Abstract
FadR is a dimeric acyl coenzyme A (acyl CoA)-binding protein and transcription factor that regulates the expression of genes encoding fatty acid biosynthetic and degrading enzymes in Escherichia coli. Here, the 2.0 Å crystal structure of full-length FadR is described, determined using multi-wavelength anomalous dispersion. The structure reveals a dimer and a two-domain fold, with DNA-binding and acyl-CoA-binding sites located in an N-terminal and C-terminal domain, respectively. The N-terminal domain contains a winged helix–turn–helix prokaryotic DNA-binding fold. Comparison with known structures and analysis of mutagenesis data delineated the site of interaction with DNA. The C-terminal domain has a novel fold, consisting of a seven-helical bundle with a crossover topology. Careful analysis of the structure, together with mutational and biophysical data, revealed a putative hydrophobic acyl-CoA-binding site, buried in the core of the seven-helical bundle. This structure aids in understanding FadR function at a molecular level, provides the first structural scaffold for the large GntR family of transcription factors, which are keys in the control of metabolism in bacterial pathogens, and could thus be a possible target for novel chemotherapeutic agents.
Keywords: acyl CoA/fatty acid/protein structure/transcription
Introduction
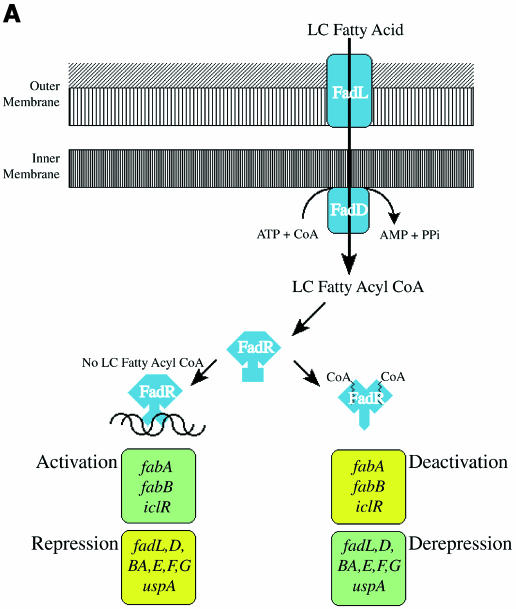
Fatty acids are essential molecular components for all forms of life. They are synthesized for incorporation into phospholipid membranes and can be used as an energy source when coupled to the tricarboxylic acid (TCA) cycle via β-oxidation. To regulate the balance between these catabolic and anabolic pathways, precise transcriptional control of the genes involved is required. Such control has been shown to exist in bacteria and mammals, and is sensitive to acyl coenzyme A (acyl CoA) concentrations. In mammals, for example, the hepatocyte nuclear transcription factor 4α (HNF-4α) has been shown to be responsive to long-chain acyl CoA (Hertz et al., 1998). Binding of the acyl CoA is thought to affect the dimerization and thus the DNA-binding properties of the protein (Black et al., 2000). In Escherichia coli, the transcription factor FadR, first described in the 1960s (Overath et al., 1969), is a central mediator that acts to balance the anabolic and catabolic fatty acid pathways (Figure 1A). This transcription factor negatively regulates genes encoding the fatty acid transport system (encoded by fadL and fadD) (DiRusso et al., 1999), enzymes of fatty acid catabolism (encoded by fadE, fadF, fadG, fadBA and fadH) and the universal stress protein gene (encoded by uspA) (Farewell et al., 1996). Simultaneously, FadR activates expression of the iclR gene, which encodes the negative regulator of the glyoxylate shunt enzymes (Gui et al., 1996). FadR also controls membrane fluidity by activating transcription of two genes essential for unsaturated fatty acid biosynthesis, fabA and fabB. These two genes encode proteins of the bacterial type II fatty acid synthase system. This system is unique to bacteria, plants and protozoan parasites. It is an essential component of the metabolism of these organisms and, as such, has been the target of novel chemotherapeutic agents, such as thiolactomycin (Jackowski et al., 1989). Recent studies have confirmed that FadR-dependent alterations in regulation of fatty acid metabolism occur during stress (Farewell et al., 1996; Spector et al., 1999), stasis survival (Farewell et al., 1996) and pathogensis in both E.coli and Salmonella typhimurium (Mahan et al., 1995; Farewell et al., 1996; Spector et al., 1999).
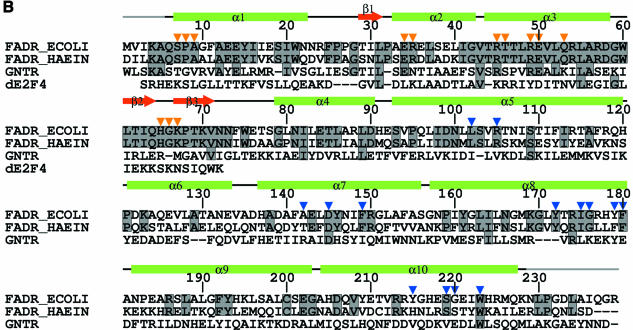
Fig. 1. (A) Schematic overview of FadR control of fatty acid metabolism in E.coli. The proteins playing a role in fatty acid import and activation are coloured blue. FadR is coloured blue. Positively regulated genes are coloured green and negatively controlled ones yellow. (B) Sequence alignment (using FadR numbering) with Clustal-W (Thompson et al., 1994) of E.coli FadR (FADR_ECOLI), a putative FadR from H.influenzae (FADR_HAEIN), GntR (GNTR) and the DNA-binding domain of E2F-4 (dE2F4). Residues conserved between the two FadRs are indicated by shaded boxes, which also cover residues in GNTR or dE2F4 for identities to the FadRs. Orange arrows indicate residues within contact distance of the DNA molecule in a model of the FadR–DNA complex (see Figure 3B). Blue arrows indicate residues that border the putative acyl-CoA-binding cavity within the C-terminal domain (see Figure 4). Below the sequences, the secondary structure of FadR is indicated by green bars (α-helices) and red arrows (β-strands); black lines indicate lack of secondary structure. The residues for which no interpretable density was observed at the termini are indicated by a grey line.
