Abstract
The interaction between fMet-tRNAfMet and Bacillus stearothermophilus translation initiation factor IF2 has been characterized. We demonstrate that essentially all thermodynamic determinants governing the stability and the specificity of this interaction are localized within the acceptor hexanucleotide fMet-3′ACCAAC of the initiator tRNA and a fairly small area at the surface of the β-barrel structure of the 90-amino acid C-terminal domain of IF2 (IF2 C-2). A weak but specific interaction between IF2 C-2 and formyl-methionyl was also demonstrated. The surface of IF2 C-2 interacting with fMet-tRNAfMet has been mapped using two independent approaches, site- directed mutagenesis and NMR spectroscopy, which yielded consistent results. The binding site comprises C668 and G715 located in a groove accommodating the methionyl side-chain, R700, in the vicinity of the formyl group, Y701 and K702 close to the acyl bond between fMet and tRNAfMet, and the surface lined with residues K702-S660, along which the acceptor arm of the initiator tRNA spans in the direction 3′ to 5′.
Keywords: NMR spectroscopy/protein–RNA interaction/site-directed mutagenesis/translation initiation
Introduction
The specific recognition of fMet-tRNAfMet by initiation factor IF2 represents one of the most important interactions occurring during translation initiation in bacteria (for reviews see Spurio et al., 1993; Schmitt et al., 1996; Gualerzi et al., 2000). This interaction determines the accuracy in the selection of the correct initiation site of both leadered (containing a 5′-UTR) and unleadered mRNAs (Grill et al., 2000 and references therein), in the speed and efficiency of both 30S and 70S initiation complex formation (Gualerzi et al., 1986) and in the formation of the first peptide bond (initiation dipeptide), which marks the transition from the initiation to the elongation phase of translation (La Teana et al., 1996; Tomšic et al., 2000).
Important progress in the elucidation of the IF2 structure was made recently. The molecular dissection of Bacillus stearothermophilus IF2 (82 kDa) allowed the identification of three domains in the molecule (the N-terminal domain, the central G-domain and the carboxyl-terminal C-domain), the site responsible for the recognition and binding of fMet-tRNAfMet being located in the 24.5 kDa C-terminal part of the protein (IF2 C) (Gualerzi et al., 1991). Further studies have shown that IF2 C is constituted by two domains of approximately equal size (IF2 C-1 and IF2 C-2) (Misselwitz et al., 1997). IF2 C-2, of 110 residues, was found to contain all the structural determinants involved in the recognition of fMet-tRNAfMet, and its complex with fMet-tRNAfMet displayed the same stability and properties as those formed by intact IF2 and IF2 C (Krafft et al., 2000; Spurio et al., 2000). The three dimensional (3D) solution structure of B.stearothermophilus IF2 C-2 determined by multinuclear NMR spectroscopy consists of a compact β-barrel, structurally homologous to domains II of elongation factors EF-Tu and EF-G, despite the lack of any significant sequence homology (Meunier et al., 2000). Based on this similarity, a recognition site for fMet-tRNAfMet was proposed. The goal of the present study is to characterize this interaction experimentally.
Chemical shifts are sensitive to the environment (Ramsey, 1950) and this property can be used to study even weak interactions between molecules (Wüthrich, 2000). The amide proton and nitrogen atoms are especially useful in localizing the region of a molecule involved in ligand binding and methods based on this property were proposed to design high affinity ligands for proteins (Shuker et al., 1996) or characterize biomolecular interactions in association with other experimental approaches (Foster et al., 1998; Takahashi et al., 2000). Similarly, we have used NMR spectral changes, in combination with site-directed mutagenesis, to characterize the binding site of IF2 to fMet-tRNAfMet.
Our data demonstrate that IF2 C-2 can form a stable complex with the aminoacylated and formylated acceptor arm of the initiator tRNA (fMet-3′ACCAAC) and that it interacts specifically, albeit with low affinity, with N-formyl-methionine. Titrations of IF2 C-2 with ligands of increasing complexity (from formyl-methionine to fMet-tRNAfMet), monitored by NMR spectroscopy and inactivation of the fMet-tRNA binding of the protein following site-directed mutagenesis, yielded consistent results, allowing the identification of the site of IF2 involved in the interaction with the initiator tRNA.
Results
Identification of two clusters of amino acids involved in fMet-tRNAfMet binding to IF2 within IF2 C-2
Earlier studies highlighted the importance of particular residues (C668, C714, K699 and R700) of IF2 C-2 for its binding to the initiator tRNA. Evidence was presented to demonstrate the role of the cysteines while attempting to introduce fluorescent tags: most replacements of C714 and some of C668 caused severe alteration of the fMet-tRNAfMet binding capacity of the protein (Misselwitz et al., 1999). Involvement of the two other amino acids was revealed by the suppression of a dominant lethal mutant in the GTP/GDP binding site of B.stearothermophilus IF2 due to the loss of the fMet-tRNAfMet binding of the factor following the double mutation K699N-R700G (Gualerzi et al., 2000; J.Tomšic, A.Smorlesi, C.L.Pon and C.O.Gualerzi, manuscript in preparation).
