Abstract
Several computer programs are available for detecting copy number variants (CNVs) using genome-wide SNP arrays. We evaluated the performance of four CNV detection software suites—Birdsuite, Partek, HelixTree, and PennCNV-Affy—in the identification of both rare and common CNVs. Each program's performance was assessed in two ways. The first was its recovery rate, i.e., its ability to call 893 CNVs previously identified in eight HapMap samples by paired-end sequencing of whole-genome fosmid clones, and 51,440 CNVs identified by array Comparative Genome Hybridization (aCGH) followed by validation procedures, in 90 HapMap CEU samples. The second evaluation was program performance calling rare and common CNVs in the Bipolar Genome Study (BiGS) data set (1001 bipolar cases and 1033 controls, all of European ancestry) as measured by the Affymetrix SNP 6.0 array. Accuracy in calling rare CNVs was assessed by positive predictive value, based on the proportion of rare CNVs validated by quantitative real-time PCR (qPCR), while accuracy in calling common CNVs was assessed by false positive/false negative rates based on qPCR validation results from a subset of common CNVs. Birdsuite recovered the highest percentages of known HapMap CNVs containing >20 markers in two reference CNV datasets. The recovery rate increased with decreased CNV frequency. In the tested rare CNV data, Birdsuite and Partek had higher positive predictive values than the other software suites. In a test of three common CNVs in the BiGS dataset, Birdsuite's call was 98.8% consistent with qPCR quantification in one CNV region, but the other two regions showed an unacceptable degree of accuracy. We found relatively poor consistency between the two “gold standards,” the sequence data of Kidd et al., and aCGH data of Conrad et al. Algorithms for calling CNVs especially common ones need substantial improvement, and a “gold standard” for detection of CNVs remains to be established.
Introduction
Copy number variation (CNV) is loosely defined as a deletion, duplication or inversion of a DNA sequence longer than one kilobase. CNVs have recently attracted considerable interest as a source of genetic variation because they may play an important role in the etiology of complex diseases and in evolution [1]–[14]. Because association studies of CNV and disease have become popular, genome-wide oligonucleotide arrays are now designed to detect both single nucleotide polymorphisms (SNPs) and CNVs [15]. The Affymetrix Genome-Wide Human SNP Array 6.0, for example, includes 906,600 SNP probes and 940,000 CNV probes.
A number of computer programs have been designed to detect CNVs using the intensity of the hybridization of sample DNA to the array probes. The underlying detection algorithm types are generally Hidden Markov model (HMM) [16], [17], or circular binary segmentation (genomic segmentation) [18]. PennCNV and QuantiCNV were first developed on an HMM-based algorithm for an Illumina platform [17], [19], then later modified to be compatible with Affymetrix platforms as well. Birdseye, another HMM-based approach, was developed to detect CNVs in SNP genotyping arrays specifically for Affymetrix platforms [16]. Two commercial software, Partek (Partek Inc., St. Louis, MO) and HelixTree (Golden Helix, Inc.), have implemented circular binary segmentation method.
Baross et al found considerable variation among the outputs from different programs, as well as substantial false call rates for CNVs, when they compared four CNV detection programs [20]: Copy Number Analyser for GeneChip® arrays (CNAG) [21], DNA-Chip Analyzer (dChip) [22], Affymetrix GeneChip® Chromosome Copy Number Analysis Tool (CNAT)[20], [23] and Gain or Loss Analysis of DNA (GLAD) [24]. Winchester et al. reviewed 12 programs and assessed 7 of them using published CNV data in HapMap samples [25]. The programs they assessed were Birdsuite, CNAT, Genome Alteration Detection Algorithm (GADA) [26], PennCNV, QuantiSNP, CNVPartition (Illumina Inc. CA), and Nexus (BioDiscovery, Inc. CA). Like the other papers, they observed large variation among different programs as well as different platforms (Illumina vs. Affymetrix). However, Winchester et al did not draw any conclusions on the performance of the programs, and it is difficult to judge which program was superior based on the data they provided.
Korn et al compared their program, Birdsuite, with two commercial tools, Partek and Nexus [16]. They tested the three programs using eight HapMap samples in which 893 CNVs had been previously identified by Kidd et al using fosmid end-pair sequencing (EPS) and validated by array comparative genomic hybridization or full-length resequencing [13]. They calculated and compared the rates at which the programs recovered these CNVs [16]; a reference CNV was considered recovered if the program called a CNV that shared at least 25% of the length spanned by the reference and called CNV together. The recovery rate of the three programs ranged from 11.6% for Partek to 93.8% for Birdsuite for regions containing more than 20 probes. However, note that Birdsuite was originally developed on HapMap data.
None of the previous studies evaluated rare and common CNV calls separately, even though common CNVs are more difficult to call accurately than rare ones. This difference may arise because some programs use all the samples in a given batch to create a reference signal at a given SNP, to which individual signals from that batch can be compared. For example, HelixTree and PennCNV-Affy establish a reference in this way, then use the log2 of the ratio of the hybridization intensity of an individual subject to the intensity of the reference (log2 R (subject/reference)) for further segmentation or for modeling copy number status [17]. As a result, frequently-occurring variations make it harder to establish a true reference signal. The same problem occurs in Partek if the option to use the log2 ratio for segmentation is selected.