The gene encoding FadR has been sequenced and the protein purified and characterized (DiRusso, 1988; DiRusso et al., 1992, 1998). There are several distinct functions associated with the FadR protein (DiRusso et al., 1999). It is a DNA-binding protein, with 11 binding sites confirmed by DNA–protein gel shift assays and DNase 1 footprinting studies (DiRusso et al., 1999). FadR negatively controls (or represses) at least 12 genes and operons and activates the transcription of at least three genes, fabA, fabB and iclR. FadR directly binds long-chain acyl CoA [demonstrated using isoelectric focusing, fluorescence quenching and isothermal titration calorimetry (Raman and DiRusso, 1995; Raman et al., 1997; DiRusso et al., 1998)]. Binding of long-chain acyl CoA to FadR is presumed to result in a conformational change that inhibits or prevents FadR–DNA binding, leading to transcriptional activation and repression (Figure 1A) (DiRusso et al., 1992, 1993).
Sequence comparisons have detected homology between the N-terminus of FadR (residues 1–80) and the prokaryotic helix–turn–helix (HTH) DNA-binding motif. This DNA-binding domain is representative of the GntR family of DNA-binding domains (Haydon and Guest, 1991) [PROSITE PS00043 (Hoffman et al., 1999)], a family that includes transcription factors key to regulation of important bacterial metabolites (Haydon and Guest, 1991), for which structural information is not currently available. Mutagenesis studies (Raman and DiRusso, 1995), construction of chimeras (Raman and DiRusso, 1995) and photolabelling experiments (DiRusso et al., 1998) have demonstrated that long-chain acyl CoAs bind with high affinity to the C-terminal domain, defined as amino acids 102–239. It is thus likely that the DNA-binding function in the N-terminus is linked via a conformational change to the acyl-CoA-binding event at the C-terminus.
A FadR homologue has been identified in the genome of Haemophilus influenzae (see Figure 1B) with 48% amino acid identity to the E.coli protein. Given the central role of FadR in the control of the type II fatty acid synthase system and in pathogenesis and stasis survival, the protein may well prove to be a potential target for the design of novel agents against pathogens. Here, we describe the structure of FadR, revealing a two-domain structure with a novel C-terminal fold; we describe its function at the molecular level by combining the structure with currently available molecular genetic and biophysical data, and show that it can serve as a structural scaffold for the GntR family of bacterial transcription factors.
Results and discussion
Overall structure
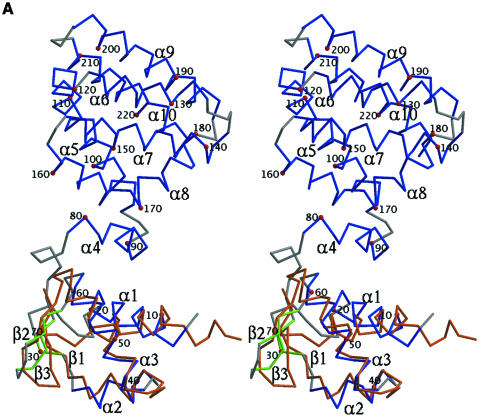
Native FadR was solved and refined to 2.0 Å resolution in spacegroup C2221, using initial experimental phases to 1.65 Å. The native structure (Figure 2A) was refined to an R of 0.200, Rfree 0.237, with proper stereochemistry (Table II). Analysis with PROCHECK (Laskowski et al., 1993) revealed that 95.4% of the residues have backbone conformations in the allowed regions of the Ramachandran plot, with the remaining 4.6% assigned to the additional allowed regions. The asymmetrical unit contains a monomer, and a tight dimer is formed by crystallographic symmetry. The protein consists of an α/β N-terminal domain (residues 1–72) and an all-helical C-terminal domain (residues 79–228), connected via a loop (see Figures 1B and 3A). This connection creates a cleft between the two domains. The N-terminal domain is structurally homologous to prokaryotic HTH DNA-binding motifs, whereas the C-terminal domain is of a type not reported previously. Both these domains are discussed in more detail below.
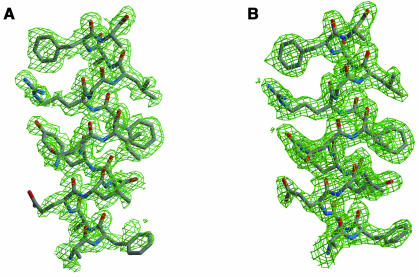
Fig. 2. Comparison of experimental and final electron density maps, for residues 139–154. A 1.65 Å Fo,φMLPHARE map contoured at 1.0 σ is shown (A), calculated using the structure factor amplitudes of the HgAc-II derivative in P21 and the phases at the end of heavy-atom refinement with MLPHARE. A 2.0 Å 2Fo – Fc,φcalc σA weighted map at the end of refinement of the native structure in C2221 with CNS, is shown (B), contoured at 1.0σ.
Table II. Refinement statistics of the native structure.
| R | 0.200 |
| Rfree | 0.237 |
| Number of molecules in the asymmetric unit | 1 |
| Number of protein atoms | 1930 |
| Number of water molecules | 159 |
| Number of sulfate ions | 2 |
| Data/parameter ratio | 2.3 |
| r.m.s.d. bonds (Å) | 0.011 |
| r.m.s.d. bond angles (°) | 1.44 |
| B-factor r.m.s.d. bonded atoms (Å2) | |
| main chain | 1.35 |
| side chain | 2.22 |
| Average B-factor (A2) | |
| main chain | 25.1 |
| side chain | 28.2 |
| water | 35.7 |
Data between 25 and 2.0 Å were included in the refinement without any F/σF cut-offs.