To characterize further the environment of the cysteine residues and their role in fMet-tRNAfMet binding and to determine whether the inactivation of the K699N-R700G double mutant was due to the substitution of K699, R700 or both, a large number of amino acid substitutions were introduced in these regions of the molecule. In addition to several mutants with wild-type activity, these experiments led to the identification of two clusters of four (K699-R700-Y701-K702) and three (E713-C714-G715) consecutive amino acid residues important or essential for the interaction of IF2 with fMet-tRNAfMet.
Residual fMet-tRNAfMet binding of the mutants was measured by their capacity to protect the initiator tRNA from spontaneous hydrolysis. Results are presented in Figure 1. Aside from some conservative substitutions (e.g. K699R, Y701F, K702R, E713D) and the E713A mutant, all other mutations strongly reduced the fMet-tRNAfMet binding capacity of the protein. Particularly drastic are the effects of the G715X mutations: all seven substitutions introduced at this position completely abolished the fMet-tRNAfMet binding of IF2. Inactivation due to replacement of C714 by Val, Asp, Arg, Lys or Glu is also severe, while substantial binding activity remains for the C714S and C714Y mutants (Misselwitz et al., 1999).
Fig. 1. Effect of amino acid replacements on the fMet-tRNAfMet binding of IF2. fMet-tRNAfMet binding of IF2 mutants was evaluated by measuring the protection of the tRNA from deaminoacylation as a function of the protein concentration. Protection levels are normalized to the value obtained using wild-type IF2 under the same conditions. Mutagenized residues are (A) K699, (B) R700, (C) Y701, (D) K702, (E) E713 and (F) G715, and the amino acids by which they were replaced are indicated in each panel. Additional experimental details are provided in Materials and methods.
Localization of the amino acids involved in fMet-tRNAfMet binding within the 3D structure of IF2 C-2
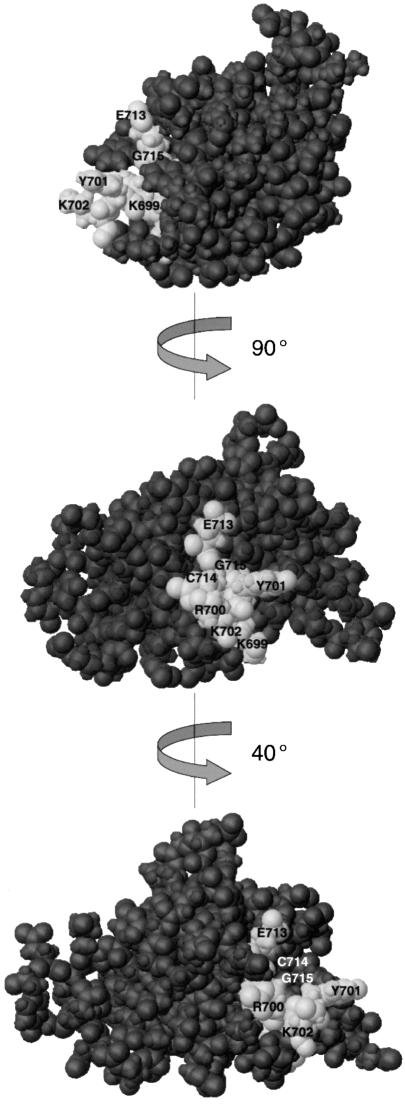
The seven amino acids affecting the interaction of IF2 with fMet-tRNAfMet are localized within the 110 residue C-terminal domain of IF2 (IF2 C-2). None of the many other mutations, site-directed or generated randomly, within the N-terminal domain of IF2 C were found to alter the initiator tRNA binding (data not shown), consistent with the finding that isolated IF2 C-2 binds fMet-tRNAfMet with specificity and affinity equal to that of native IF2 (Spurio et al., 2000). Figure 2 displays a space filling representation of the 3D structure of IF2 C-2 (Meunier et al., 2000) in which the identified residues are colored using a lighter shade: they constitute a continuous, well defined area at the surface of the protein, forming a shallow cleft which could accommodate the acceptor end of the initiator tRNA.
Fig. 2. Location on IF2 C-2 structure of the amino acid residues involved in the interaction with fMet-tRNAfMet, as identified by mutagenesis. Space filling representation of the 3D structure of IF2 C-2 (Meunier et al., 2000); rotations of 90° and 40° were operated about the y-axis. The amino acid residues identified by site-directed mutagenesis as being involved in the interaction with fMet-tRNAfMet are colored with a lighter shade. This figure, as well as Figure 5B, D and F, were generated using the program MOLMOL (Koradi et al., 1996).