In the Birdsuite package, on the other hand, the Canary program is designed to call common CNVs previously established by McCarroll et al [15]. The designers constructed a series of prior models based on summarized intensity measurements of HapMap samples with different copy number states for the established common CNVs. This approach avoids the common variant reference problem, but it is still problematic because the HapMap sample is not necessarily an appropriate reference for a given study population. For rare or de novo CNVs, Birdsuite has an entirely different program, Birdseye, which establishes a reference intensity similar to that of HelixTree or PennCNV-Affy. It extracts the intensity of all samples in a given batch, then excludes those regions already determined to be copy-variable via Canary, and also excludes the 10% of samples with the highest intensities and the 10% of samples with the lowest intensities [16]. It uses the results to estimate the mean and variance of the normal distribution of two copies.
In this study, we assessed four currently used CNV detection software programs for their accuracy in detecting both rare and common CNVs in the Affymetrix 6.0 platform. We used Affymetrix SNP Array 6.0 data in 270 HapMap samples and 1001 bipolar cases and 1033 controls of European ancestry from Bipolar Genome Study (BiGS). The software packages tested were Birdsuite (version 1.5.2), PennCNV-Affy (a trial version), HelixTree (Version 6.4.2), and Partek (Version, 6.09.0129). We assessed their recovery rate per Korn et al's method with modifications as described below [16]. In addition, based on qPCR, we estimated their false positive and false negative call rates for three common CNVs, as well as their positive predictive values for singleton CNVs, i.e., those that occur once in a dataset.
Results
Recovery test based on sequencing data of eight HapMap samples
We used the same 893 CNVs used by Korn et al. as a reference to compare the recovery rates of Birdsuite, Partek, PennCNV-Affy and HelixTree [16]. Korn et al.'s Birdsuite recovered CNVs spanning more than 20 markers at a rate (88.5%) comparable to that of Korn et al (93.8%) when using their criteria (File S1: Table S1) [16]. With our additional requirement of copy number consistency (the CNV status identified by the programs should be in the same direction as the reference CNV), the recovery rate of Birdsuite decreased from 88.5% to 70%, whereas PennCNV-Affy, Partek and HelixTree had relatively smaller decreases (5–6%). Consistent with the results of Korn et al [16], the recovery rate increased with the number of probes spanned by the CNV for all algorithms (Table 1). The sensitivity of PennCNV-Affy increased from 55.4% to 58.5% when pedigree information was used, which is a unique option of PennCNV-Affy.
Table 1. The recovery rates of each of the CNV-calling programs depending on CNV length based on data of Kidd et al.
| # markers | # CNVs in the reference list from Kidd et al. | # CNVs recovered by Birdsuite | # CNVs recovered by Partek | # CNVs recovered by PennCNV-Affy_trios* | # CNVs recovered by PennCNV-Affy | # CNVs recovered by HelixTree |
| 1 | 329 | 6 (1.8%) | 0 | 0 | 0 | 2 (0.6%) |
| 2–5 | 249 | 71 (28.5%) | 0 | 3 (1.2%) | 2 (0.8%) | 19 (7.6%) |
| 6–10 | 112 | 47 (42.0%) | 10 (8.9%) | 20 (17.9%) | 11 (9.8%) | 28 (25%) |
| 10–20 | 73 | 32 (43.8%) | 26 (35.6%) | 27 (37.0%) | 24 (32.9%) | 17 (23.3%) |
| >20 | 130 | 91 (70.0%) | 70 (53.8%) | 76 (58.5%) | 72 (55.4%) | 54 (41.5%) |
*The pedigree information was incorporated with the calling of CNVs.
Recovery rate was calculated with the requirement of copy number consistency.
Closer inspection of the frequency of 130 CNVs (112 regions containing more than 20 markers) revealed that 100 of these CNV regions occurred only once in the eight HapMap samples. The recovery rate of individual CNVs decreased as the frequency of the CNVs increased (File S1: Figure S1). We further looked at the size of CNVs that spanned more than 20 markers and calculated the recovery rate in each size bin. There was no correlation between the size and the recovery rate when the analysis was limited to CNVs containing more than 20 markers (data not shown).
Recovery test based on array CGH data of 90 CEU HapMap samples
We used 51,440 CNVs detected and validated in 90 CEU samples by Conrad et al as a reference to compare the recovery rate of Birdsuite, Partek, PennCNV-Affy and HelixTree [14]. The recovery rate also increased with the number of probes spanned by the CNV for all algorithms (Table 2). However, the highest recovery rate for any program on detection of CNVs with >20 markers was only 47.69% by Birdsuite.
