Do photosynthetic and respiratory electron transport chains share redox proteins? Trends Biochem. Sci., 15, 458–462. [DOI] [PubMed] [Google Scholar]
- Schmid U., Schimz,K.-L. and Sahm,H. (1989) Determination of intracellular pyridine nucleotide levels by bioluminescence using anaerobic bacteria as a model. Anal. Biochem., 180, 17–23. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Endo,T., Yamada,Y., Hashimoto,T., Asada,K. and Yokota,A. (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl Acad. Sci. USA, 95, 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton D.K., Gendel,S.M. and Atherly,A.G. (1993) DNA sequence and shuttle vector construction of plasmid pGL3 from Plectonema boryanum PCC6306. Nucleic Acids Res., 21, 746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. (1988) Preparation of genomic DNA from bacteria. In Current Protocols in Molecular Biology. John Wiley & Sons, New York, pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]
- Woo K.C., Anderson,J.M., Boardman,N.K., Downtown,W.J.S., Osmond,C.B. and Thorne,S.W. (1970) Deficient photosystem II in agranal bundle sheath chloroplasts of C4 plants. Proc. Natl Acad. Sci. USA, 67, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]