The acceptor end of fMet-tRNAfMet forms a stable interaction with IF2 C-2
Spurio et al. (2000) showed that IF2 C-2 could be trimmed at the N- and C-termini to yield a shortened form of the domain (90 versus 110 amino acids), which corresponds to the structured part of the molecule (Meunier et al., 2000) and can be considered as the minimal size polypeptide still retaining full biological function since it binds fMet-tRNAfMet with properties equal to those of intact IF2.
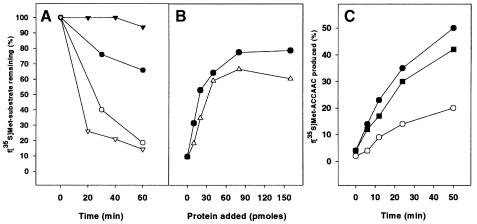
Conceptually similar experiments were carried out to identify the minimal fragment of the initiator tRNA still capable of interacting with IF2. Figure 3 shows that the acceptor end of the initiator tRNA (fMet-3′ACCAAC) is protected by IF2 C-2 from spontaneous hydrolysis with almost the same kinetic (Figure 3A) and protein concentration dependence (Figure 3B) as full-length fMet-tRNAfMet. The dissociation constant of the IF2 C-2–fMet-3′ACCAAC complex is ∼2-fold higher (Kd ≈ 1.8 µM as estimated from the results of Figure 3B) than the 0.9 µM value estimated for the complexes formed by the intact fMet-tRNAfMet and either IF2 C or IF2 C-2 (Krafft et al., 2000; Spurio et al., 2000). Additional experiments demonstrate that fMet-3′ACCAAC displays the same affinity for IF2 C-2 and IF2 C (data not shown).
Fig. 3. Interaction of the acceptor end of fMet-tRNAfMet with IF2 C-2. (A) Time course of the hydrolysis of 10 pmol of intact f[35S]Met-tRNAfMet (inverted triangles) and f[35S]Met-3′ACCAAC (circles) in the absence (open symbols) and in the presence (filled symbols) of 120 pmol of IF2 C-2. (B) Protection from hydrolysis of intact f[35S]Met-tRNAfMet (circles) and f[35S]Met-ACCAAC (triangles) as a function of the amount of IF2 C-2. The reaction was carried out at 37°C for 70 min. (C) Time course of f[35S]Met-ACCAAC production by RNase T1 cleavage of the G70–C71 diester bond in the presence of IF2 C-2 (filled circles), IF2 C (filled squares) and in the absence of protein (open circles). Additional experimental details are given in Materials and methods.
To characterize further the interaction between IF2 C-2 and the acceptor stem of fMet-tRNAfMet, RNase T1 digestion was performed in the presence and in the absence of IF2 C-2. The time course of fMet-3′ACCAAC production from the intact fMet-tRNAfMet indicates that the phosphodiester bond 5′G70–C71 is not protected from RNase T1 cleavage in the presence of IF2 C-2 (Figure 3C); on the contrary, the amount of 3′ acceptor fragment produced is significantly increased in the presence of the protein (or equivalent amounts of IF2 C). This suggests that the interaction of fMet-tRNAfMet with these proteins destabilizes the terminal base pairs (G2-C71,C3-G70) of the initiator tRNA. An interaction between IF2 C and the 5′ end (e.g. with the 5′ phosphate) of fMet-tRNAfMet could be responsible for destabilizing the acceptor stem duplex which, in turn, may favor the formation of the first peptide bond; the same interaction could contribute somewhat to the thermodynamic stability of the complex and thereby account for the slightly higher affinity (2- to 3-fold) displayed by IF2 C-2 for intact fMet-tRNAfMet compared with the fMet-3′ACCAAC acceptor hexanucleotide (see Discussion).
The present results expand on previous reports (Spurio et al., 2000) by demonstrating that essentially all thermodynamic determinants governing the stability and the specificity of the fMet-tRNAfMet–IF2 complex are localized within two fairly small fragments: the 90-amino acid IF2 C-2 and the fMet-3′ACCAAC fragment of the initiator tRNA.