Table 2. The recovery rates of each of the CNV-calling programs depending on CNV length based on data by Conrad et al. in 90 CEU samples.
| # markers | # CNVs in the reference list from Conrad et al. * | # CNVs recovered by Birdsuite | # CNVs recovered by Partek | # CNVs recovered by PennCNV-Affy_trios** | # CNVs recovered by HelixTree |
| 1 | 28366 | 88 (0.31%) | 2 (0.007%) | 111 (0.39%) | 110 (0.39%) |
| 2–5 | 11837 | 1362 (11.51%) | 8 (0.068) | 138 (1.17%) | 486 (4.11%) |
| 6–10 | 3142 | 926 (29.47%) | 209 (6.65%) | 599 (19.06%) | 720 (22.92%) |
| 10–20 | 2754 | 973 (35.33%) | 507 (18.41%) | 747 (27.12%) | 711 (25.82%) |
| >20 | 5341 | 2547 (47.69%) | 1400 (26.21%) | 1883 (35.26%) | 1770 (33.14%) |
*5,341 CNVs spanned by >20 markers were included in this analysis.
**The pedigree information was incorporated with the calling of CNVs.
We further calculated the average recovery rate of CNVs with different frequency spanned by more than 20 makers. The average recovery rate decreased with the increased frequency of CNVs. The average recovery rate of CNVs with a frequency ≤20% ranged from 65.62% by Partek to 85.95% by PennCNV-Affy with pedigree information incorporated (See Table 3). CNVs with a frequency larger than 80% were difficult to recover (average recovery rate: 10.81%–30.50%).
Table 3. The average recovery rates of each of the CNV-calling programs depending on CNV frequency based on data by Conrad et al. in 90 CEU samples.
| Frequency(a) | # CNVs in the reference list from Conrad et al.* | # CNVs recovered by Birdsuite | # CNVs recovered by Partek | # CNVs recovered by PennCNV-Affy_trios** | # CNVs recovered by HelixTree |
| a< = 20% | 669 | 537 (80.27%) | 439 (65.62%) | 575 (85.95%) | 528 (78.93%) |
| 20%<a< = 40% | 488 | 270 (55.33%) | 221 (45.29%) | 329 (67.41%) | 314 (64.34%) |
| 40%<a< = 60% | 793 | 579 (73.01%) | 210 (26.48) | 373 (47.04%) | 442 (55.74%) |
| 60%<a< = 80% | 765 | 352 (46.01%) | 105 (13.73%) | 139 (18.17%) | 123 (16.08%) |
| 80%<a< = 1 | 2626 | 801 (30.50%) | 284 (10.81%) | 366 (13.94%) | 342 (13.02%) |
*CNVs spanned by more than 20 markers were included in this analysis.
**Pedigree information was incorporated.
CNV regions spanned by more than 20 markers were compared across programs. Birdsuite, PennCNV-Affy and Helixtree can recover about 50% of CNV regions with a sensitivity ≥90% and 30% of CNV regions with a sensitivity ≤10%. For example, Birdsuite recovered 58.21% of CNV regions with a very high sensitivity (>90%), and 34.83% regions with a sensitivity ≤10%. The percentage of CNV regions, with a recovery rate higher than 90%, decreased from 66.39% to 28.13% when the frequency of CNVs increased from ≤20% to >80% (File S1: Table S2). 21 highly frequent (>80%) but poorly recovered (≤10%) CNVs were all duplications. Seven out of nine highly frequency regions with high recovery rates were deletions (File S1: Table S2, Birdsuite data).
Separately, we compared the consistency of the CNVs reported by Kidd et al and by Conrad et al in the same 8 HapMap individuals. Two CNVs identified by Conrad et al and Kidd et al were considered as the same if they shared at least 25% of the total length spanned. 3,024 out of 4,537 CNVs (66.65%) from Conrad et al were not larger than 5 kb while 89.33% of CNVs from Kidd et al were larger than 10 kb. This is expected because of the greater density of probes in Conrad et al. But overall, there was not an impressive degree of overlap between the two studies, even for the larger CNVs (Table 4). Yet each of these studies has been considered as a potential “gold standard” for CNV calling.
Table 4. CNVs detected in 8 HapMap individuals shared between two studies.
| Size (kb) | # of CNVs in Conrad et al (total 4,537) | # detected by Kidd et al (Percentage) * | # of CNVs in Kidd et al (total 9,513) | # detected by Conrad et al (Percentage) |
| ≤5 | 3024 | 38 (1.27%) | 441 | 1 (0.23%) |
| 5to 10 | 647 | 149 (23.02%) | 574 | 13 (2.26%) |
| 10 to 50 | 514 | 146 (28.40%) | 8174 | 300(3.67%) |
| 50 to 100 | 180 | 19 (10.56%) | 217 | 39 (17.97%) |
| 100 to 1000 | 172 | 11 (6.40%) | 107 | 11 (10.28%) |
*Criterion: Two CNVs from Conrad et al and Kidd et al were considered as the same if they shared at least 25% of the total length spanned. One CNV from Conrad et al can share with more than one CNV from Kidd et al, vice versa.
