Abstract
Dosage compensation in Drosophila is mediated by a multiprotein, RNA-containing complex that associates with the X chromosome at multiple sites. We have investigated the role that the enzymatic activities of two complex components, the histone acetyltransferase activity of MOF and the ATPase activity of MLE, may have in the targeting and association of the complex with the X chromosome. Here we report that MLE and MOF activities are necessary for complexes to access the various X chromosome sites. The role that histone H4 acetylation plays in this process is supported by our observations that MOF overexpression leads to the ectopic association of the complex with autosomal sites.
Keywords: ATP-dependent helicase/chromatin remodeling/dosage compensation/Drosophila/histone acetylation
Introduction
Dosage compensation is a regulatory process that ensures that males and females have equal amounts of X chromosome gene products. In Drosophila, where it is achieved by a doubling of X-linked gene transcription in males relative to females, dosage compensation involves a complex of gene products of at least five known genes (for review see Lucchesi, 1998): maleless (mle), male-specific lethal 1, 2 and 3 (msl1, msl2, msl3) and males absent on the first (mof). The complex is preferentially associated with numerous sites on the X chromosome in somatic cells of males but not of females and its presence on the X chromosome is correlated with a significant increase of a specific histone isoform: histone H4 acetylated at Lys16 (for review see Turner, 1998). In addition, two untranslated RNAs, RNA on the X1 and 2 (roX1 and roX2), are found only in males or in transgenic females that have been induced to form an MSL complex (for review see Stuckenholz et al., 1999). The roX RNAs associate with the X chromosome with a distribution that mirrors that of the MSLs, and one of them (roX2) is present in a purified MSL complex isolated from cultured cells (Smith et al., 2000). Recently, a kinase (JIL-1) has been found to be enriched on the X chromosome of male salivary gland nuclei (Jin et al., 1999).
Although the ubiquitous association of each of the five MSLs with the X chromosome, seen in wild-type males, depends on the presence and availability of the other gene products, loss-of-function mutations of mle and msl3 (Palmer et al., 1994; Bashaw and Baker, 1995) or mof (Gu et al., 1998) allow the residual binding of MSL1 and MSL2 to ∼30–40 sites. Approximately 30 of these sites have been mapped (Lyman et al., 1997) and are thought to be chromatin entry points for the complex. The order of assembly at these sites was shown to be MSL1/MSL2, MLE, MOF and lastly MSL3 (Gu et al., 1998). Kelley et al. (1999) have reported that the roX1 and roX2 genes themselves are located within two entry sites where the formation of complexes containing roX RNA occurs.
Here we report that, in order to become stably associated with the numerous other sites along the X chromosome where it is normally found, the MSL complex requires the histone acetyltransferase activity of MOF as well as the ATPase activity of MLE. If either of these activities is impaired, complexes containing the known MSLs are formed but are unable to access X-chromosome chromatin beyond the entry sites. Finally, we report that overexpression of MOF leads to the acetylation of numerous autosomal sites and to the autosomal association of the MSL complex. This study represents the first demonstration that the enzymatic activities of a chromatin remodeling complex are required for its targeting within the genome.
Results
The effect of the mof1 mutation on the distribution of the complex is not due to protein instability
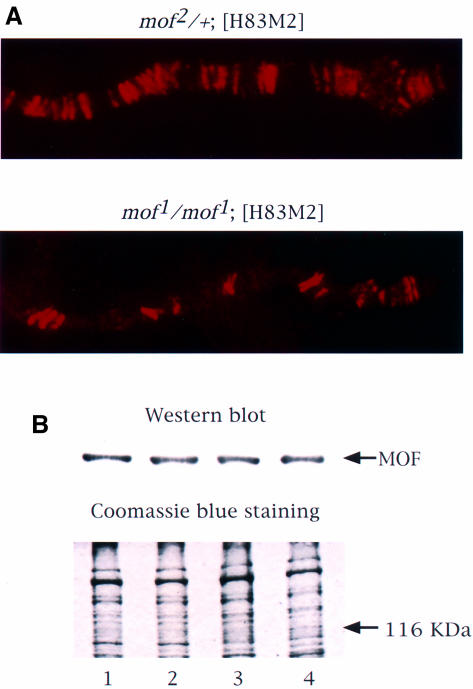
We have shown previously that in the presence of the mof1 mutation, MSL complexes are targeted only to the entry sites (Gu et al., 1998; Figure 1A). Note that, because the salivary glands of dying mutant males are unfavorable for cytological studies, these observations were made on female larvae carrying an msl2+ transgene (H83M2) where the ectopic expression of MSL2 leads to the formation of the MSL complex (Kelley et al., 1995). The mof1 mutation, which is a glycine to glutamic acid replacement at the most highly conserved residue of the acetyl coenzyme A (acetyl CoA)-binding domain (G691E; Hilfiker et al., 1997), results in the loss of the histone acetyltransferase activity for both recombinant MOF (Akhtar et al., 2000) and the MSL complex (Smith et al., 2000). Its effect on the distribution of the complex could be due to loss of histone acetyltransferase activity or to a decrease in the level of MOF1 protein due to instability. To determine whether the latter is the case, protein extracts from the heads of adult flies were resolved by SDS–PAGE and exposed to antibodies against MOF. The amount of protein loaded was monitored by Coomassie Blue staining. As shown in Figure 1B, the levels of MOF in wild-type males and females, and in [H83M2] transgenic females homozygous for mof1 or carrying a mof+ wild-type allele over a null allele, were very similar.
Fig. 1. MSL complexes are targeted only to the entry sites on the X chromosome in the presence of mof1. (A) Polytene chromosomes from control mof2/+; [H83M2] and mof1/mof1; [H83M2] female larvae were immunolabeled with antiserum against MOF. The distribution of the MOF protein is the same as that observed for all of the other MSLs (Gu et al., 1998). (B) MOF is expressed at a similar level in wild-type males (lane 1), wild-type females (lane 2), mof1/mof1; [H83M2] females (lane 3) and mof2/+; [H83M2] females (lane 4). Protein extracts prepared from adult fly heads were resolved by 7.5% SDS–PAGE. The proteins were transferred to nitrocellulose membrane and incubated with anti-MOF serum. A parallel gel was stained with Coomassie Brilliant Blue R-250 to monitor the amount of protein loaded.
In the presence of the mof1 mutation, complexes are formed but cannot access the X chromosome beyond the entry sites
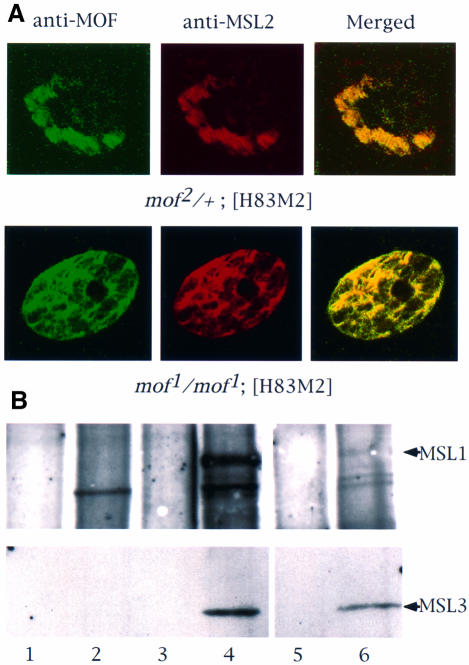
To determine whether the MOF1 protein that is not found along the X chromosome is free or is present in association with any of the other MSLs, we stained whole salivary glands with antibodies against MSL2 and MOF (Figure 2A). In mof2/+; [H83M2] control females, these two proteins are found only on the X chromosomes, as expected (the mof2 allele encodes a truncated protein; Gu et al., 1998). In mof1/mof1; [H83M2] females, there were some bands with strong staining for MOF and MSL2 due to the presence of MSL complexes at the entry sites, as shown previously on polytene chromosome spreads (Figure 1A). Besides these strong bands on the X chromosome, MSL2 and MOF were also co-localized elsewhere in the nuclei. Since there is no evidence of the association of these complex components with autosomal chromatin in polytene chromosome spreads of mof1 males (Hilfiker et al., 1997) or in mof1/mof1; [H83M2] females such as the one used in Figure 1A (data not shown), our observations suggest the presence of free, untargeted MSL complexes in the nucleoplasm. Co-immunoprecipitation of MSL1 and MSL3 presented in Figure 2B established that such complexes do exist.