Mapping the fMet-tRNAfMet binding site of IF2 C-2 by NMR spectroscopy
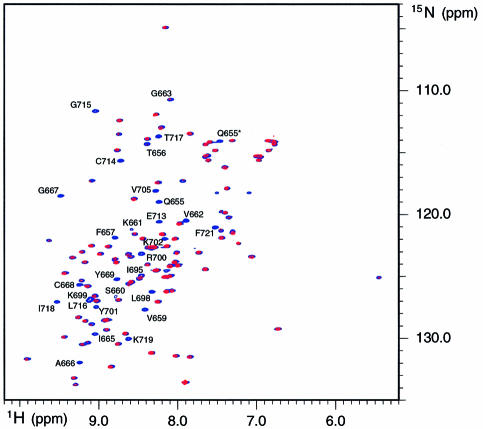
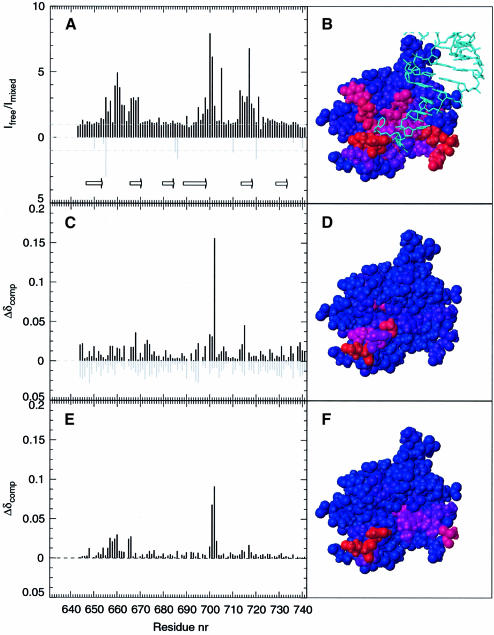
Comparison of two dimensional (2D) NMR spectra of IF2 C-2 recorded in the presence and in the absence of fMet-tRNAfMet (Figure 4) does not reveal any noticeable chemical shift changes. However the intensity of specific correlation peaks is clearly affected by the presence of fMet-tRNAfMet; the ratio of intensities measured in the presence and in the absence of tRNA are displayed in Figure 5A as a function of the protein sequence. The strongest intensity decreases are observed for residues that are located in the β1–β2 loop (Q655, T656, F657, V659, S660, K661, V662, G663, I665), in the β2 strand (A666, G667, C668, Y669), at the edge of the β4 strand, at the beginning of the β4–β5 loop (I695, L698, K699, R700, Y701, K702, V705) and in the β5 strand (E713, C714, Q715, L716, T717, I718, N719 and F721), and that constitute a rather continuous region at the surface of the protein (Figure 5B). Since two structural elements of this interacting surface, namely the β1–β2 and β4–β5 loops, belong to a disordered and flexible part of the protein in solution (Meunier et al., 2000), they could undergo a conformational change upon binding to fMet-tRNAfMet. The observed intensity decreases upon interaction with fMet-tRNAfMet can then be due either to intermediate exchange broadening caused by dynamic events at the protein–RNA interface or to an equilibrium with free protein.
Fig. 4. Overlay of the 1H/15N HSQC spectra of IF2 C-2 in the presence and in the absence of fMet-tRNAfMet. The spectra recorded in the presence and in the absence of fMet-tRNAfMet are colored in blue and red, respectively. The weakened correlation peaks are labeled according to the corresponding residue number. Side-chains are indicated by an asterisk (*).
Fig. 5. Titration of IF2-C2 with fMet-tRNAfMet, N-formyl-methionine and dACCAAC. (A) Effect of the addition of fMet-tRNAfMet on the intensity of the 1H/15N HSQC peaks for the backbone (black bars) and side chains (gray bars) of IF2 C-2 displayed as a function of the protein primary sequence. Arrows indicate the limits of the six β strands as described in Meunier et al. (2000). (C) Chemical shift variations observed upon addition of 100 mM N-formyl-methionine (black bars) or methionine (gray bars). The value of Δδcomp is defined in the text. (E) Chemical shift variations observed upon addition of 2 mM dACCAAC. (B), (D) and (F) display a space filling representation of IF2 C-2 3D structure (Meunier et al., 2000) color coded from blue to red according to the intensity of the variation observed in (A), (C) and (E), respectively. (B) also displays the acceptor arm of yeast Phe-tRNAPhe as observed in its complex with T.aquaticus EF-Tu (Nissen et al., 1995) after superimposing IF2 C-2 with domain II of EF-Tu as indicated in Meunier et al. (2000).
To identify the residues of IF2 C-2 in contact with different parts of the initiator tRNA, we titrated this protein with the initiator amino acid in its formylated (N-formyl-methionine) and unformylated (methionine) forms, as well as with a DNA hexanucleotide (dACC AAC) mimicking the 3′ acceptor arm of the initiator tRNA. Solubility limits restricted the amino acid concentrations to ∼100 mM, while the hexanucleotide was added up to 2 mM.