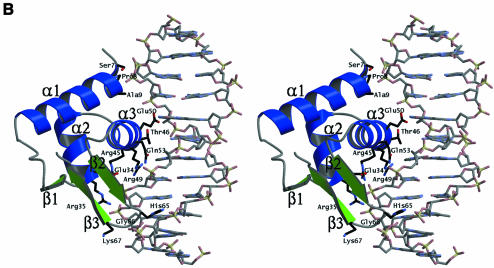
Fig. 3. (A) Structure of the FadR monomer. FadR is shown as a stereo Cα trace with every tenth residue labelled and indicated by a red sphere. Helices are coloured blue, strands green and loops grey. Secondary structure elements, determined with DSSP (Kabsch and Sander, 1983) followed by visual inspection, are labelled α1–10 and β1–3 corresponding to the assignment in Figure 1B. The superimposed structure of dE2F4 is also shown as an orange Cα trace. (B) Stereo model of FadR–DNA complex. The DNA from the dE2F4–DNA complex (Zheng et al., 1999) is shown as a stick model. The FadR DNA-binding domain was superimposed on the dE2F4 structure and is shown as a ribbons model with helices coloured blue and strands green. Labels indicate the assignments of these secondary structure elements (see also Figure 1B). The model of the FadR–DNA complex was inspected for intermolecular contacts: side chains interacting with the DNA (defined as interatomic distances being smaller than the sum of their van der Waals radii + 1.0 Å) are shown as sticks with black carbons and are labelled. During crystallographic model building and refinement, the side chains of residues Arg35, Gln64 and His65 were truncated to alanine because of poor density, here they are shown as standard rotamers.
The DNA-binding domain
Mutagenesis experiments have shown that the DNA-binding function of FadR resides in the N-terminal domain (Raman et al., 1997). This domain, encompassing residues 1–72, contains a small β-sheet core formed by three strands, and three α-helices (see Figures 1B and 3A). An HTH motif of the ‘winged’ type is formed by helices α2 and α3, connected via a tight turn, together with the β-sheet. A search for structural homologues with DALI (Holm and Sander, 1993) identified several ‘hits’, all DNA-binding domains, of which the most significant ones were two mammalian proteins: the DNA-binding domain of RNA-specific adenosine deaminase ADAR1 (Schwartz et al., 1999) [DALI Z-score, 8.1; root mean square deviation (r.m.s.d.) on aligned Cα positions, 2.2 Å] and the DNA-binding domain of the transcription factor E2F-4 (dE2F4) (Zheng et al., 1999) (DALI Z-score, 6.9; r.m.s.d. on aligned Cα positions, 2.2 Å). In both cases, analysis of superimposed structures shows that all secondary structure elements have been conserved and are in similar relative positions. The structures of the FadR DNA-binding domain and dE2F4 are compared in Figure 3A. The HTH motif runs from Ala33 to Asp58 (helices α2 and α3) in FadR and superimposes with an r.m.s.d. of 1.1 Å on equivalent Cα carbons of residues 42–68 in dE2F4. The turn and β-hairpin that follow α3 structurally align residues Gly59–Asn72 in FadR with residues 69–82 in dE2F4. Although the β1–β2 loops are of equal length in both structures, they have a rather different conformation. The resulting overall structure-based sequence alignment thus covers residues Ser7–Asn72 in FadR and residues 21–82 in dE2F4. Analysis of the aligned sequences reveals that five of the 66 residues in FadR are identical to dE2F4 (Gly10, Leu31, Val43, Leu55 and Gly59), which additional conservative substitutions, such as lysine to arginine, for residues interacting with the DNA backbone.
The dE2F4 structure was solved in the presence of a B-DNA duplex. Since the dE2F4 and FadR backbones overlap well, this allows the construction of a model for the FadR–DNA interaction. Figure 3B shows the proposed FadR–DNA binding mode, obtained by superimposition of the FadR DNA-binding domain on to dE2F4, through the structural alignment obtained with DALI (Holm and Sander, 1993). Using WHAT IF (Vriend, 1990) a list of putative FadR–DNA contacts was generated from this model, where a contact was defined as an interatomic distance that was shorter than the sum of the van der Waals radii + 1.0 Å. No severe clashes were observed between the superimposed FadR and the dE2F4 DNA. Residues contacting the DNA in the model of the complex are shown in Figures 1B and 3B, and are all conserved in the putative protein from H.influenzae. FadR could interact with the DNA in four distinct regions (Figures 1B and 3B). At the N-terminus, the side chains of residues Ser7, Pro8 and, to a lesser extent, Ala9 are within contact distance of the DNA backbone. Residues 1–5, which could not be seen in the electron density maps, could also be involved in further contacts (residue 5 is a lysine). At the beginning of helix α2, the start of the HTH motif, Glu34 (which coordinates Arg45 and Arg49) and Arg35 are positioned to interact with the DNA model. Such contacts are frequently observed in DNA-binding proteins containing the HTH motif. Helix α3, often termed the recognition helix in the HTH motif, is positioned in the major groove of the DNA helix. Residues Arg45, Arg49 and Gln53 can interact with the backbone of the DNA model. Residues Thr46, Arg49 and Glu50 point from the α3 helix into the major groove containing the exposed Watson–Crick base pairs, with which they could make several contacts. The final area of putative interaction is at the tip of the β2–β3 hairpin, where Gly66 and Lys67 interact with the DNA backbone, whereas His65 could be positioned to point into the minor groove. This area of interaction is less certain, however, because the FadR–dE2F4 structure comparison (Figure 3A) shows significant conformational differences in that region.
Previously, the functional importance of residues in the FadR DNA-binding domain has been assessed using hydroxylamine random mutagenesis and alanine mutagenesis scanning of the HTH motif (Raman et al., 1997). In total, mutations at six positions were identified that gave a dominant negative phenotype: Ala9→Val, Arg35→Cys/Ala, Arg49→Ala, His65→Tyr/Ala, Gly66→Asp/Ser/Ala and Lys67→Ala. These residues were subsequently shown to affect DNA binding directly in vitro (Raman et al., 1997). Using the FadR structure, these data can now be interpreted in a three-dimensional framework (Figure 3B). The mutations appear to fall in all of the four separate FadR–DNA contact areas described above, and are all in contact with the DNA as evaluated from the model for the FadR–DNA complex (Figures 1B and 3B). Mutation of these residues would indeed disrupt electrostatic or specific contacts between FadR and DNA. Obviously, the model of the FadR–DNA complex does not allow a detailed analysis of side-chain–base contacts for analysis of binding specificity, which is only possible from a high-resolution crystal structure of such a complex. However, the model allows interpretation of the mutational data available, which support the model, pointing to four separate regions forming a semi-continuous binding surface where FadR is likely to interact with the DNA helix.