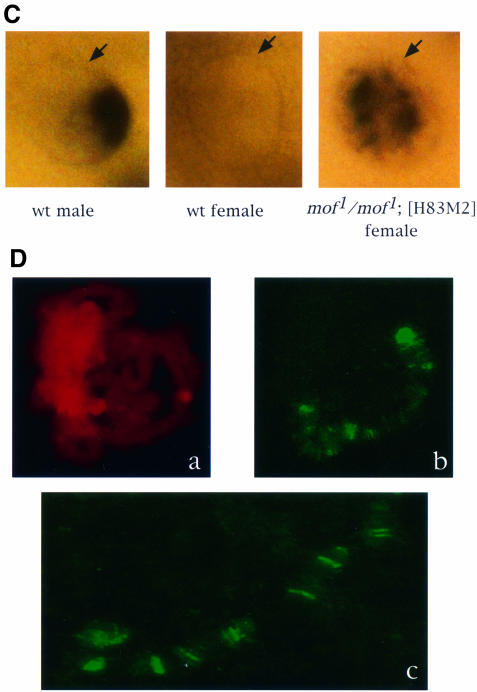
Fig. 2. In the absence of MOF activity, MSL complexes are found in the nucleoplasm. (A) Whole salivary glands from mof1/mof1; [H83M2] and mof2/+; [H83M2] larvae were stained for MSL2 and MOF by indirect immunofluorescent labeling. In the nuclei of larvae expressing mof+, the two antigens are co-localized along the paired X chromo somes. In mof1 homozygotes, the MSL complex appears in the interchromosomal spaces. Note that it must also be present at the entry sites on the X chromosomes although these cannot be resolved in this material. (B) Co-immunoprecipitation of MSL1 and MSL3. Protein extracts prepared from wild-type female (lanes 1 and 2), wild-type male (lanes 3 and 4) and mof1/mof1; [H83M2] female (lanes 5 and 6) flies were immunoprecipitated with pre-immune (lanes 1, 3 and 5) or MSL1 antisera (lanes 2, 4 and 6). The precipitate was analyzed by western blotting using MSL1 and MSL3 antisera. (C) Localization of roX1 RNA, determined by in situ hybridization in whole salivary glands. The RNA is localized to a limited area corresponding to the X chromosome in male nuclei and is absent in the nuclei of wild-type females. In mof1/mof1; [H83M2] females, roX1 RNA is dispersed in a pattern that mimics that of the MSLs. The arrows mark the nuclear envelope. (D) In situ hybridization of roX1 RNA on the polytene chromosomes of mof1/mof1; [H83M2] female larvae. (a) Propidium iodide staining of a nucleus; (b) roX1 is present at the entry sites on the X chromosome in the same nucleus; (c) a stretched region of X chromosome from another nucleus.
Since transcribed roX RNAs do not accumulate in the absence of the MSL complex (Meller et al., 2000), we reasoned that the presence of these RNAs would confirm the presence of the MSL complex in the nucleoplasm of mof1 mutant individuals. To study the distribution of roX1 RNA, salivary glands from wild-type males and females, and from mof1/mof1; [H83M2] females were hybridized with roX1 DNA probes. As expected, roX1 RNA is present in wild-type male but not female nuclei (Figure 2C). roX1 RNA was also observed in mof1/mof1; [H83M2] females. However, in contrast to the wild-type male pattern, in which roX1 is localized only to the X chromosome, in mof1 mutant individuals roX1 RNA was dispersed throughout the nuclei. In order to distinguish whether this RNA is associated with all the chromosomes or is present in the nucleoplasm, we performed in situ hybridization on polytene chromosome spreads. As shown in Figure 2D, roX1 RNA was present only at the X chromosome entry sites. Thus, the dispersed pattern of roX1 RNA in the whole nuclei indicates its presence in the nucleoplasm. Since roX1 RNA requires the MSL complex for stabilization, this result confirmed that assembled MSL complexes unable to be targeted to the X chromosome are present.
We performed similar experiments on Schneider 2 (S2) cells transfected with mof1 cDNA under the control of the Mtn promoter. After induction with CuSO4, in most transfected cells, MSL1 is dispersed in interphase nuclei, although occasionally, because of the variability in the degree of transfection, the level of MOF1 is not sufficient to compete fully with endogenous MOF and staining of the X chromosome by active complex can be seen. Transfected cells clearly overexpressing MOF1 had very significantly reduced levels of histone H4 acetylated at Lys16 (H4Ac16) (Figure 3). The absence of complex on the autosomes in salivary gland preparations of mof1/mof1; [H83M2] female larvae strongly suggests that in these S2 cells the complex is not associated with autosomal chromatin; rather, it is present in the nucleoplasm. Since propidium iodide stains both DNA and RNA, it is not possible to determine the distribution of the MSL complex on the basis of MSL1 staining in interphase nuclei. For this reason, we searched for mitotic figures in our preparations and were able to establish that, during cell division, the level of MSL1 appears to be substantially lower, perhaps due to disassembly and degradation.