Figure 5C shows extremely limited and relatively uniform chemical shift variations upon addition of up to 100 mM methionine (Δδcomp <0.025). The absence of any specific interaction is consistent with the moderate influence played by the nature of the amino acid loaded on the initiator tRNA on its affinity for IF2, methionine being replaceable by valine or glutamine (Wu and RajBhandary, 1997). In contrast, formylation plays a major role in the specificity and affinity of the interaction between fMet-tRNAfMet and IF2 (RajBhandary and Chow, 1986; Gualerzi and Pon, 1990) and significant chemical shift displacements are observed upon titration with N-formyl-methionine. The residues most strongly affected by this ligand are K702 (Δδcomp = 0.16) and G715 (Δδcomp = 0.045) as well as C668, R700 and Y701, although at a lower level (Δδcomp ∼0.035). Altogether these residues form a continuous patch at the surface of the protein, defining an area that is contained within the fMet-tRNAfMet binding site of IF2 C-2 (Figure 5D). An interaction between IF2 and N-formyl-methionine had never been reported before; this is not surprising since we estimate the Kd to be in the 100 mM range. Nevertheless, the observed interaction is very specific, as judged from the comparison with the results obtained with methionine, from the evidence that the interacting residues are also sensitive to the presence of fMet-tRNAfMet (Figure 5A and B) and from the fact that the same amino acids were identified independently by site-directed mutagenesis as being involved in IF2–fMet-tRNAfMet interaction (Figures 1 and 2). Since IF2 can also protect the acyl bond of NacPhe-tRNA from hydrolysis (Spurio et al., 2000), we conducted titrations using phenylalanine and N-acetyl-phenylalanine as well. It was found that only the α-amino blocked amino acid affects the chemical shifts of residues in the same region of IF2 C-2 as fMet (data not shown).
Chemical shift variations are also observed upon titration with 3′dACCAAC (Figure 5E). The Kd of the complex between IF2 C-2 and the DNA hexanucleotide was estimated to be ∼10 mM, although the complete titration curve was not obtained. The IF2 residues involved in the interaction with the hexanucleotide belong to the β1–β2 loop (F657, K658, V659, S660), to the β2 strand (I665, A666) and to the β4–β5 loop (Y701, K702, D703), the largest chemical shift changes being observed for Y701 and K702. These residues are also clustered at the surface of the protein, in an area contained within the surface of IF2 C-2 implicated in the binding to fMet-tRNAfMet (Figure 5F) but only partly overlapping (residues Y701 and K702) with the region interacting with N-formyl-methionine.
Discussion
It has recently been shown that the site of IF2 responsible for its interaction with fMet-tRNAfMet is entirely located within the C-terminal 90 amino acid portion of the protein (Spurio et al., 2000), and the 3D structure of this domain (IF2 C-2) was determined using multidimensional NMR spectroscopy (Meunier et al., 2000). The present study contributes further to our knowledge of the fMet-tRNAfMet–IF2 interaction with the demonstration that this interaction involves almost exclusively the acceptor hexanucleotide fMet-3′ACCAAC of the initiator tRNA, whose affinity for IF2 is no more than two to three times lower than that displayed by intact fMet-tRNAfMet. Furthermore, taking advantage of the sensitivity of the NMR spectra to changes of the environment by which even weak interactions can be monitored, we have shown that specific, albeit feeble interactions, occur between IF2 C-2 and N-formyl-methionine, and between IF2 C-2 and 3′dACCAAC. Titrations of IF2 C-2 with these two ligands, monitored by chemical shift differences of individual amino acid residues and comparison of the HSQC spectra of the protein recorded in the presence and in the absence of fMet-tRNAfMet, also allowed us to map the initiator tRNA binding site of IF2 at the level of the individual amino acid residues. This spectroscopic approach provided results remarkably consistent with the parallel study involving extensive random and site-directed mutagenesis of infB.
Three groups of amino acids directly implicated or at least selectively affected upon binding of IF2 to fMet-tRNAfMet (Figure 5A and B) were identified by NMR. These residues belong to specific structural elements of the protein, namely the β1–β2 loop, the β2 and β5 strands, the edge of the β4 strand and the beginning of the β4–β5 loop. Among these amino acids, seven, grouped in two clusters, were also shown by site-directed mutagenesis: K699, R700, Y701, K702 (β4–β5 loop) and E713, C714, Q715 (β5 strand). Concerning the first group of residues, our results suggest that the positive charges of K699 and K702 (both functionally replaceable by Arg) are necessary for fMet-tRNAfMet binding while R700, which can be partly replaced by Lys, Gln, Asn and Thr, could act as an H-bond donor. Finally, the residual activity displayed by the Y701F mutant hints at an important role of the aromatic ring or of the hydrophobic properties of Y701. Interpretation of the mutagenesis data for residues belonging to the second cluster (E713, C714 and G715) is somewhat less straightforward. Although the NMR data clearly indicates the involvement of these residues in the binding to fMet-tRNAfMet, the binding loss observed following their substitutions could result from a structural alteration of the β strand. It is noteworthy, however, that none of the seven G715X mutants displayed any detectable binding to fMet-tRNAfMet; this may arise from the steric encumbrance imposed by larger side-chains.
Three additional residues shown by NMR spectroscopy, C668, F657 and F721, had previously been subjected to mutagenesis (Misselwitz et al., 1999; Spurio et al., 2000). Substitutions of C668 with Asp, Tyr and Arg severely reduced fMet-tRNAfMet binding (Misselwitz et al., 1999), while the conservative F→W substitution at positions 657 and 721 had different consequences: the F721W mutant displayed a reduced affinity for fMet-tRNAfMet (3-fold) unlike the F657W mutant which retained wild-type activity (Spurio et al., 2000).