Evaluation of CNV calls in the BiGS data set
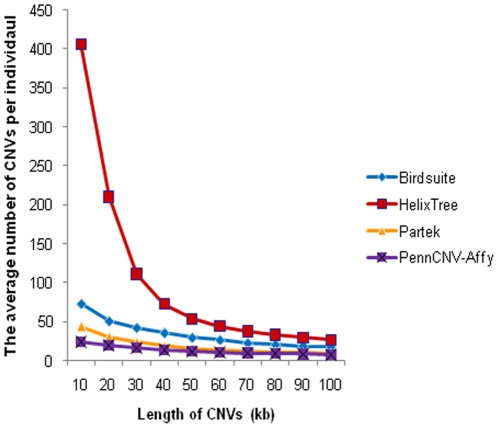
The average number of CNVs called per individual varied greatly among programs (Figure 1); for example, for CNVs larger than 100 kb, PennCNV-Affy called an average of 8, while HelixTree called an average of 27. The differences declined when the size of CNVs increased, so when selecting CNVs for qPCR validation, we chose CNVs larger than 100 kb.
Figure 1. The average number of CNVs per individual called by each of the four CNV-calling programs.
The average number of CNVs per individual varies greatly among programs, especially for CNVs less than 10 kb. The X axis represents length of CNVs. The Y axis is the average number of CNVs per individual.
Singleton deletions and duplications in the BiGS data set
The largest number of program-specific singleton deletion calls was made by HelixTree (Table 5), at a surprisingly higher frequency than Birdsuite, Partek and PennCNV-Affy, which tended to agree with each other. Although software agreement does not necessarily correspond with the validity of the calls, the program-specific calls tended not to be validated, as demonstrated below. More singleton duplications were identified by each program than singleton deletions (Table 5); however, substantial variation among programs was observed in the number of singleton duplications. HelixTree produced the highest percentage of program-specific singleton duplications.
Table 5. The number of singleton CNVs called by each program, and percentage by which they overlap with calls made by the other programs.
| Program | Birdsuite | HelixTree | Partek | PennCNV-Affy | |
| Deletions | Shared ** | 306(90.5%)* | 250(38.8%) | 279(90.0%) | 228(80.6%) |
| Program-specific *** | 32(9.5%) | 394(61.2%) | 31(10.0%) | 55(19.4%) | |
| Total | 338 | 644 | 310 | 283 | |
| Duplications | Shared | 401(74.7%) * | 332(41.7%) | 354(90.8) | 289(85.3%) |
| Program-specific | 136(25.3%) | 465(58.3%) | 36(9.2%) | 50(14.7%) | |
| Total | 537 | 797 | 390 | 339 |
*Data format: number of events (percentage of events shared by other programs).
**Shared singleton deletions or duplications were defined as CNVs called by one program that overlapped at all with singleton deletions or duplications called by any other program.
***Program-specific CNVs are those that did not overlap at all with any singleton deletions or duplications called by any other program.
qPCR validation of selected singleton CNVs in the BiGS data set
The positive predictive values of Birdsuite and Partek for the sampled deletions were 100%. This finding agrees with our previous finding of a positive predictive value for Birdsuite using 19 singleton deletions [10]. Strikingly, none of the five sampled deleted regions called by HelixTree were confirmed by qPCR. Three out of five deleted regions called by PennCNV-Affy were confirmed as deletions by qPCR, a positive predictive value of 60% (Table 6).
Table 6. Positive predictive value for rare CNVs of each program, based on qPCR validation of their program-specific singletons.
| Programs | Birdsuite | HelixTree | Partek | PennCNV-Affy |
| Deletions | 5/5 = 100%* | 0/5 = 0% | 5/5 = 100% | 3/5 = 60% |
| Duplications | 2/5 = 40% | 2/5 = 40% | 2/6 = 33.3% | 4/6 = 66.7% |
*Positive predictive value: true positive/(true positive + false positive). For each region, five samples were tested, one with a putative deletion/duplication, the other four with two putative).
For each program, we also randomly selected five to six program-specific singleton duplications for qPCR validation. None of the programs had all the selected program-specific duplications validated: positive predictive values ranged from 40% to 66.7%.
Common CNVs in the BiGS dataset
We observed that unlike rare CNV calls, common CNVs calls were influenced by plate effects. Common CNVs called by Birdsuite's Canary program were tested for plate effects by doing a series of GWASs plate by plate. We compared the CNV frequencies measured for each plate against those of all other plates in each analysis, using PLINK. About 44% of common CNVs showed a plate effect (data not shown).
Three common CNV regions frequently called by Canary were randomly selected for qPCR investigation: CNP1293, which showed no plate effect, and CNP2157 and CNP2057, which both had plate effects; these CNVs are located on chr8:39354760-39506122 (56 CN and 3 SNP probes), chr16:22465433-22612022 (61 CN and 16 SNP probes), and chr15:19803370-20089386 (104 CN and 60 SNP probes), respectively. The frequencies of duplication and deletion of these three regions as called by each program are shown in Table 7.