FadR belongs to the GntR family of ∼35 transcription factors, whose HTH motifs and, to a certain extent, complete DNA-binding domains can be aligned by sequence (Haydon and Guest, 1991) [PROSITE PS00043 (Hoffman et al., 1999), PFAM PF00392 (Bateman et al., 2000)]. No structural information is currently available for this family, whose members are important for control of several key bacterial metabolic pathways (Haydon and Guest, 1991). Apart from FadR, the repressor of the gnt operon in Bacillus subtilis, GntR is the only well characterized member of this family (Miwa and Fujita, 1988). GntR negatively regulates transcription of the enzymes involved in the gluconate catabolism in B.subtilis. In a response reminiscent of the FadR–acyl CoA interaction, repression by GntR is switched off by direct binding of gluconate to the protein. Using hydroxylamine mutagenesis, four mutants were isolated that showed a negative transdominant phenotype, Ser43→ Leu, Ala66→Thr, Glu74→Lys and Arg75→Gln (Yoshida et al., 1993). These positions align with residues Pro32, Ala56, Gln64 and His65, respectively, in FadR. Pro32 and Ala56 contribute to the hydrophobic core of the DNA-binding domain and may be important for structural integrity. Ala56 (which is conserved between FadR and GntR) on the winged-HTH recognition helix is in contact with the hydrophobic residue at position 63 (isoleucine in FadR, leucine in GntR) on the β2–β3 hairpin, and may thus be important for fixing the relative orientations of these DNA-binding regions. Gln64 and His65 (the latter also tested by mutation in FadR), lie on the tip of the β2–β3 hairpin, which interacts directly with the DNA (Figure 3B). Thus, it appears that the FadR structure can serve as a structural scaffold for the GntR family and could be useful for interpretation of new biochemical data and evaluation of their function at the molecular level.
The acyl-CoA-binding domain
Previous mutagenesis experiments have suggested that the acyl-CoA-binding function of FadR resides in the C-terminus (Raman and DiRusso, 1995). The structure (Figure 2A) indeed reveals a separate C-terminal domain, starting around residue 79, which consists of seven α-helices (α4-α10) with short connecting loops, packed together to form a bundle. The helices are not arranged in a simple circular fashion in this bundle. Rather, looking down the first helix from the N- to the C-terminus (Figure 2A), the clockwise arrangement is α4–α5–α10–α9–α6–α7–α8, running down–up–down–up–down–up–down. All stackings of the helices with their neighbours are thus antiparallel, except the first (α4) and last (α8) helices, which stack together in a parallel fashion. Although the short helix α4 has many contacts with its neighbours (α5 and α8), it is more loosely associated with the bundle. The helices are straight, with two exceptions: helices α5 and α8. In α5 there is a bend at residue Ser109, which changes the direction of the helical axis to allow it to point more to the other side of the bundle, which has to be crossed before descending into α6. Helix α8 contains a more severe kink at residue Lys169, allowing it to cross the bundle, connecting from α7 to α9, which are at opposite ends of the bundle. The turns between the helices are tight, ranging from one to three residues in length (Figure 1B). The core of the bundle consists mainly of aromatic and aliphatic side chains, with some notable exceptions, which will be discussed in more detail. A search through the database of known structures using DALI (Holm and Sander, 1993) revealed no significant structural homology. The best hits concerned only fragments consisting of two or three helices identified in other helical proteins, with an r.m.s.d. on superimposed Cα atoms of >3.5 Å. A search through the hierarchically organized CATH (Orengo et al., 1997) and SCOP (Murzin et al., 1995) structural databases also did not reveal structural homology. The seven-helical bundle in FadR is superficially reminiscent of the bundle seen in the structure of bacteriorhodopsin (Pebay-Peyroula et al., 1997). However, these structures cannot be directly superimposed, because of their different helical topology and overall shape, which is probably related to their diverse functions. In summary, it appears that the FadR structure contains a novel three-dimensional fold, of which the apparent function is to interact with acyl CoA.
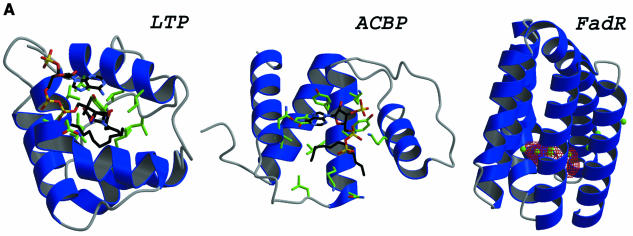
FadR has previously been crystallized in the presence of CoA or octanoyl CoA (van Aalten et al., 2000). Using the FadR structure described here as a search model, we have been able to solve these structures by molecular replacement. Unfortunately, subsequent refinement and analysis revealed that neither of the ligands had co-crystallized with the protein (data not shown). However, careful analysis of the present structure together with available biochemical data gives a consistent picture as to where the acyl CoA would bind. Analysis of cavities using VOIDOO (Kleywegt and Jones, 1994) revealed a large cavity within the seven-helical bundle (Figure 4). This cavity has an empty volume of 142 Å3, using a 1.4 Å probe and the ‘probe-accessible’ definition. Both VOIDOO (Kleywegt and Jones, 1994) and WHAT IF (Vriend, 1990) find additional small cavities neighbouring this large one, but these were not consistent between the two programs and are therefore not discussed here. WHAT IF, however, was able to find a tunnel leading from the outside of the helical bundle up to the large cavity. The cavity is filled with five ordered water molecules; three additional water molecules can be found in the tunnel (average B-factor of these eight water molecules, 29.6 Å2). Two other structures of acyl-CoA-binding proteins in complex with acyl CoA are currently available. The lipid transfer protein (LTP) (Lerche et al., 1997) and the acyl-CoA-binding protein (ACBP) (D.M.F.van Aalten, K.G.Milne, J.Y.Zou, G.J.Kleywegt, T.Bergfors, J.Knudsen and T.A.Jones, submitted) are compared with the structure of the FadR acyl-CoA-binding domain in Figure 4A. Interestingly, these acyl-CoA-binding domains are all-helical structures, albeit with different topology. LTP is a small four-helical bundle, ACBP also contains four helices, but these are arranged in a bowl shape. In LTP and ACBP the acyl chains are bound in hydrophobic cavities formed by the helices with apolar side chains (Figure 4A), whereas the adenosine 3′ phosphate and pyrophosphate moieties are more exposed to the solvent. A similar mode of acyl CoA binding could be possible for FadR, with the mainly hydrophobic cavity within the helical bundle binding the acyl chain, whereas the other moieties could interact with the solvent-exposed outer surface of the helical bundle.