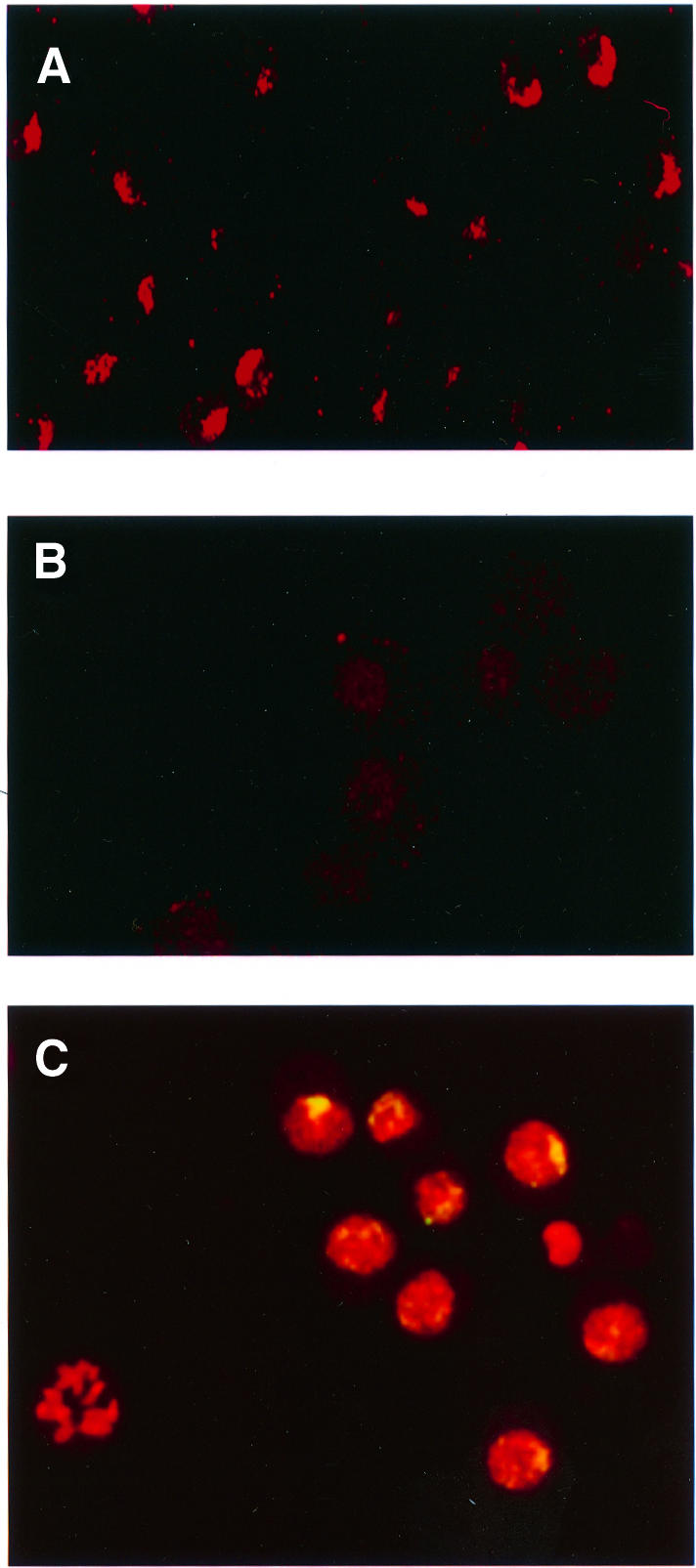
Fig. 3. Overexpression of MOF1 in S2 cells causes a reduction in the level of H4Ac16 present on the X chromosomes and relocation of MSL complexes. Histone H4Ac16 in S2 cells transfected with the vector (A) or with mof1 cDNA (B) and induced with CuSO4. (C) Merged image of induced S2-mof1 cells stained for MSL1 (green) and counterstained for nucleic acids with propidium iodide (red). The yellow color indicates the presence of the MSL1 protein.
Overexpression of MOF causes association of the MSL complex at ectopic sites
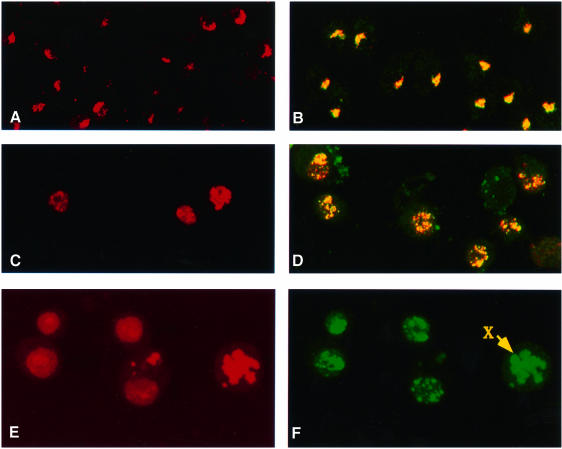
The results reported above demonstrate that the histone acetyltransferase activity of MOF plays an important role in spreading the complex from the entry sites to other sites on the X chromosome. The acetylation of histone H4 at Lys16, for which MOF is responsible (Smith et al., 2000), may render the X-chromosome chromatin accessible to MSL complexes. If this is the case, an induced ectopic acetylation of histone H4 at Lys16 could lead to the association of the MSLs at some autosomal sites. To investigate this possibility, we used S2 cells transfected with mof cDNA and selected for stable integration of the construct. Cells overexpressing MOF had a very high level of the H4Ac16 isoform in comparison with mock-transfected S2 cells, where H4Ac16 was only observed on the X chromosome (Figure 4A and C). When MOF-overexpressing cells were stained for MSL1 and MSL3, these two proteins were co-localized on the X chromo somes as well as at many other sites in the nuclei (Figure 4D). To determine whether these complexes are present free in the nucleoplasm or are bound to autosomes, we stained cells with MSL1 antiserum and counterstained them with propidium iodide to identify nuclei with morphologically distinct chromosomes. In cells whose nuclei had entered mitosis, MSL1 was not only present on the X chromosome, but was also clearly associated with the autosomes; little if any complex appeared free in the nucleoplasm (Figure 4E and F). Although there is an X chromosome entry site mapped at 5C where mof is located, the autosomal binding in induced cells is unlikely to be due to the creation of ectopic entry sites by the integration of the mof cDNA construct at multiple autosomal locations, because (i) this type of autosomal association is absent in transfected but non-induced cells, where the MSL complex is limited to the X chromosome (data not shown) and (ii) in salivary gland preparations of larvae carrying a mof cDNA transgene known to rescue the mof1 mutation at an autosomal location there is no evidence of complex association with the transgene (data not shown).
Fig. 4. MSL complexes are targeted to autosomal sites in S2 cells overexpressing MOF. (A) Histone H4Ac16 staining is restricted to the X chromosome in cells transfected with the vector induced with CuSO4. (B) Co-localization to the X chromosome is also seen in the merged image of immunofluorescent labeling for MSL1 (red) and MSL3 (green) in similar cells. (C) In contrast, H4Ac16 is widespread throughout the nuclei in cells transfected with mof cDNA and induced with CuSO4. (D) Merged images of immunofluorescent labeling for MSL1 (red) and MSL3 (green) in similar cells. The complex distribution generally follows the distribution of H4Ac16. (E) Induced S2-mof cells were stained for nucleic acids with propidium iodide. (F) The same cells were immunolabeled with anti-MSL1 serum. The distribution of MSL1 and, by inference, of the MSL complex is clearly autosomal. The arrow points to a chromosome that is more intensely stained than the others and is presumably the X chromosome.