Meunier et al. (2000) have recently reported a striking structural similarity between IF2 C-2 and domain II of EF-Tu and have proposed that IF2 may interact with the initiator tRNA in a manner similar to that by which EF-Tu interacts with the elongator tRNAs. The present results support this premise, at least to some extent. After aligning the 3D structures of domain II of EF-Tu and of IF2 C-2, we have drawn the acceptor stem of Phe-tRNAPhe as observed in its complex with Thermus aquaticus EF-Tu (Nissen et al., 1995) above the structure of IF2 C-2 (Figure 5B). In this way the phenylalanine is situated above the IF2 residues found to be affected by the addition of N-formyl-methionine while the acceptor stem of the tRNAPhe is in close proximity to the IF2 residues affected by the DNA hexanucleotide sharing the sequence of the acceptor arm of tRNAfMet. The similarity between IF2 C-2 and EF-Tu in their interactions with their respective ligands is somewhat less satisfactory in other parts of the molecule, however. In fact, the model proposed in Figure 5B predicts that other residues of IF2 should also be affected by fMet-tRNAfMet. In particular, while K725 and E726 (corresponding to R295 and E299 of EF-Tu) should be very close to the 5′ end of tRNAPhe (Figure 5B), the NMR data (Figure 5A) do not support their involvement in the interaction with fMet-tRNAfMet. This could be taken to mean that unlike EF-Tu, which forms a salt bridge with the 5′ end of tRNAPhe via R300, IF2 does not interact with the 5′ end of initiator tRNA. Nevertheless, a weak interaction between IF2 C-2 and the 5′ end of the initiator tRNA is likely in light of the increased accessibility of the G70–C71 diester bond to RNase T1 induced by IF2 C-2 (Figure 3C) and of the somewhat higher affinity displayed by IF2 C-2 for the intact tRNA compared with its 3′ acceptor fragment (Figure 3B). In conclusion, our data suggest that the interaction of the initiator tRNA with IF2 C-2, although overall similar to that observed in the aminoacyl-tRNA–EF-Tu complexes, might involve a somewhat different orientation of the acceptor arms of the two tRNAs. Indeed, a rotation of approximately –45° about the z-axis of the acceptor arm of fMet-tRNA centered on fMet would account more satisfactorily for our NMR results (Figure 5B). As to the reason for the different orientations, the participation of the three domains of EF-Tu in aminoacyl-tRNA binding is likely to impose restraints on the orientation of the acceptor end on domain II; on the contrary, these restraints are not present in IF2 whose molecular determinants for the interaction with fMet-tRNAfMet are all confined within IF2 C-2 (Spurio et al., 2000).
Finally, we should also point out that while the conformation of the bound state of IF2 C-2 remains unknown, the identified interacting surface of the domain comprises the most flexible parts of the protein (the β1–β2 and β4–β5 loops) and that structural changes or restricted flexibility may be imposed by the interaction. However, such conformational changes can only be of restricted magnitude (Krafft et al., 2000) due to the compactness of the β-barrel structure of the domain.
Materials and methods
Introduction of unique restriction sites at the 3 ′ end region of infB
Three independent modules that can be easily moved in and out of both mutagenesis and expression vectors were used as targets for localized mutagenesis within the sequence encoding IF2 C-2. These modules were generated by oligonucleotide-directed mutagenesis to create three unique restriction sites within the desired DNA sequence. The wild-type amino acid sequence of the protein was preserved choosing restriction sequences corresponding to silent mutations. Oligonucleotides used for this purpose were: 5′dATGAGTGCGGGTTAACGATCAAAA (HpaI site at L716), 5′dCCGGCTGCTACGTAACCGACGGC (SnaBI site at V670) and 5′dTTGCAAAAAATTGATGTCGAA (the C→T transition results in the loss of the ClaI site at D565, thus leaving a single ClaI site within IF2 C at D696). Base changes are indicated in bold and the newly created restriction sites are underlined. The B.stearothermophilus infB gene containing the above sequence modifications was named B.st infB H-S-C and the corresponding expression vector pIM401 H-S-C.
Site-directed mutagenesis
Substitution of individual residues within the sequence of IF2 C-2 was carried out by site-directed mutagenesis as described previously (Spurio et al., 2000). The mutations giving rise to the amino acid changes listed below were introduced in the corresponding DNA modules with appropriately designed mutagenic oligonucleotides (not indicated here): K699→N, I, R, T, L, Q, G; R700→G, Q, L, T, N, K; Y701→F, N, R, D; K702→G, Q, R, I, D, E; E713→A, D, K; C714→Y, R, S, K, D, E; G715→E, V, Y, K, D, P, Q. Introduction of the desired mutations was confirmed by DNA sequencing and the DNA modules were transferred into the expression vector to overproduce the mutant proteins, which were subsequently purified and tested for their fMet-tRNAfMet binding capacity (Spurio et al., 2000).