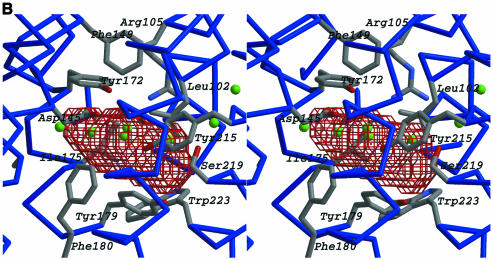
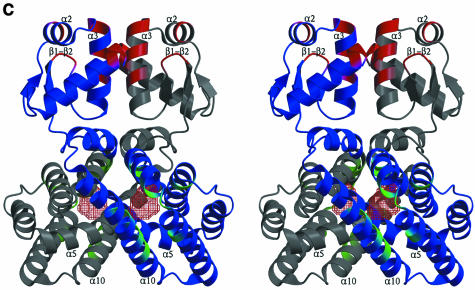
Fig. 4. (A) Comparison of acyl-CoA-binding proteins. The structures of the LTP (Lerche et al., 1997) and ACBP (D.M.F.van Aalten, K.G.Milne, J.Y.Zou, G.J.Kleywegt, T.Bergfors, J.Knudsen and T.A.Jones, submitted) are shown together with the acyl-CoA binding domain of FadR (residues 79–227). Structures are drawn as ribbons, with helices coloured blue and loops coloured grey. The acyl-CoA ligand is shown in LTP and ACBP as a stick model, with carbons coloured black. Side chains interacting with the acyl-CoA ligand in LTP and ACBP are shows as sticks, with carbons coloured green. In FadR, the cavity that is discussed in the text is shown in red chickenwire and water molecules occupying the cavity and the tunnel are shown as green spheres. (B) The putative FadR acyl-CoA-binding site. Helices (blue) and residues bordering the cavity (sticks coloured according to atom type) are shown in stereo, together with the cavity, which is represented by red chickenwire. Water molecules occupying and leading to the cavity are shown as green spheres. (C) Structure of the dimer. The structures of two crystallographically related FadR molecules are shown as blue and grey ribbon models in stereo. The acyl-CoA-binding cavity is shown as red chickenwire in both molecules. Mutations discussed in the text that affected DNA binding are shown in red, and those that affected acyl CoA binding are shown in green. The crystallographic 2-fold axis relating these molecules runs from the bottom to the top of the image on the plane of the paper. The helices in the HTH motif (α2,α3), the β2–β hairpin and the α5,α10 helices are labelled.
A detailed view of the cavity is shown in Figure 4B. The following residues together form this cavity: Leu102, Arg105, Ala142, Asp145, Phe149, Tyr172, Ile175, Gly176, Tyr179, Phe180, Tyr215, Ser219, Gly229 and Trp223. Ten of these residues are conserved and the other four conservatively substituted in the putative FadR from H.influenzae (Figure 1B). They are mainly hydrophobic; the most notable exceptions are Arg105 and Asp145, which are within 4.7 Å of each other. These two residues and the polar groups on the other residues (i.e. the hydroxyl groups on the tyrosines and serine) all appear to have their hydrogen-bonding potential fulfilled by side chain–side chain/backbone hydrogen bonds, rather than by extensive hydrogen bonding to the water molecules in the cavity. Thus, the side-chain surfaces contacting the cavity are mainly hydrophobic. In a similar fashion, structures of the fatty acid- and retinoid-binding proteins in their apo form show several water molecules buried deep inside a hydrophobic cavity (Sacchettini et al., 1989; Winter et al., 1993).
The hypothesis that the acyl chain binds in the buried cavity is confirmed by analysis of the available biochemical data. Genetic experiments in which the DNA-binding domain of LexA was fused to the C-terminal domain (residues 102–239) of FadR resulted in a protein that retained the LexA DNA-binding specificity and was inducible by long-chain acyl CoAs (Raman et al., 1997), thus locating the acyl-CoA-binding function to the C-terminal domain. Measurements of the intrinsic tryptophan fluorescence have demonstrated a significant quenching effect upon binding of acyl CoA to FadR (Raman and DiRusso, 1995). The C-terminal domain contains only one tryptophan (Trp223), which borders the cavity (Figure 4B) and could indeed show a decrease in intrinsic fluorescence upon interaction with an acyl chain. Photoaffinity labelling and subsequent tryptic digests with the palmitoyl analogue 9-p-azidophenoxy(9-3H)nonaoic acid–CoA ester (DiRusso et al., 1998), which contains a photoreactive group at the ω-end of the acyl chain, identified the peptide SLA LGFYHK as being part of the acyl-binding pocket. This peptide, which maps to residues Ser187–Lys195 on helix α9, does not show any direct interaction with the cavity. However, Ala189, Phe192 and Tyr193 (the latter two conserved in the H.influenzae homologue, Figure 1B) all point towards the cavity. Tyr193 hydrogen bonds to the cavity-forming Asp145, whereas Phe192 makes a stacking interaction with the cavity-forming Tyr215. Slight rearrangements of side chains, which are likely to occur upon acyl CoA binding, would thus put Ala189, Phe192 and Tyr193 in direct contact with the ligand. Mutagenesis studies using hydroxylamine and position-specific alanine scanning identified four mutations, Tyr179→Ala, Gly216→Ala, Ser219→Asn and Trp223→Ala, which resulted in a so-called super-repression phenotype (Raman and DiRusso, 1995). These proteins can still bind DNA tightly, but this interaction can no longer be disrupted by the addition of acyl CoA. Three of these amino acids are conserved in the H.influenzae homologue (Figure 1B). Tyr179 and Trp223 lie next to each other and form the bottom half of the cavity (Figure 4B). Mutation of these residues to Ala would significantly alter the shape and character of the cavity. Ser219 contributes to the cavity with its Cβ atom, whereas Oγ points towards the tunnel leading to the exterior, and makes a water-mediated hydrogen bond to Arg105, which also lies at the mouth of the tunnel. Mutation of Ser219 to the bulkier asparagine could either seal off most of the tunnel, or disrupt the hydrophobic character of the cavity. Gly216 is not conserved (Figure 1B) and is changed to asparagine in the H.influenzae homologue. It is not clear why the Gly216→Ala would have such a large effect, although it, like Ser219 one helix turn away, points towards the more polar tunnel, and mutation to alanine could disrupt this polar character.