Loss of MLE function prevents the spreading of the complex by affecting its stability and/or function
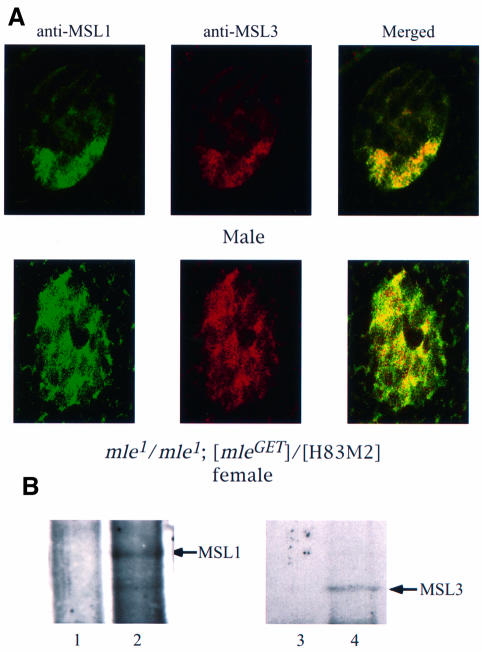
The MLE protein is required for assembly of the MSL complex, as demonstrated by the observation that only MSL1/MSL2 can be found at the entry sites in its absence (Palmer et al., 1994; Gu et al., 1998). Using a point mutation that abolishes the helicase activity of MLE (mleGET), Lee et al. (1997) observed that the mutant protein was able to bind to a reduced number of sites on the X chromosome, as well as to a number of ectopic sites on the autosomes, in mutant males. MSL1 was found to co-localize with MLEGET at a number of the X chromosome sites, the so-called entry sites. As the authors suggested, the MLEGET binding pattern may have been influenced by the moribund state of the mutant male larvae examined. We wished, therefore, to ascertain the role of the mleGET mutation in transgenic females and, thereby, to be able to compare its effect with that of the mof1 mutation. We stained polytene chromosomes of mle1/mle1;mleGET/[H83M2] females for the presence of MSL1 and MSL3 (the mle1mutant allele has a stop codon that truncates the protein after the first 125 amino acids, so that the only MLE present in these females is the mutant MLEGET protein). We observed that the MSL1 and MSL3 proteins (Figure 5) as well as MOF (data not shown) are co-localized at the entry sites. Once again, we used the presence of roX RNA as an indicator of MSL complexes to determine whether free, untargeted complexes are present in these nuclei. Surprisingly, no roX1 RNA was detected in whole salivary gland nuclei by in situ hybridization with roX1 DNA probes (Figure 6A). Since this result is different from those obtained with [H83M2] transgenic females homozygous for the mof1 loss-of-function mutation (Figure 2C), we confirmed the absence of roX1 RNA signal by RT–PCR using roX1-specific primers (Figure 6B). Furthermore, no roX2 RNA was detected either by in situ hybridization in whole salivary glands (Figure 6A) or by RT–PCR (Figure 6B). Thus, it appears that in the absence of the ATP-dependent function of MLE, MSL complexes can be assembled but, once assembled, these complexes no longer contain roX RNA.
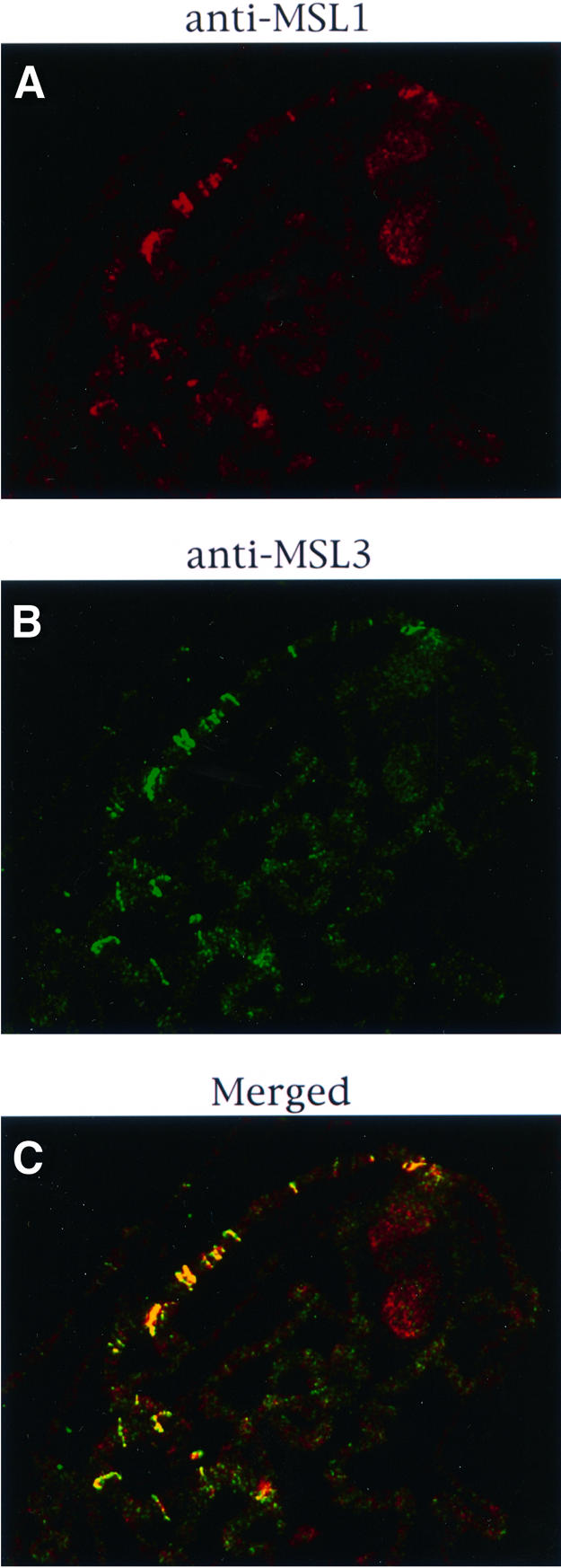
Fig. 5. Effect of the mleGET mutation on the localization of the MSL complex. Polytene chromosomes from mle1/mle1; [mle-GET]/[H83M2] larvae were stained for MSL1 (A) or MSL3 (B). As seen in the merged image (C), these two MSLs are co-localized at the entry sites.
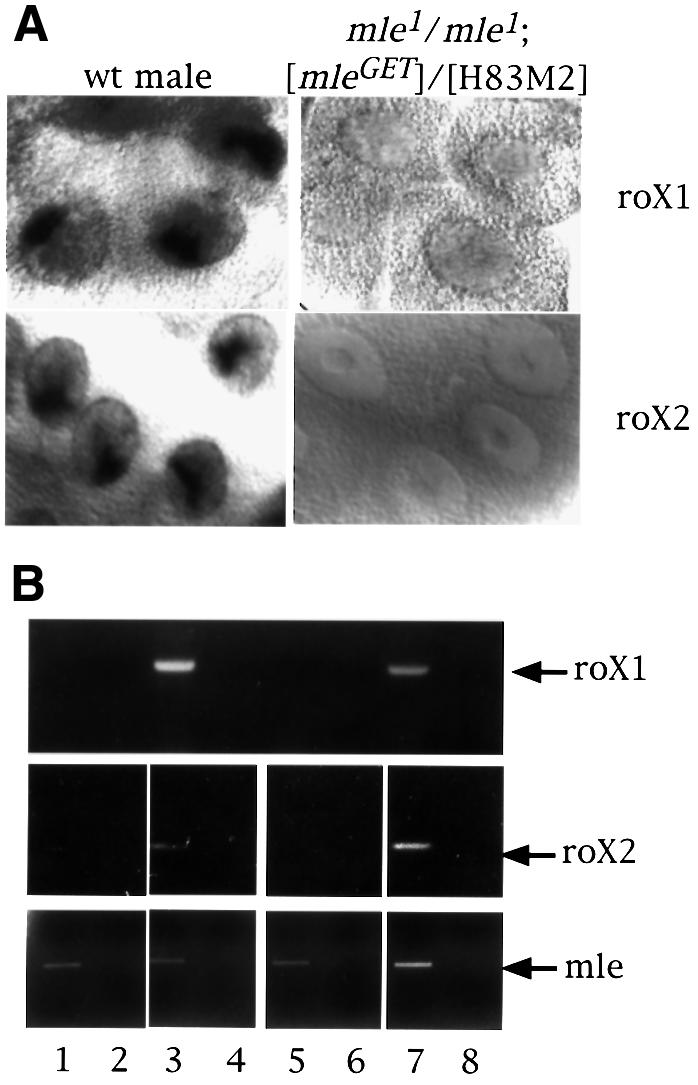
Fig. 6. roX RNA is absent in females that produce only the MLEGET mutant protein. (A) Localization of roX1 and roX2 RNA by in situ hybridization in whole salivary glands from a wild-type male and a mle1/mle1; [mle-GET]/[H83M2] female larva. (B) RT–PCR for roX RNA. Total RNA from adult flies of wild-type females (lanes 1 and 2), wild-type males (lanes 3 and 4), mle1/mle1; [mle-GET]/[H83M2] females (lanes 5 and 6) and mle1/+; [mle-GET]/[H83M2] females (lanes 7 and 8) were reverse transcribed (lanes 1, 3, 5 and 7) or not (lanes 2, 4, 6 and 8) and then amplified for roX1 or roX2 RNA by PCR. The lower panel shows the amplification of MLE RNA by RT–PCR in the same samples.