Table 7. Frequencies with which each program calls three common CNVs identified by Canary in the BiGS dataset.
| ID | Birdsuite | HelixTree | Partek | PennCNV-Affy | |
| CNP2157 | Frequency of Duplications | 1833(90.1%) | 29 (1.4%) | 24 (1.2%) | 15 (0.7%) |
| Frequency of Deletions | 1 (0.05%) | 187 (9.2%) | 42 (2.1%) | 38 (1.9%) | |
| CNP1293 | Frequency of Duplications | 1 (0.05%) | 664 (32.6%) | 619 (30.4%) | 508 (25.0%) |
| Frequency of Deletions | 1277(62.8%) | 449 (22.1%) | 344 (16.9%) | 262 (12.9%) | |
| CNP2057 | Frequency of Duplications | 170(8.4%) | 254 (12.5%) | 197 (9.7%) | 103 (5.1%) |
| Frequency of Deletions | 653(32.1%) | 248 (12.2%) | 142 (7.0%) | 188 (9.2%) |
To validate these CNV calls, we validated 3 CNPs with qPCR. We assayed 69 samples with qPCR for CNP2157 and found that CNV calls by all of the programs in this region were imprecise (Table 8). For CNP2057, we quantified 87 samples; none of the programs performed satisfactorily. When 85 samples were studied for CNP1293, which had no plate effect, we found that, strikingly, Birdsuite achieved a 0% false positive rate and a 1.9% false negative rate, while the other three programs all had a low sensitivity (i.e., 1 - the false positive rate) and specificity (i.e., 1 - the false negative rate) for this CNV. The performance of each program varied greatly from CNV to CNV; none performed consistently. The results observed here were consistent with the recovery rate evaluated in HapMap samples especially for Birdsuite (see above).
Table 8. qPCR validation of the calls made by each program for three common CNVs identified by Canary in the BiGS dataset.
| CNP | Birdsuite | HelixTree | Partek | PennCNV-Affy | |
| CNP2157 | False positive rate | 100% | 8.5% | 0.0% | 0.0% |
| False negative rate | 100% | 66.7% | 66.7% | 66.7% | |
| CNP1293 | False positive rate | 0.0% | 96.9% | 71.9% | 71.9% |
| False negative rate | 1.9% | 80.8% | 80.8% | 80.8% | |
| CNP2057 | False positive rate | 55.1% | 0.0% | 0.0% | 0.0% |
| False negative rate | 62.5% | 46.9% | 62.5% | 62.5% |
Discussion
The sensitivity and specificity of CNV identification is an essential component of association studies of CNVs with disease. In our evaluation of two commercial programs and two publicly available programs widely used for CNV identification in GWAS data, we found considerable variation among the programs in the number of CNVs called. The differences declined when the size of CNVs increased, but substantial variation still existed in the accuracy of CNV calls as determined by the recovery test and by qPCR validation, even for large CNVs containing more than 20 markers or larger than 100 kb in length.
For recovery of CNVs detected by Korn et al in eight HapMap samples, Birdsuite was superior to the other three programs, as it recovered more CNVs in each category. However, the sensitivity of all four programs was poor when the number of probes spanned by a CNV was small. For CNVs containing more than 20 markers, Birdsuite recovered 88.5% of CNVs, which is comparable to Korn et al's finding of 93.8% for Birdsuite [16]. Some of the discrepancy may be explained by the fact that Korn et al. analyzed the same samples but with different. CEL files produced by different labs; the files used in the present study were obtained from Affymetrix, while Korn et al. used. CEL files produced in their own lab [16]. Also, their Birdsuite recovery rate was inflated since no agreement on CNV state (deletion or duplication) was required by Korn et al. just after the development of Birdsuite; we found that only 70% of CNVs containing 20 or more markers were recovered by Birdsuite with an agreement on CNV state.
Birdsuite's recovery rate of common CNVs containing more than 20 markers varied across genomic regions. For example, there were eight CNV regions carried by any two of the eight HapMap individuals, and therefore common. Six of those eight regions were recovered in both individuals by Birdsuite, but the other two regions were not recovered. Poor recovery rates of CNVs with high frequency (i.e., carried by more than three of the eight HapMap samples), was also observed (File S1: Figure S1).
As expected, the sensitivity of Partek showed a 5.2-fold increase, to 60.7%, after using quantile normalization, as compared to Korn et al.'s finding of 11.2% without using quantile normalization. A very recent study has shown that normalization can improve performance in analysis of miRNA array data and that quantile normalization is the most robust normalization method [27]. Like all microarrays, Affymetrix SNP arrays are affected by systematic sources of experimental variation. Normalization can help reduce or remove noise that distorts the distribution of observed array data, and thus improve the accuracy of genotyping calls and copy number calls.
For our recovery test in 90 CEU HapMap samples, the highest recovery rate was only 47.91% from Birdsuite, in detecting CNVs spanned by more than 20 markers. Consistent with the recovery rate shown above, the average recovery rate decreased with increased CNV frequency. On closer inspection, it is clear that Birdsuite's recovery rate of most CNVs containing more than 20 markers was either ≤10% or >90% (File S1: Table S2). For example, there were 32 CNV regions that showed a very high frequency in CEU HapMap samples (>80%). Six out of 32 CNV regions were 100% recovered but the recovery rates of 21 CNV regions were less than 10%, which is consistent with our qPCR results on tested common CNVs. Regardless the recovery rate, the number of CNVs identified by different programs varied greatly. The most CNVs were identified by Helixtree, almost 4 times the number of CNVs detected by Birdsuite and 7 times that of CNVs detected by PennCNV-Affy and Partek.