Alignment of the full-length GntR and FadR is possible and gives an overall sequence identity of 16%. The proteins are of about the same length (239 and 243 residues, respectively), and secondary structure prediction with the program PHD (Rost et al., 1994) gives an all-helical C-terminal domain with helices in approximately the same positions as in FadR. Residues Tyr179 and Trp223, which border the cavity in the FadR structure and have been shown to be crucial for proper function, are conserved in GntR. Several GntR mutants have been isolated with diminished ability to bind gluconate (Yoshida et al., 1995), which were thus not able to dissociate from the gnt promoter, a situation similar to the super-repressors in FadR. The mutations were a C-terminal deletion of 23 amino acids and two point mutations at Met209 and Ser230. These are in the same region as the mutations affecting acyl CoA binding in FadR. Thus, it is possible that the FadR structure might be able to serve as a structural scaffold not only for the winged-HTH DNA-binding domains of the GntR family but also for the C-terminal effector molecule-binding domains.
The FadR dimer
Previous experiments employing gel filtration and ultracentrifugation have shown that FadR exists as a homodimer in solution and binds DNA as a dimer (Raman et al., 1997). The crystal structure reveals a dimer interaction, generated by a crystallographic 2-fold axis (Figure 4C). The consensus DNA-binding site for FadR consists of 17 base pairs: ANCTGGTCNGACNTNTT (DiRusso et al., 1999). This sequence is pseudo-palindromic, and in some FadR-binding sites the dyad symmetry is more precise, such as in the fadB gene: ATCTGGTACGACCAGAT. The symmetry in the binding site supports binding of FadR in dimeric form to DNA. The FadR structure presented here does not contain DNA or acyl CoA, and is taken to represent the conformation prior to binding DNA (Figure 1A). The interaction of a FadR monomer with DNA as shown in Figure 3B is consistent with the biochemical data discussed here, yet such interactions are not possible in the structure of the dimer shown in Figure 4C because of steric clashes of the second monomer with the DNA molecule.
Dimerization of FadR results in a reduction of the solvent-accessible surface by 1634 Å2. This surface consists of two main contributions. Dimerization results in the formation of two buried salt bridges between the monomers on the DNA-binding domain: Glu13–Arg57 (2.96 Å) and Arg57–Asp58 (2.55 Å). The larger dimerization surface on the C-terminal domain has a more apolar character, with the most significant side-chain–side-chain contacts being Trp75–Phe154/Pro159/Leu163, Ile82–Leu163, Leu101–Ile160 and Ile108–Ile108. The rather polar and more exposed interface between the DNA-binding domains suggests that these could dissociate relatively easily, whereas the more hydrophobic nature of the C-terminal domain interface may represent a tighter interaction. The putative acyl-CoA-binding pockets lie at different ends of the dimer (Figure 4C), in agreement with the observation that the binding stoichiometry is two molecules of acyl-CoA per FadR dimer. Although a putative binding site for the acyl chain could be identified using our structure and the available biochemical data, the precise conformation of the acyl-CoA molecules in the dimer and the associated conformational change remains uncertain, until a detailed structure of a complex is available.
Conclusions
The structure of the FadR dimer described here provides a first view, at the molecular level, of the transcriptional regulation of fatty acid metabolism in bacteria. The structure contains spatially separated DNA- and acyl-CoA-binding sites, which are readily identifiable using structural analyses and interpretation of the available biochemical data. A winged-HTH motif provides the DNA-binding site, whereas a cavity buried inside a novel seven-helical bundle fold provides a binding site for the acyl chain. This represents one of the few examples of a high-resolution transcription factor structure in which both the DNA and effector-binding domains can be observed, yet its architecture is significantly different from other such examples [e.g. catabolite gene-activating protein (Schultz et al., 1991), lac repressor (Lewis et al., 1996) and modE (Hall et al., 1999)]. The structure is the first one available for the GntR class of prokaryotic transcription factors, which control important bacterial metabolic pathways such as fatty acid synthesis (DiRusso et al., 1999), gluconate synthesis (Miwa and Fujita, 1988), glycolate oxidation (Pellicer et al., 1996), rhizopine catabolism (Rossbach et al., 1994) and trehalose metabolism (Schoeck and Dahl, 1996). The FadR structure gives the sequences of these transcription factors a structural scaffold that can be used to interpret biochemical data, and may provide an inroad into design of novel chemotherapeutical agents that would interfere with these important metabolic pathways. FadR itself controls expression of enzymes in the type II fatty acid synthase pathway, which has already been validated as a target for rational drug design (Jackowski et al., 1989).
Materials and methods
Purification and crystallization
FadR was overexpressed in E.coli and purified as described previously (DiRusso et al., 1998). The resulting protein solution contained 4.6 mg/ml FadR in 50 mM KH2PO4, 10% glycerol pH 8.0. Previously described crystallizations from polyethylene glycol solutions, in the presence of CoA or octanoyl CoA (van Aalten et al., 2000), were not suitable for derivative searches, so a new crystallization protocol was developed. Hanging drop vapour diffusion crystallization experiments were set up using 1 µl of protein solution and an equal volume of well solution containing 2.3 M (NH4)2SO4, 0.75 M Li2SO4 and 100 mM Tris–HCl pH 9.0. Crystals were grown at room temperature and reached a size of 0.2 × 0.2 × 0.2 mm within 24 h.