To determine whether fully or partially formed complexes lacking roX RNA exist free in the nucleoplasm of mle1/mle1;mleGET/[H83M2] females, we stained whole salivary gland nuclei for MSL1 and MSL3 by indirect immunofluorescent labeling (Figure 7A). We also performed co-immunoprecipitation of MSL1 and MSL3 from whole mutant female extracts (Figure 7B). The results of these experiments provided evidence for the existence of MSL complexes in the nucleoplasm of mleGET mutant larvae.
Fig. 7. In the absence of MLE ATPase activity, MSL complexes lacking roX RNA are found in the nucleoplasm. (A) Whole salivary glands from mle1/mle1; [mle-GET] and wild-type male larvae were stained for MSL1 and MSL3 by indirect immunofluorescent labeling. In the nuclei of male larvae expressing only mleGET, the two antigens are co-localized along the X chromosome. In the nuclei of female larvae from mle1/mle1; [mle-GET], although some MSL1 (green) and MSL3 (red) proteins appear to be independent of each other, a substantial amount of these proteins are associated (yellow, in the merged image), leading to the conclusion that the MSL complex exists in the interchromosomal spaces. Note that, as stated in the legend of Figure 5, the complex must also be present at the entry sites on the X chromosomes, although these cannot be resolved in this material. (B) Co-immunoprecipitation of MSL1 and MSL3. Protein extracts prepared from mle1/mle1; [mle-GET] female flies were immuno precipitated with pre-immune (lanes 1 and 3) or MSL1 antisera (lanes 2 and 4). The precipitate was analyzed by western blotting using MSL1 and MSL3 antisera. Note that the lanes presented in this figure were obtained from the gel illustrated in Figure 2, which includes the wild-type male and female controls.
Discussion
It has long been known that eukaryotic transcription is regulated at initiation by a large group of factors that associate with RNA polymerase II, target it to promoter regions with spatial and temporal specificity, and allow it to clear the promoter. Recently, another level of transcriptional regulation, involving specialized multiprotein aggregates that interact with chromatin components to control the rate of transcription, has come to light. At present, these aggregates can be grouped into two broad categories: (i) complexes that use the energy of ATP hydrolysis to alter nucleosomal conformation (SWI/SNF in yeast and in mammals; RSC, Sfh1p in yeast; NURF, CHRAC and ACF in Drosophila); and (ii) complexes that target specific histone acetyltransferases to their site of action and alter chromatin conformation via the acetylation of histones (GCN/ADA, NuA4 and SAGA in yeast). All of these complexes interact with nucleosomal proteins (i.e. histones); they may interact with components of the initiation complex, although they do not activate silent genes, and they enhance the level of transcription of large groups of activated genes (for review see Grant and Berger, 1999; Muchardt and Yaniv, 1999). The dosage compensation complex (or MSL complex) of Drosophila is particularly interesting because it includes both an ATP-dependent helicase (MLE) and a histone acetyltransferase (MOF).
Meller et al. (2000) have shown that the MLE protein is required very early in the process of assembly of the MSL complex at the sites of roX RNA synthesis. MLE is a member of the DEAH/DEAD RNA helicase family (Kuroda et al., 1991; Lee and Hurwitz, 1993; for review see Eisen and Lucchesi, 1998). The recombinant protein has been shown to have nucleic acid binding and DNA/RNA helicase activity, and the MLE/roX RNA interaction may be similar to that demonstrated for putative DEAD-box helicases during ribosome biogenesis in yeast (Daugeron and Linder, 1998; de la Cruz et al., 1998, 1999; Kressler et al., 1998). A point mutation generated by in vitro mutagenesis in the ATP-binding domain (mleGET) abolishes the helicase activity in vitro while the nucleic acid-binding affinity is unaffected (Lee et al., 1997). The mleGET gene product cannot rescue the lethality of the mle-null genotype, indicating that its helicase activity is required for dosage compensation (Lee et al., 1997). Our results in transgenic females that express only the MLEGET protein show that the RNA helicase activity of MLE may not be required for the assembly of MSL complexes. This activity, though, appears to be essential for the stability of the complexes with respect to their ability to retain their roX RNA component. roX RNA is essential for the assembly of the MSL complex, but once that is accomplished it can be lost and the other complex components are presumably held together by protein–protein interactions. This is consistent with the observation of Akhtar et al. (2000) that MSL complexes immunoprecipitated from RNase-treated S2 cell extracts remain assembled in the absence of demonstrable roX2 RNA. Our results suggest that roX RNA-less complexes can access the entry sites but are unable to spread to their sites of action along the X chromosome.
MOF is a member of the MYST family of histone acetyltransferases (Borrow et al., 1996; Reifsnyder et al., 1996; Hilfiker et al., 1997; Neal et al., 2000). In vitro assays with full-length proteins or truncated fragments have shown that the members of this family acetylate histones H2A and H3, but show a strong preference for H4. The specific residues acetylated on histone H4 have been determined for yeast Esa1p (as a recombinant protein or as part of the NuA4 complex), human Tip60 (as a recombinant protein) and Tetrahymena p80. All three of these MYST family members acetylate lysines 5, 8, 12 and 16 (Kimura and Horikoshi, 1998; Smith et al., 1998; Ohba et al., 1999). In contrast, MOF—both as a recombinant protein (Akhtar and Becker, 2000) and as a member of the MSL complex (Smith et al., 2000)—shows a clear, marked preference for Lys16. Since database monitoring has revealed that several MYST family acetyl transferases other than MOF exist in Drosophila, it is likely that some level of acetylation of histone H4 at Lys16 occurs throughout the chromatin of both males and females. The enrichment of the monoacetylated H4Ac16 form along the X chromosome in males, though, is due to the specific targeting of MOF to this chromosome by the MSL complex. In females, MOF is present but is completely dispensable (Hilfiker et al., 1997; Gu et al., 1998), leading to the conclusion that it has no function in this sex or that its function is assumed by some other MYST family member. We wish to point out that available data do not allow us to establish whether the specific acetylation mediated by the MSL complex is responsible per se for the doubling in transcription of X-linked genes or whether it renders the X chromosome more accessible to other factors, such as the JIL-1 kinase (Jin et al., 1999), which are the actual effectors of the enhancement. We also do not know whether components of the complex itself are acetylated by MOF.