Preparation of f[35S]Met-3 ′ACCAAC fragment and analysis of the IF2–tRNA interaction
Three hundred picomoles of f[35S]Met-tRNAfMet (2500 c.p.m./pmol) were digested with 30 U of RNase T1 for 2 h at 37°C in 60 µl of a buffer containing 20 mM imidazole–HCl, 50 mM NaCl and 1 mM β-mercaptoethanol at pH 7.5. After the RNase T1 digestion, the tRNA was used directly in a standard protection assay (Misselwitz et al., 1999); the amount of intact f[35S]Met-tRNAfMet and f[35S]Met-3′ACCAAC fragment remaining after Tris–HCl catalyzed hydrolysis were resolved on 18% polyacrylamide-urea gel run in a 20 mM MOPS–NaOH buffer at pH 7.0 and quantified by Molecular Imager.
The time course of the RNase T1 digestion of f[35S]Met-tRNAfMet in the presence or in the absence of IF2 fragments was carried out at 37°C in 35 µl of a buffer containing 20 mM imidazole–HCl pH 7.5, 50 mM NaCl, 70 mM NH4Cl, 3% glycerol, 1 mM β-mercaptoethanol, 150 pmol f[35S]Met-tRNAfMet, 6.5 U of RNase T1 (Sigma) and, when indicated, 2 nmol of either IF2 C or IF2 C-2. This time course was monitored loading 5 µl aliquots on 18% polyacrylamide-urea gel as described above.
NMR spectroscopy and titration experiments
NMR experiments were carried out on Bruker AMXT/DRX 600 spectrometers operating at 600 MHz 1H frequency. The temperature was set to 312 K. The protein samples used to study the fMet-tRNAfMet interaction were composed of uniformly 15N-labeled IF2 C-2 dissolved in a 90% H2O/10% D2O solution containing 200 mM KCl and either 50 mM deuterated sodium acetate pH 5.5 or, for the titration experiments, 20 mM KPi (pH 5.2). For these experiments, stock solutions of l-methionine (Sigma), N-formyl-l-methionine (Sigma) and dACCAAC (Genset oligos) were prepared in the same buffer at concentrations of 800, 300 and 4 mM, respectively. Titrations with these compounds were performed by adding increasing amounts of the appropriate stock solution to a protein sample at an original concentration of 1.5 mM. To study the interaction of fMet-tRNAfMet with IF2 C-2, the spectra of the protein (0.1 mM) were recorded in the absence and in the presence of 0.05 mM of tRNA. The 1H/15N HSQC spectra (Kay et al., 1992) were recorded using spectral width of 13.3 and 34.5 p.p.m. for the 1H and 15N dimensions, respectively. Fourier transformation was applied after gaussian apodization and zero filling to yield 1024 × 256 2D matrices. Data processing and spectral analysis was performed using the program Felix 97.0 (MSI, San Diego). Peak intensities were estimated by measuring the peak volumes. Chemical shift variations were monitored using an index Δδcomp defined as Σλ = HN,N(ωλΔδλ)2/Σλ = HN,Nωλ2. The weighting factors ωλ were chosen so that ωN/ωHN = Rscale with Rscale being defined as suggested in Mulder et al., 1999. Rscale = σN/σHN where σN and σHN are the amide nitrogen and proton chemical shifts average variances observed for the common amino acid residues (excluding proline) in proteins. Using data deposited in the BioMagResBank (URL http://www.bmrb.wisc.edu), a value of 6.5 for Rscale, irrespective of the amino-acid type, is found.
Acknowledgments
Acknowledgements
This work was supported by grants from the EC Biotechnology Program to R.B. and C.O.G., and from the Italian C.N.R. (Progetto strategico ST 74) and MURST-C.N.R. (95/95) to C.O.G.