Heavy-atom search and data collection
Crystals were transferred into mother liquor containing 10% ethylene glycol and, after 60 s, frozen in a nitrogen gas stream (100 K) for data collection. A suitable mercury acetate heavy-atom derivative was identified after extensive screening. For the first derivative crystal (HgAc-I), 20 mM mercury acetate was present in the cryoprotecting solution. For the second derivative (HgAc-II), the crystal was soaked for 12 h in mother liquor containing 20 mM mercury acetate and then cryoprotected using a solution without the heavy atom. Data were collected on MAR CCD detectors at beamlines X11 at the EMBL outstation at the DESY synchrotron, Hamburg, and beamline BM14 at the ESRF synchrotron, Grenoble. On the latter beamline, a multi-wavelength anomalous dispersion experiment was carried out using HgAc-I, close to the mercury LIII edge. Data were collected at three wavelengths, determined from an X-ray fluorescence scan, to obtain optimal anomalous and dispersive differences. λ3 corresponds to the maximum of f” (peak wavelength), λ2 to the minimum of f′ (inflection point) and λ1 to a high-energy remote wavelength.
Images were processed using DENZO and reflections merged using SCALEPACK, of the HKL suite (Otwinowski and Minor, 1997). Data collection and processing statistics are summarized in Table I.
Table I. Details of data collection and heavy atom-refinement.
| Native 1 | HgAc-I |
HgAc-II | |||
|---|---|---|---|---|---|
| λ1 | λ2 | λ3 | |||
| Wavelength (Å) | 0.9535 | 0.9537 | 1.0085 | 0.9999 | 0.9073 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 |
| Space group | C2221 | P21 | P21 | P21 | P21 |
| Unit cell (Å) | |||||
| a | 59.71 | 58.98 | 58.97 | 58.95 | 58.82 |
| b | 101.97 | 89.04 | 89.02 | 88.99 | 88.64 |
| c | 87.51 | 58.96 | 58.95 | 58.93 | 58.82 |
| β (°) | 118.46 | 118.46 | 118.46 | 118.56 | |
| Resolution (Å) | 25–2.0 | 25–1.8 | 25–1.8 | 25–1.8 | 25–1.65 |
| Highest bin (Å) | 2.07–2.00 | 1.86–1.80 | 1.86–1.8 | 1.86–1.8 | 1.71–165 |
| Redundancy | 3.2 (2.0) | 2.5 (2.1) | 2.5 (2.1) | 2.5 (2.1) | 3.4 (3.0) |
| Completeness (%) | 95.2 (73.8) | 88.7 (88.7) | 89.3 (90.4) | 89.4 (92.3) | 99.5 (99.2) |
| Rmerge | 0.070 (0.448) | 0.054 (0.336) | 0.060 (0.362) | 0.069 (0.345) | 0.067 (0.466) |
| I/σI | 9.7 (2.6) | 13.4 (2.7) | 12.4 (2.7) | 12.9 (3.0) | 15.7 (2.7) |
| Riso | – | – | 0.067 | 0.098 | 0.208 |
| Number of sites | – | – | 5 | 5 | 5 |
| Phasing power | |||||
| acentric | – | – | 0.78 | 1.26 | 0.86 |
| centric | – | – | 0.63 | 1.01 | 0.63 |
| Rcullis | |||||
| acentric | – | – | 0.87 | 0.77 | 0.89 |
| centric | – | – | 0.88 | 0.75 | 0.84 |
| anomalous | – | – | 0.93 | 0.94 | 0.93 |
HgAc, mercury acetate; Rmerge = ΣhΣi|Ii(hkl) – <I(hkl)>|/ΣhΣi<I(hkl)>; Riso = Σh|Fp – Fph|/Σh|Fp|; phasing power = Σhfc/Σhε; Rcullis = Σ||Fph – |Fp + fc||/Σ||Fph – |Fp|| where fc is the calculated heavy atom factor, ε is the lack-of-closure error, I is the intensity, Fp is the native structure factor and Fph is the heavy atom derivative structure factor.
Structure determination and refinement
The data of a native crystal, and a MAD dataset on the HgAc-I derivative, collected at the tunable beamline BM14 in Grenoble, were processed and scaled well in C2221 with one molecule in the asymmetrical unit, and appeared to be similar to one of the crystal forms identified previously (van Aalten et al., 2000) (Table I). From Patterson maps calculated from the anomalous and dispersive signal, the same strong peaks were identified, which appeared to correspond to two sites that were both in special positions. Although favourable statistics were obtained after refinement of the heavy-atom sites and phasing with MLPHARE (Collaborative Computational Project, 1994), SHARP (de la Fortelle and Bricogne, 1997) or SOLVE (Terwilliger and Berendzen, 1999), the maps were uninterpretable and did not even reveal solvent boundaries. Derivative HgAc-II was soaked for 12 h, but back-soaked during the 60 s of cryoprotection. The resulting 1.65 Å data, however, did not scale in C2221, as with the previous crystals, but in P21, with two FadR molecules in the asymmetric unit. The self-rotation function, calculated with the REPLACE package (Tong and Rossmann, 1997), identified a peak at φ = 180.0°, ψ = 59.5°, κ = 180.0°, suggesting that the crystallographic 2-fold symmetry present in C2221 had broken down to pseudosymmetry due to the soaking procedure. Pattersons calculated from the anomalous signal identified at least two strong (16σ and 10σ) peaks. When the HgAc-I MAD dataset, processed previously in C2221, was reprocessed in P21, the same sites were identified; there also appeared to be an isomorphous signal with respect to λ1 HgAc-I due to the different soaking procedures. The heavy-atom parameters were refined in MLPHARE (CCP4, 1994) with data to 1.8 Å (Table II), resulting in phases with a figure-of-merit of 0.46. Solvent boundaries and some secondary structure elements could be identified; after solvent flattening and phase extension to 1.65 Å with DM (Cowtan, 1994), a well defined trace appeared with clearly identifiable side chains and holes in the rings of aromatic residues (Figure 2A). The phases were then used as input for the autobuilding procedure in warpNtrace (Perrakis et al., 1999). After 200 cycles of building and refinement, 415 of the 478 residues in the asymmetrical unit were built. Using the side-chain building algorithm in warpNtrace, side chains for all these residues were built. After deletion of water molecules in the model and the assignment of a test-set for calculation of an Rfree (750 reflections), the model was subjected to a simulated annealing run in CNS (Brunger et al., 1998) (R = 0.286, Rfree = 0.307). The Fo – Fc map showed interpretable density for three missing loops, and allowed extension by two or three residues on the termini, which were built with WHAT IF (Vriend, 1990). The map also showed a 60σ peak for each monomer, at ∼2.2 Å from the Sγ atom of the only cysteine (Cys200) in FadR, corresponding to the sites of two mercury atoms, which were included in the refinement. Further iterations of model building in O (Jones et al., 1991) and refinement in CNS (Brunger et al., 1998) reduced the R-factor to 0.216 (Rfree = 0.232). A monomer from this partially refined model was then used as search model for molecular replacement calculations with AMoRe (Navaza, 1994) using 8.0–4.0 Å data from a native dataset in C2221. The structure was solved with an R-factor of 0.337 (correlation coefficient 0.683). After simulated annealing in CNS (R = 0.282, Rfree = 0.312), this structure was used as a starting point for further refinement and model building, which included placement of water molecules and some alternative side-chain conformations, truncation of some side chains to alanine due to poor density, and refinement of weakly restrained individual B-factors. The resulting final model has an R-factor of 0.200 (Rfree = 0.237) using all data between 25 and 2.0 Å, with good stereochemistry (see Table II) and continuous density for the backbone from residues 6 to 228 (Figure 2B).