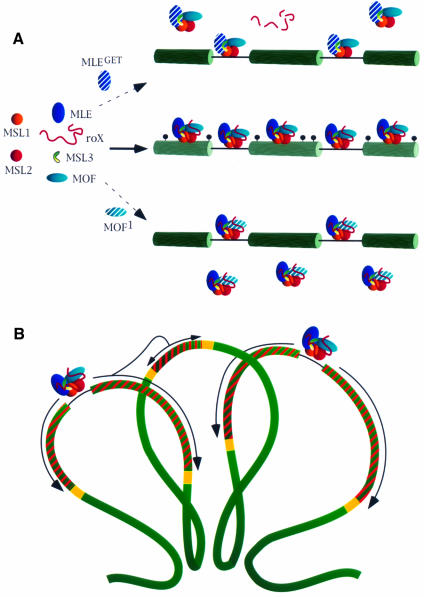
The normal association of the MSL complex at hundreds of sites along the X chromosome appears be a process with at least three major steps (Figure 8A). The first is the formation of functional complexes at the two entry sites where the roX RNAs are transcribed (Kelley et al., 1999). It should be noted that although the MSL1 and MSL2 proteins are able to access the X chromosome at the entry sites and to recruit MLE, further complex assembly can only occur in the presence of the roX RNAs. This contention is supported by the observation that, in the absence of the two roX genes, no complex is seen to form in embryonic stages where it is normally evident (Franke and Baker, 1999). A caveat is that removal of roX2 was accomplished by using a deletion of such size that other roX-like genes or other unidentified components of the complex or genes whose product is required for complex stability, closely linked to roX2, may have been deleted as well. In any event, since the roX RNAs are unstable unless they are associated with the complex (Meller et al., 2000), the process of assembly can proceed only at the entry sites containing the roX genes. Once complexes are formed, they access the X chromosome through all of the entry sites, presumably via the affinity of their MSL1/MSL2 components for these sites. Finally, the complexes spread from the entry sites to the many other sites along the X chromosome where they are normally found. This last step requires the histone acetyltransferase activity of MOF. We suggest that the spreading process involves the acetylation of neighboring nucleosomes, thereby altering the conformation of adjacent chromatin and rendering it more accessible to the entry of additional MSL complexes. The latter may require the presence of acetylated histone H4 tails in order to stabilize their chromatin association. This conclusion is consistent with our observation that, in S2 cells overexpressing MOF, the resulting abnormal ectopic acetylation of histone H4 at Lys16 leads to the association of the MSL complex along autosomal chromatin. This may mirror the normal situation in vivo where complexes, initially attracted to the entry sites, acetylate histone H4 at Lys16 and thereby make adjacent chromatin regions accessible to more complexes. The affinity of the MSL complex for histone H4 tails implied in our model is reminiscent of a similar role played by histone tails in the spreading of complexes containing SIR2, 3 and 4 during the silencing of mating type loci and telomeric heterochromatin formation in yeast (Hecht et al., 1996; Stone and Pillus, 1998). Although critical to the spreading process, the role played by the ATPase function of MLE, either directly or in conjunction with roX RNA, is not sufficiently understood to be incorporated in the model.
Fig. 8. A model for the association and spreading of the MSL complex along the X chromosome. (A) In wild type, the MSL complex assembles at the roX-bearing entry sites, moves to other sites and acetylates histone H4 of neighboring nucleosomes at Lys16 (indicated as black circles). In the absence of the ATPase activity of MLE (MLEGET), the association of the complex with roX RNA is unstable. Free complexes devoid of roX RNA and unable to access the X chromosome are present in the nucleoplasm. If the acetylase function of MOF is impaired by mutation (MOF1), complexes are formed; although they can access other entry sites, they cannot spread along the X chromosome and are found, unbound, in the nucleoplasm. (B) Wild-type MSL complexes present at the entry sites spread along the X chromosome by acetylating nucleosomes. Spreading of the complexes and concomitant acetylation (indicated by the striped regions) may be interrupted by some type of boundary elements (yellow boxes). The looping of the chromatid fiber into rosettes may enable spreading of the complex from a region where acetylation initiated at an entry site to a region where no entry sites are present (from the left to the central loop in the diagram).
We believe that the process just described can provide the following explanations for the gaps in MSL binding that occur along the X chromosome, or at ectopic autosomal sites where the complex has been caused to form at the site of a roX transgene (Kelley et al., 1999). It is possible that the spread of H4 acetylation and complex association may be stopped by some insulator or some as yet uncharacterized boundary elements. This would not necessarily require that the entry sites be entirely responsible for the pattern seen along the X chromosome. As illustrated in Figure 8B, the interphase chromosome is believed to consist of a series of rosettes formed by loops of the chromatid fiber anchored to a central core by dispersed regions that have affinity for one another (Pirrotta, 1997, 1998; Ostashevsky, 1998; Solovjeva et al., 1998; Munkel et al., 1999). In such an arrangement, a cluster of complexes that have been stopped by some boundary element could acetylate the nucleosomes on a neighboring loop, initiating a spreading process on the other side of a gap.
The above considerations raise a number of questions that remain to be resolved. Is the pattern of complex association on the X chromosome tissue specific? Is it dependent on a tissue-specific distribution of the entry sites (other than those containing the roX loci, which must remain invariant in all tissues)? Is the tissue-specific distribution established when the complex first forms in early embryogenesis (Rastelli et al., 1995; Franke et al., 1996; McDowell et al., 1996) and is the pattern perpetuated through the mitotic divisions that give rise to a particular tissue (Lavender et al., 1994)? To answer these questions will require a thorough melding of cytological and biochemical approaches.
Materials and methods
Fly stocks
Flies were reared on standard cornmeal–sugar–yeast–agar medium containing propionic acid and methylparaben as mold inhibitors. The [w+ H83M2] transgene is a P-element construct that contains the msl2 open reading frame under hsp83 promoter control. The insert lacks the SXL-binding sites present in the 5′ and 3′ UTR of msl2 and, therefore, can give rise to the MSL2 protein in transgenic females (Kelley et al., 1995). The [w+ mle-GET] transgene is similar but the mle open reading frame contains a lysine → glutamic acid substitution in the ATP-binding motif GKT (Lee et al., 1997). Females of the w cv mof1/w cv mof1; [w+ H83M2] CyO genotype were generated by crossing w cv mof1/w cv mof1 virgin females to w cv mof1/Y; 18H1 Bc/[w+ H83M2] CyO males. Control y w mof2/+; [w+ H83M2] CyO females were produced by crossing y w/y w virgin females to y w mof2/Y; 18 H1 Bc/[w+ H83M2] CyO males. In these stocks, 18H1 designates the presence of an insert that covers the mof mutation. Larvae were recognized by the lack of black cells and adults by the presence of curly wings. To produce females expressing only the mleget mutant protein, w; pr mle1/w; pr mle1; msl3 [w+ H83M2]/msl3 [w+ H83M2] females were crossed to w; pr mle1/CyO; [w+ mle-GET] males. The resulting w; pr mle1/w; pr mle1; [w+ mle-GET]/msl3 [w+ H83M2] and w; pr mle1/CyO; [w+ mle-GET]/msl3 [w+ H83M2] larvae were distinguished by the absence or presence of the rearranged CyO chromosome in polytene chromosome spreads of salivary glands. A full description of the mutants listed and of the CyO balancer chromosome can be found in FlyBase (http://flybase.bio.indiana.edu).
In situ hybridization of roX RNA
The insert from pZero-2/roX1 DNA, which contains a 1.9 kb roX1 DNA fragment amplified from genomic DNA using primers GTTACGTTC GGAGTGGAAAATGG and GTTTCTTCTGGGTGTAGCTTCTTGG, was used as the probe. The probe for roX2 RNA was a 1.1 kb roX2 DNA fragment amplified from genomic DNA using primers CTCCGA TTGCCTTGCACTCG and AAGTGTCAGTTCTGGTCACCCTGG. Probe was random-prime labeled with digoxigenin using Klenow DNA polymerase (Roche Molecular Biochemicals). In situ hybridization of whole salivary glands was performed according to Meller et al. (1997). In situ hybridization of polytene chromosomes was performed using digoxigenin-labeled antisense roX1 RNA, which was transcribed in vitro with SP6 RNA polymerase (Roche Molecular Biochemicals) using pZero-2/roX1 cut with EcoRV as the template. The hybridization was performed according to Kelly et al. (1999) except that the signal was detected using the TSA Fluorescein System (NEN Life Science Products).