One possible reason for the discrepancy of recovery rate in two datasets (Kidd et al and Conrad et al) is that the detection method: very dense microarrays used by Conrad et al vs. sequencing used by Kidd et al [13], [14]. Conrad et al's experiment design aimed to discover CNVs of size greater than 500 bp. However, the median size of insert clone is around 40 kb in paired-end sequencing by Kidd et al., which would make detection of small CNVs more challenging. Consequently, more small CNVs (≤5 kb) were validated by Conrad et al, and more median size CNVs (10–50 kb) were reported by Kidd et al. This may also partially explain the poor consistency rate of CNVs between the two studies.
There was significant variation in singleton deletion calls among programs: HelixTree detected about two-fold more singleton deletions in the BiGS data set (644 singleton deletions) than Birdsuite, Partek, or PennCNV-Affy. However, more than half of those detections were specific to HelixTree (59.3%), and the majority of HelixTree program-specific calls of deletions were not validated by qPCR. This meant that for program-specific singleton deletions, the positive predictive value of HelixTree was zero based on the qPCR validation of sampled regions. For Partek and Birdsuite, on the other hand, all selected singleton deletions were validated by qPCR. In the recovery test and in qPCR results on singletons, Birdsuite had the best performance among the tested programs.
We checked closely the program-specific calls because some authors have proposed to combine CNV calls from multiple independent software calls to improve accuracy. The program-specific calls in our data tended not to be validated (most likely to be in error). Positive predictive value could not be calculated in an unbiased manner in the present study since we only selected program-specific CNVs for qPCR validation. A sampling from all CNVs (both program-specific and shared calls) would provide a more accurate measure of predictive values.
More singleton duplications were called by each program in the BiGS dataset than singleton deletions, and each program called singleton duplications that were not called by any of the other three programs. The average positive predictive value for these program-specific singleton deletions for all four programs (65%) was higher than for singleton duplications (45%) in our tested regions. We speculate that the performance of programs in the detection of deletions is better than the detection of duplications because a deletion represents a 2-fold change in copy number while a duplication produces only a 1.5-fold change. As might be expected, 21 highly frequent (>80%) but poorly recovered (≤10%) CNVs were all duplications (File S1: Table S2 Birdsuite's recovery data).
For common CNV regions identified by Canary in the present study, we observed striking variation in the frequency of calls by different programs. Each program had substantial false positive and false negative rates in at least one of the three CNV regions tested. The high false positive and negative rates observed here may be due to the fact that common CNVs will affect the mean and variance of hybridization intensity over the region included and thus affect the observed log2 ratio of the CNV. The fact may also lead to the poor recovery rate of highly frequent CNVs.
Plate effects may play a significant role in the accuracy of common CNVs called by Canary. CNP1293, which showed no plate effect, had a high sensitivity and specificity, but CNP2157 and CNP2057 had plate effects and showed a very low sensitivity and specificity. According to our limited qPCR results, the algorithms evaluated here for calling common CNVs all need improvement. We would conclude that without independent experimental genotyping, software-called common CNVs based on GWAS array data are not suitable for association studies. In contrast, rare CNVs called by Birdsuite and Partek are of substantially better quality. Similarly, Marenne et al concluded that further validation was required to assess CNVs as risk factors in complex diseases when they evaluated CNVpartition, PennCNV and QuantiSNP using Illumina Infinium Human 1 Million SNP array data[28].
Recently, Mei et al developed two new methods to identify common CNV regions [29]. They evaluated their methods with sequencing-based results from Kidd et al. However, the lowest discordance rate was 55% after excluding individual regions with a confidence score (as developed by them) below the 80th percentile. Two previously published methods, STAC and GISTIC had similar performance at identifying CNVs with high frequency and moderate confidence [30], [31]. These reports further confirmed our observation that common CNV detection methods still have much room for improvement.
Winchester et al recommended using a second program to generate the most informative results [25]. This recommendation seems to be based on the assumption that the second program performed similarly to the first one, and that their overlap increases the reliability. This might not be a safe assumption when not all software suites perform equally well. Birdsuite is a better choice for identifying rare CNVs than the others in our evaluation.
One limitation of the present study is the small number of CNVs tested by qPCR, particularly for the common CNVs. Although the number of qPCR tests performed for validation was limited, the overall trend of frequent non-validation is in agreement with other results from larger datasets (the recovery tests on HapMap samples). These two independent lines of evidence support our concerns regarding the validity of CNV calls based on GWAS data.
The intention of this study is to identify potential traps of current practice in the GWAS-based CNV analysis, rather than an attempt to provide a solution. It is possible that program tweaking would improve accuracy, but it appeared reasonable to start with the default parameters recommended by each program's provider.
We evaluated the reproducibility of the two “gold standards” used in this study, the paired-end sequence data of Kidd et al, and the very high density array Comparative Genomic Hybridization (aCGH) in the same 8 HapMap individuals. We found relatively poor consistency between the two “gold standards.” The lack of a standard sets a limit on much of the recovery of GWAS-based CNV calls, particularly for common CNVs since they would be over-represented when 8 individuals are studied. Next-generation sequencing of whole genome of population samples might be able to provide an ultimate gold standard for identification of common CNVs.