References
- Foster M.P., Wuttke,D.S., Clemens,K.R., Jahnke,W., Radhakrishnan,I., Tennant,L., Reymond,M., Chung,J. and Wright,P.E. (1998) Chemical shift as a probe of molecular interfaces: NMR studies of DNA binding by the three amino-terminal zinc finger domains from transcription factor IIIA. J. Biomol. NMR, 12, 51–71. [DOI] [PubMed] [Google Scholar]
- Gold L. (1988) Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem., 57, 199–233. [DOI] [PubMed] [Google Scholar]
- Grill S., Gualerzi,C.O., Londei,P. and Bläsi,U. (2000) Selective stimulation of leaderless mRNA by IF2: evolutionary implications for translation. EMBO J., 19, 4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O., Pon,C.L., Pawlik,R.T., Canonaco,M.A., Paci,M. and Wintermeyer,W. (1986) Role of the initiation factors in Escherichia coli translational initiation. In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer-Verlag, New York, NY, pp. 621–641. [Google Scholar]
- Gualerzi C.O., Severini,M., Spurio,R., La Teana,A. and Pon,C.L. (1991) Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J. Biol. Chem., 266, 16356–16362. [PubMed] [Google Scholar]
- Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A., Spurio,R., Tomšic,J. and Pon,C.L. (2000) Translation initiation in bacteria. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 477–494. [Google Scholar]
- Kay L.E., Keifer,P. and Saarinen,T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc., 114, 10663–10665. [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Krafft C., Diehl,A., Lättig,S., Behlke,J., Heinemann,U., Pon,C.L., Gualerzi,C.O. and Welfle,H. (2000) Interaction of fMet-tRNA(fMet) with the C-terminal domain of translational initiation factor 2 from Bacillus stearothermophilus. FEBS Lett., 471, 128–132. [DOI] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1996) Late events in translation initiation. Adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol., 256, 667–675. [DOI] [PubMed] [Google Scholar]
- Meunier S., Spurio,R., Czisch,M., Wechselberger,R., Guenneugues,M., Gualerzi,C.O. and Boelens,R. (2000) Solution structure of the fMet-tRNA(fMet)-binding domain of B.stearothermophilus initiation factor 2. EMBO J., 19, 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz R., Welfle,K., Krafft,C., Gualerzi,C.O. and Welfle,H. (1997) Translational initiation factor IF2 from Bacillus stearothermophilus: a spectroscopic and microcalorimetric study of the C-domain. Biochemistry, 36, 3170–3178. [DOI] [PubMed] [Google Scholar]
- Misselwitz R., Welfle,K., Krafft,C., Welfle,H., Brandi,L., Caserta,E. and Gualerzi,C.O. (1999) The fMet-tRNA binding domain of translational initiation factor IF2: role and environment of its two Cys residues. FEBS Lett., 459, 332–336. [DOI] [PubMed] [Google Scholar]
- Mulder F.A., Schipper,D., Bott,R. and Boelens,R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol., 292, 111–123. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F.C. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNA(Phe), EF-Tu and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- RajBhandary U.L. and Chow,C.M. (1986) Initiator tRNAs and initiation of protein synthesis. In Söll,D. and RajBandary,U.L. (eds), tRNA. Structure, Biosynthesis and Function.ASM Press, Washington, DC, pp. 511–528. [Google Scholar]
- Ramsey N.F. (1950) Magnetic shielding of nuclei in molecules. Phys. Rev., 78, 699–703. [Google Scholar]
- Schmitt E., Guillon,J.M., Meinnel,T., Mechulam,Y., Dardel,F. and Blanquet,S. (1996) Molecular recognition governing the initiation of translation in Escherichia coli. A review. Biochimie, 78, 543–554. [DOI] [PubMed] [Google Scholar]
- Shuker S.B., Hajduk,P.J., Meadows,R.P. and Fesik,S.W. (1996) Discovering high-affinity ligands for proteins: SAR by NMR. Science, 274, 1531–1534. [DOI] [PubMed] [Google Scholar]
- Spurio R., Severini,M., La Teana,A., Canonaco,M.A., Pawlik,R.T., Gualerzi,C.O. and Pon,C.L. (1993) Novel structural and functional aspects of translational initiation factor IF2. In Nierhaus,K.H., Franceschi,R., Subramanian,A.R., Erdmann,V.A. and Wittmann-Liebold,B. (eds), The Translational Apparatus: Structure, Function, Regulation, Evolution. Plenum Press, New York, NY, pp. 241–252. [Google Scholar]
- Spurio R., Brandi,L., Caserta,E., Pon,C.L., Gualerzi,C.O., Misselwitz,R., Krafft,C., Welfle,K. and Welfle,H. (2000) The C-terminal sub-domain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J. Biol. Chem., 275, 2447–2454. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Nakanishi,T., Kami,K., Arata,Y. and Shimada,I. (2000) A novel NMR method for determining the interfaces of large protein–protein complexes. Nature Struct. Biol., 7, 220–223. [DOI] [PubMed] [Google Scholar]
- Tomšic J., Vitali,L.A., Daviter,T., Savelsbergh,A., Spurio,R., Striebeck,P., Wintermeyer,W., Rodnina,M.V. and Gualerzi,C.O. (2000) Late events of translation initiation in bacteria: a kinetic analysis. EMBO J., 19, 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.-Q. and RajBhandary,U.L. (1997) Effect of the amino acid attached to Escherichia coli initiator tRNA on its affinity for the initiation factor IF2 and on the IF2 dependence of its binding to the ribosome. J. Biol. Chem., 272, 1891–1895. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. (2000) Protein recognition by NMR. Nature Struct. Biol., 7, 188–189. [DOI] [PubMed] [Google Scholar]