Antisera
Anti-MSL antibodies were raised against various fragments of MSL proteins fused to glutathione S-transferase (GST) as follows: rabbit anti-MSL1 (amino acids 423–1029), guinea pig anti-MSL2 (amino acids 78–529), guinea pig anti-MSL3 (full-length) and rabbit anti-MOF (amino acids 748–827). Secondary antisera were donkey or goat anti-rabbit IgG conjugated with Cy5 or anti-guinea pig IgG conjugated with fluorescein isothiocyanate (FITC; Jackson Immunoresearch Laboratories). For immunofluorescent staining, primary antisera were diluted at 1:200 and secondary antisera at 1:250 in phosphate-buffered saline (PBS; 10 mM NaH2PO4 pH 7.2, 130 mM NaCl), 1.0% bovine serum albumin (BSA) and 0.2% Tween-20.
Immunofluorescent staining
Polytene chromosome spreads were prepared as previously reported (Gu et al., 1998). Chromosome preparations were washed three times with PBS and blocked for 1 h with PBS, 1% BSA, 0.2% Tween-20. Antibody was added and the slides were incubated at 4°C overnight. The slides were then washed with PBS and blocked once again with PBS, 1% BSA, 0.2% Tween-20 and 2% normal goat or donkey serum. Fluorochrome-conjugated secondary antibody was added and the slides maintained at room temperature for 2 h or at 4°C overnight. After extensive washing with PBS, the slides were mounted with Slow-Fade mounting medium (Molecular Probe) and observed with a Bio-Rad confocal microscope.
Whole salivary glands were fixed and permeabilized according to Bone et al. (1994). Immunofluorescence procedures were the same as described above for polytene chromosomes.
Wild-type or transfected S2 cells were seeded on the slides the night before staining. Slides were washed briefly with PBS and fixed in 4% formaldehyde in PBS for 15 min at room temperature. After washing three times with PBS, cells were permeabilized for 5 min at –20°C with pre-cooled acetone. Cells were then washed with PBS and processed for antibody staining as described for polytene chromosomes.
Western blot analysis for MOF
Heads from 20–30 adult flies were homogenized in 100 µl of sample buffer (0.1 M Tris pH 6.8, 4% SDS, 0.2% β-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue) and heated at 100°C for 3 min. After centrifugation, 2.5–10 µl of supernatant were loaded on a 7.5% SDS–polyacrylamide gel. The gel was either stained with Coomassie Blue R-250 to monitor the amount of protein loaded or transferred to a nitrocellulose membrane which was exposed to anti-MOF serum.
Immunoprecipitation
About 100–150 flies were ground in liquid nitrogen and then homogenized 15 times in 1 ml of lysis buffer [50 mM Tris–HCl pH 8.8, 300 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 µg/ml leupeptin, 2 µg/ml antipain, 2 µg/ml aprotinin, 1 mM phenyl methylsulfonyl fluoride (PMSF)]. The homogenate was incubated for 40 min on ice after the addition of 1 ml of lysis buffer. The protein extract was then separated from the debris after centrifugation for 20 min at 4°C. The protein extract was cleared by incubation with 20 µl/ml of protein A-coupled agarose beads and then incubated at 4°C overnight with 4 µl of MSL1 antiserum or 6 µl of MSL1 pre-immune antiserum bound to 10 µl of protein A-coupled agarose beads. After washing the beads five times with the buffer (20 mM HEPES pH 7.2, 10% glycerol, 300 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.2% Tween-20, 1 mM PMSF), immunoprecipitated proteins were eluted from the agarose beads with 60 µl of sample buffer and 20 µl were resolved on a 7.5% SDS–polyacrylamide gel. The gel was transferred to nitrocellulose membrane, which was probed with MSL1 and MSL3 antiserum and then incubated with the corresponding secondary antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories). The secondary antibody was detected using ECL western blotting detection reagents (Amersham–Pharmacia Biotech).
Transfection of S2 cells
The coding regions for MOF and MOF1 were amplified from genomic DNA prepared from wild-type or w cv mof1 females, using primers caaagatatcctcgagTCTGAAGCGGAGCTGGAA and aaaccattcctaggtc gacATGCGATGATAGCAGAACGG (nucleotides in lower case were added as linkers for the purpose of cloning). The amplified DNA was cut with XhoI and NheI, and the vector pMt/HA (a derivative of pMK322) was cut with XhoI and SpeI. The restriction fragments were ligated to generate pMt-MOF and pMt-MOF1, which will express MOF and MOF1, respectively, after induction with CuSO4. S2 cells were transfected as described (Di Nocera and Dawid, 1983). Briefly, 10–15 µg of DNA in 250 µl of 250 mM CaCl2 were added dropwise to 250 µl of 2× HEPES-buffered saline while mixing. The solution was added to S2 cells after 40 min at room temperature. After 24 h, cells were washed with medium and allowed to grow for another 24 h before selection with 200 µg/ml hygromycin. Lines of stably transfected cells were established after continuous selection for ∼2 weeks.
RT–PCR
Total RNA was prepared from 60–80 adult flies using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Total RNA was eluted in 50 µl of H2O and then treated with RNase-free DNase I (Promega). After purification, ∼100 ng of total RNA were reverse transcribed for 1 h at 37°C using Senscript reverse transcriptase (Qiagen) with primer RevoX1 (GGTCACCCTATCAGTAGCAGTACCACAC) (roX1), primer r2CPr3 (GAGCGAGATGACAATAGAGAGG) (roX2) or primer mleP3r (ACAGAGTGTGAAGCAGAAGC) (mle). Amplification of cDNA was performed in a 50 µl reaction with one-tenth of the reverse transcription reaction, 100 µM dNTPs, 1 U of Perfect-Match PCR-Enhancer (Stratagene) and 10 pmol of each forward and reverse primer. The cycling conditions were 94°C for 5 min followed by 30 cycles (roX1) or 25 cycles (roX2 and mle) of 94°C for 1 min, 52°C for 1 min, 72°C for 45 s. The primers were as follows. roX1: RevoX1 (see above) and DiroX1 (CATCGTGCAACAATCCCAAAG); roX2: r2CPr3(see above) and r2CPd2 (GCCATCGAAAGGGTAAATTGG); mle: mleP3r (see above) and mleP3d (CTACTCGGTGCGATTGAG).
Acknowledgments
Acknowledgements
We thank Dr Mitzi Kuroda for generously providing us with the transgenic lines bearing the ectopic msl2 and mleGET alleles under heat-shock promoter control, and Dr Bryan Turner for the gift of the H4Ac16 antiserum. We acknowledge the excellent technical assistance of Hisahiro Tajima and Krista Fehr. Finally, we are grateful to Edwin Smith, Arri Eisen, Karama – and Geogette Sass for many fruitful discussions and comments. This research was supported by grant GM15691 from the National Institutes of Health.