A more extensive list of independently validated CNV regions, and the raw hybridization or other data files used to detect them, should be made publicly available. A greatly expanded version of Kidd et al's or Conrad et al's HapMap data set used in this and previous studies [13], with all CNVs confirmed by high coverage sequencing, and with the addition of parental data, might provide an acceptable resource. The public data from dbGaP and similar sources can also be used for this purpose, as can the CNV validation data we have produced in this study and plan to produce in future studies. Once these datasets are available, independent validation studies must be performed, even though they require the expenditure of valuable time and funds.
Materials and Methods
Subjects
To test the programs' recovery rates, we obtained the Affymetrix Genome-Wide Human SNP Array 6.0 data (SNP Array 6.0) from 270 HapMap samples (90 CEPH, 90 YRI and 45 CHB and 45 JPT) from Affymetrix. To test the programs' false positive/negative rates for common CNVs and positive predictive values for singletons, we obtained Bipolar Genome Study (BiGS) genotype data from 1001 bipolar cases and 1033 controls of European ancestry via dbGaP (phs000017.v1.p1); those data were collected using the Affymetrix Genome-wide Human SNP Array 6.0, details described elsewhere [32].
Software algorithms and parameters
We performed plate-wise quantile normalization, then identified CNVs for each plate using each of the four programs. The settings of the software packages are presented in Table 9. Since the optimal parameters for a given dataset may not be optimal for another dataset, we chose to mainly use the programs' default parameters to make the comparisons as fair as was possible. To better compare HelixTree and Partek, log2 ratios were created by normalizing raw intensity data against a set of reference samples from a given batch in HelixTree, and then were called by Partek and HelixTree for CNVs using segmentation. Genomic segmentation was recommended by Partek and was therefore used for evaluation here. For PennCNV-Affy, we ran the program with and without the pedigree information.
Table 9. Settings used for each of the four software suites.
| Software | Plate-wise quantile normalization | Detecting algorithm | Parameters |
| Birdsuite | Yes (APT)* | HMM | Using population-specific prior models*** |
| HelixTree | Yes (HelixTree) | Segmentation | Default |
| Partek | Yes (HelixTree) ** | Segmentation | Default |
| PennCNV-Affy | Yes (APT) | HMM | No prior models |
For HelixTree and Partek, default settings were used; normalization was done before CNV calling. For PennCNV-Affy, the standard procedure was followed without wave adjustment. For Birdsuite, population-specific prior models were employed.
Platewise normalization was done by Affy Power Tools (APT1.10.0) plug in Birdsuite/PennCNV-Affy.
Normalization was done by HelixTree.
For Canary, the appropriate prior model was selected based on the ancestry of the sample.
Measuring CNV recovery rates in eight and 90 CEU HapMap samples
For eight HapMap samples, we used the same recovery statistic as Korn et al., with one modification. Since deletions identified by one program could be called as duplications by another program, an additional criterion was included that not only a variation be called, but that it should be in the same direction as the reference CNV, i.e., a CNV called as 0 copy or 1 copy had to correspond to a deletion called in the eight validated HapMap samples in Kidd et al [13]. If it did not match, the reference CNV was not considered recovered. Similarly, another recovery rate was calculated based on 51,440 reference CNVs validated in 90 CEU samples reported by Conrad et al [14]. Normalization was done prior to segmentation when evaluating Partek. We used the same inclusion criterion for CNV calls as Korn et al [16], requiring CNV calls to have LOD scores (probability of the segment being the stated copy number versus the copy number of the flanking region) ≥5 for inclusion.
Consistency of CNVs Detected in 8 HapMap Individuals in Two Studies (Conrad et al and Kidd et al)
CNVs validated by Conrad et al were compared with CNVs reported by Kidd et al. Two CNVs from Conrad et al and Kidd et al were considered as the same if they shared at least 25% of the total length spanned. One CNV from Conrad et al can share with more than one CNV from Kidd et al, vice versa.
Comparison and validation of CNV calls in the BiGS dataset
In the BiGS dataset, all CNV calls made by all four programs were compared (Figure 1). Birdsuite is the only one of the four that filters out CNVs with LOD scores less than 10.
We are particularly interested in the role of singleton CNVs in common diseases [10], so when choosing rare CNVs to validate, we focused on singleton CNVs. Singletons were defined as deletions or duplications that occurred only once in the entire BiGS data set, including controls, and did not overlap with any other CNVs. The analysis was done by PLINK (version 1.05) [33].
To validate the singleton CNVs called by the software packages in the BiGS sample, quantitative real-time PCR (qPCR) with SYBR-green dye was used to measure the copy number of a subset of singleton CNVs using the ABI Prism® 7900HT Sequence Detection System. The copy number at target regions relative to the reference is approximately 2− ΔΔCt (for more details see elsewhere [10]), where Ct, the threshold cycle number, is a function of the amount of starting template.
We selected a total of 20 singleton deletions and 22 singleton duplications for qPCR validation. For each program, five or six duplications and five deletions were randomly selected that were uniquely called by that program, which we refer to as program-specific calls. We then designed three pairs of primers for each region and tested five samples for singleton duplications and five other samples for singleton deletions. So, in each test, one sample carried a program-specific singleton deletion or duplication, and the other four samples did not carry any CNVs called that overlapped that region, i.e., all programs called two copies for that sample.
To validate the common CNVs called by the software packages for the BiGS sample, we randomly selected one common duplication and two common deletions called by Canary for qPCR validation. We used TaqMan® Copy Number Assays from ABI (Applied Biosystems Inc, CA) on the ABI Prism® 7900HT Sequence Detection System for validation since the target and an endogenous control can be amplified in the same reaction. Only one probe was designed for each region, so CNVs called by different programs were required to share at least 50% of the total length spanned by the two calls combined to be considered the same CNV. All the primers and probes designed for qPCR are provided in supplementary data (File S1: Table S3).
To assess the relative accuracy of the programs in detecting rare CNVs in the BiGS data set, we calculated and compared their positive predictive values, i.e., the ratio of true positives to positive calls both true and false; data to calculate false negative and false positive rates were not collected, since these CNVs are, by definition, rare. To compare the programs' abilities to detect common CNVs, we calculated and compared their specificities and sensitivities.
Web resources
http://www.biodiscovery.com/index/nexus
http://www.openbioinformatics.org/penncnv/penncnv_tutorial_affy_gw6.html
http://www.ncbi.nlm.nih.gov/gap
http://pngu.mgh.harvard.edu/purcell/plink/)
Supporting Information
Includes Tables S1 to S3 and Figure S1.
(0.42 MB DOC)
Acknowledgments
We acknowledge Dr. Judith A. Badner for extensive statistical advice, Ms. Kay Grennan for her editing assistance and scientific comments and the advice of Drs. Kai Wang and Joshua M. Korn on the use of PennCNV-Affy and Birdsuite.
The BiGS collaborators
University California, San Diego: John R. Kelsoe, Tiffany A. Greenwood, Caroline M. Nievergelt, Rebecca McKinney, Paul D. Shilling; Scripps Translational Science Institute: Nicholas J. Schork, Erin N. Smith, Cinnamon S. Bloss; Indiana University: John I. Nurnberger, Jr., Howard J. Edenberg, Tatiana Foroud, Daniel L. Koller; University of Chicago: Elliot S. Gershon, Chunyu Liu, Judith A. Badner; Rush University Medical Center: William A. Scheftner; Howard University: William B. Lawson, Evaristus A. Nwulia, Maria Hipolito; University of Iowa: William Coryell; Washington University: John Rice; University California, San Francisco: William Byerley; National Institute Mental Health: Francis J. McMahon, Thomas G. Schulze; University of Pennsylvania: Wade H. Berrettini; Johns Hopkins: James B. Potash, Peter P. Zandi, Pamela B. Mahon; University of Michigan: Melvin G. McInnis, Sebastian Zöllner, Peng Zhang; The Translational Genomics Research Institute: David W. Craig, Szabolcs Szelinger; Portland Veterans Affairs Medical Center: Thomas B. Barrett; Georg-August-University Göttingen: Thomas G. Schulze.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from The National Institute of Mental Health, R01MH080425 and 1R01MH081804, The National Alliance for Research on Schizophrenia and Depression, R01MH081804, the Eklund family, and the Geraldi Norton Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 2.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 3.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 7.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D, Cheng L, Qian Y, liey-Rodriguez N, Kelsoe JR, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol Psychiatry. 2009;14:376–380. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 12.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 16.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hadley D, Liu R, Glessner J, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 19.Colella S, Yau C, Taylor JM, Mirza G, Butler H, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baross A, Delaney AD, Li HI, Nayar T, Flibotte S, et al. Assessment of algorithms for high throughput detection of genomic copy number variation in oligonucleotide microarray data. BMC Bioinformatics. 2007;8:368.:368. doi: 10.1186/1471-2105-8-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Li C, Paez JG, Chin K, Janne PA, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Wei W, Zhang J, Liu G, Bignell GR, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20:3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- 25.Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief Funct Genomic Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 26.Pique-Regi R, Monso-Varona J, Ortega A, Seeger RC, Triche TJ, et al. Sparse representation and Bayesian detection of genome copy number alterations from microarray data. Bioinformatics. 2008;24:309–318. doi: 10.1093/bioinformatics/btm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pradervand S, Weber J, Thomas J, Bueno M, Wirapati P, et al. Impact of normalization on miRNA microarray expression profiling. RNA. 2009;15:493–501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marenne G, Rodríguez-Santiago B, Closas MG, Pérez-Jurado L, Rothman N, et al. Hum Mutat. In press; 2010. Assessment of copy number variation using the illumina infinium 1M SNP-array: a comparison of methodological approaches in the Spanish bladder cancer/EPICURO study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei TS, Salim A, Calza S, Seng KC, Seng CK, et al. Identification of recurrent regions of Copy-Number Variants across multiple individuals. BMC Bioinformatics. 2010;11:147–160. doi: 10.1186/1471-2105-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diskin SJ, Eck T, Greshock J, Mosse YP, Naylor T, et al. STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Research. 2006;16:1149–1158. doi: 10.1101/gr.5076506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. PNAS. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Includes Tables S1 to S3 and Figure S1.
(0.42 MB DOC)