References
- Akhtar A. and Becker,P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 367–375. [DOI] [PubMed] [Google Scholar]
- Akhtar A., Zinc,D. and Becker,P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, in press. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker,B.S. (1995) The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex-specifically regulated by Sex-lethal. Development, 121, 3245–3258. [DOI] [PubMed] [Google Scholar]
- Bone J.R., Lavender,J., Richman,R., Palmer,M.J., Turner,B.M. and Kuroda,M.I. (1994) Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev., 8, 96–104. [DOI] [PubMed] [Google Scholar]
- Borrow J. et al. (1996) The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature Genet., 14, 33–41. [DOI] [PubMed] [Google Scholar]
- Daugeron M.C. and Linder,P. (1998) Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA, 4, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler,D., Tollervey,D. and Linder,P. (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J., 17, 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- Di Nocera P.P. and Dawid,I.B. (1983) Transient expression of genes introduced into cultured cells of Drosophila. Proc. Natl Acad. Sci. USA, 80, 7095–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A. and Lucchesi,J.C. (1998) Unraveling the role of helicases in transcription. BioEssays, 20, 634–641. [DOI] [PubMed] [Google Scholar]
- Franke A. and Baker,B.S. (1999) The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell, 4, 117–122. [DOI] [PubMed] [Google Scholar]
- Franke A., Dernburg,A., Bashaw,G.J. and Baker,B.S. (1996) Evidence that MSL-mediated dosage compensation in Drosophila begins at blastoderm. Development, 122, 2751–2760. [DOI] [PubMed] [Google Scholar]
- Grant P.A. and Berger,S.L. (1999) Histone acetyltransferase complexes. Semin. Cell Dev. Biol., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- Gu W., Szauter,P. and Lucchesi,J.C. (1998) Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet., 22, 56–64. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger,S. and Grunstein,M. (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature, 383, 92–96. [DOI] [PubMed] [Google Scholar]
- Hilfiker A., Hilfiker-Kleiner,D., Pannuti,A. and Lucchesi,J.C. (1997) mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J., 16, 2054–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Wang,Y., Walker,D.L., Dong,H., Conley,C., Johansen,J. and Johansen,K.M. (1999) JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell, 4, 129–135. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Solovyeva,I., Lyman,L.M., Richman,R., Solovyev,V. and Kuroda,M.I. (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell, 81, 867–877. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Meller,V.H., Gordadze,P.R., Roman,G., Davis,R.L. and Kuroda,M.I. (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell, 98, 513–522. [DOI] [PubMed] [Google Scholar]
- Kimura A. and Horikoshi,M. (1998) Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells, 3, 789–800. [DOI] [PubMed] [Google Scholar]
- Kressler D., de la Cruz,J., Rojo,M. and Linder,P. (1998) Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae.Mol. Cell. Biol., 18, 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M.I., Kernan,M.J., Kreber,R., Ganetzky,B. and Baker,B.S. (1991) The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell, 66, 935–947. [DOI] [PubMed] [Google Scholar]
- Lavender J.S., Birley,A.J., Palmer,M.J., Kuroda,M.I. and Turner,B.M. (1994) Histone H4 acetylated at lysine 16 and proteins of the Drosophila dosage compensation pathway co-localize on the male X chromosome through mitosis. Chromosome Res., 2, 398–404. [DOI] [PubMed] [Google Scholar]
- Lee C.G. and Hurwitz,J. (1993) Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem., 268, 16822–16830. [PubMed] [Google Scholar]
- Lee C.G., Chang,K.A., Kuroda,M.I. and Hurwitz,J. (1997) The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J., 16, 2671–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J.C. (1998) Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev., 8, 179–184. [DOI] [PubMed] [Google Scholar]
- Lyman L.M., Copps,K., Rastelli,L., Kelley,R.L. and Kuroda,M.I. (1997) Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics, 147, 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell K.A., Hilfiker,A. and Lucchesi,J.C. (1996) Dosage compensation in Drosophila: the X chromosome binding of MSL-1 and MSL-2 in female embryos is prevented by the early expression of the Sxl gene. Mech. Dev., 57, 113–119. [DOI] [PubMed] [Google Scholar]
- Meller V.H., Wu,K.H., Roman,G., Kuroda,M.I. and Davis,R.L. (1997) roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell, 88, 445–457. [DOI] [PubMed] [Google Scholar]
- Meller V.H., Gordadze,P.R., Park,Y., Chu,X., Stuckenholz,C., Kelley,R.L. and Kuroda,M.I. (2000) Ordered assembly of roX RNAs into MSL complexes on the dosage compensated X chromosome in Drosophila. Curr. Biol., 10, 136–143. [DOI] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv,M. (1999) ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol., 293, 187–198. [DOI] [PubMed] [Google Scholar]
- Munkel C., Eils,R., Dietzel,S., Zink,D., Mehring,C., Wedemann,G., Cremer,T. and Langowski,J. (1999) Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol., 285, 1053–1065. [DOI] [PubMed] [Google Scholar]
- Neal K.C., Pannuti,A., Smith,E.R. and Lucchesi,J.C. (2000) A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim. Biophys. Acta, 1490, 170–174. [DOI] [PubMed] [Google Scholar]
- Ohba R., Steger,D.J., Brownell,J.E., Mizzen,C.A., Cook,R.G., Cote,J., Workman,J.L. and Allis,C.D. (1999) A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol. Cell. Biol., 19, 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostashevsky J. (1998) A polymer model for the structural organization of chromatin loops and minibands in interphase chromosomes. Mol. Biol. Cell, 9, 3031–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer M.J., Richman,R., Richter,L. and Kuroda,M.I. (1994) Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev., 8, 698–706. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1997) PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev., 7, 249–258. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1998) Polycombing the genome: PcG, trxG and chromatin silencing. Cell, 93, 333–336. [DOI] [PubMed] [Google Scholar]
- Rastelli L., Richman,R. and Kuroda,M.I. (1995) The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech. Dev., 53, 223–233. [DOI] [PubMed] [Google Scholar]
- Reifsnyder C., Lowell,J., Clarke,A. and Pillus,L. (1996) Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nature Genet., 14, 42–49. [Erratum: Nature Genet., 16, 109.] [DOI] [PubMed] [Google Scholar]
- Smith E.R., Eisen,A., Gu,W., Sattah,M., Pannuti,A., Zhou,J., Cook,R.G., Lucchesi,J.C. and Allis,C.D. (1998) ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl Acad. Sci. USA, 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Pannuti,A., Gu,W., Steurnagel,A., Cook,R.G., Allis,C.D. and Lucchesi,J.C. (2000) The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol., 20, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovjeva L., Svetlova,M., Stein,G., Chagin,V., Rozanov,Y., Zannis-Hadjopoulos,M., Price,G. and Tomilin,N. (1998) Conformation of replicated segments of chromosome fibres in human S-phase nucleus. Chromosome Res., 6, 595–602. [DOI] [PubMed] [Google Scholar]
- Stone E.M. and Pillus,L. (1998) Silent chromatin in yeast: an orchestrated medley featuring Sir3p. BioEssays, 20, 30–40. [DOI] [PubMed] [Google Scholar]
- Stuckenholz C., Kageyama,Y. and Kuroda,M.I. (1999) Guilt by association: non-coding RNAs, chromosome-specific proteins and dosage compensation in Drosophila.Trends Genet., 15, 454–458. [DOI] [PubMed] [Google Scholar]
- Turner B.M. (1998) Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol. Life Sci., 54, 2131. [DOI] [PMC free article] [PubMed] [Google Scholar]