In addition to the native structure, the HgAc-II was also further refined to an R-factor of 0.197 (Rfree = 0.225), and deposited with the Protein Data Bank. The non-crystallographic dimer from HgAc-II superimposes with an r.m.s.d. of 0.45 Å on the native dimer, whereas the two independent monomers in HgAc-II superimpose with an r.m.s.d. of 0.2 Å. The largest conformational change induced by incorporation of the mercury atom is around the site of attachment. Since the native dimer represents the more biologically relevant structure and is refined to high resolution, the HgAc-II structure is not further discussed here.
Acknowledgments
Acknowledgements
We thank the EMBL-Hamburg outstation at the DESY synchrotron for use of beamlines X11 and X31, and the ESRF synchrotron in Grenoble for the time on beamline BM14. We thank Gordon Leonard for his help during data collection and David Komander for his assistance with refinement. D.M.F.v.A. is supported by a Wellcome Trust Career Development Research Fellowship. The coordinates and structure factors of the native and HgAc-II derivative have been submitted to the Protein Data Bank (entry 1E2X).
References
- Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonhammer,E.L. (2000) The PFAM protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black P.N., Faergeman,N.J. and DiRusso,C.C. (2000) Long-chain acyl-CoA dependent regulation of gene expression in bacteria, yeast and mammals. J. Nutr., 130, 305S–309S. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software system for macromolecular structure determination. Acta Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 34–38. [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- DiRusso C.C. (1988) Nucleotide sequence of the fadR gene, a multifuntional regulator of fatty acid metabolism in Escherichia coli. Nucleic Acids Res., 16, 7995–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRusso C.C., Heimert,T.L. and Metzger,A.K. (1992) Characterization of fadr, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. J. Biol. Chem., 267, 8685–8691. [PubMed] [Google Scholar]
- DiRusso C.C., Metzger,A.K. and Heimert,T.L. (1993) Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in E.coli by FadR. Mol. Microbiol., 7, 311–322. [DOI] [PubMed] [Google Scholar]
- DiRusso C.C., Tsvetnitsky,V., Hojrup,P. and Knudsen,J. (1998) Fatty acyl-CoA binding domain of the transcription factor FadR. J. Biol. Chem., 273, 33652–33659. [DOI] [PubMed] [Google Scholar]
- DiRusso C.C., Black,P.N. and Weimar,J.D. (1999) Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog. Lipid Res., 38, 129–197. [DOI] [PubMed] [Google Scholar]
- Farewell A., Diez,A.A., DiRusso,C.C. and Nystrom,T. (1996) Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad and fab genes. J. Bacteriol., 178, 6443–6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L., Sunnarborg,A. and LaPorte,D.C. (1996) Regulated expression of a repressor protein: FadR activates iclR. J. Bacteriol., 178, 4704–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.R., Gourley,D.G., Leonard,G.A., Duke,E.M.H., Anderson,L.A., Boxer,D.H. and Hunter,W.N. (1999) The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (modE) from Escherichia coli: a novel combination of domain folds. EMBO J., 18, 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon D.J. and Guest,J.R. (1991) A new family of bacterial regulatory proteins. FEMS Microbiol. Lett., 79, 291–296. [DOI] [PubMed] [Google Scholar]
- Hertz R., Magenheim,J., Berman,I. and Bar-Tana,J. (1998) Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature, 392, 512–516. [DOI] [PubMed] [Google Scholar]
- Hoffman K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jackowski S., Murphy,C.M., Cronan,J.E. and Rock,C.O. (1989) Acetoacetyl-acyl carrier protein synthase—a target for the antibiotic thiolactomycin. J. Biol. Chem., 264, 7264–7629. [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D, 50, 178–185. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lerche M.H., Kragelund,B.B., Bech,L.M. and Poulsen,F.M. (1997) Barley lipid transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure, 5, 291–306. [DOI] [PubMed] [Google Scholar]
- Lewis M., Chang,G., Horton,N.C., Kercher,M.A., Pace,H.C., Schumacher,M.A., Brennan,R.G. and Lu,P. (1996) Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science, 271, 1247–1254. [DOI] [PubMed] [Google Scholar]
- Mahan M.J., Tobias,J.W., Slauch,J.M., Hanna,P.C., Collier,J.R. and Mekalanos,J.J. (1995) Antibiotic based selection for bacterial genes that are specifically induced during infection of a host. Proc. Natl Acad. Sci. USA, 92, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa Y. and Fujita,Y. (1988) Purification and characterization of a repressor for the Bacillus subtilis gnt operon. J. Biol. Chem., 263, 13252–13257. [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH—a hierarchic classification of protein domain stucture. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Overath P., Pauli,G. and Schrairer,H.U. (1969) Fatty acid degradation in Escherichia coli. an inducible acyl-CoA synthetase, the mapping of old mutants and isolation of regulatory mutants. Eur. J. Biochem., 7, 559–574. [PubMed] [Google Scholar]
- Pebay-Peyroula E., Rummel,G., Rosenbusch,J.P. and Landau,E.M. (1997) X-ray structure of bacteriorhodopsin at 2.5 Å from microcrystals grown in lipidic cubic phases. Science, 277, 1676–1681. [DOI] [PubMed] [Google Scholar]
- Pellicer M.T., Badia,J., Aguilar,J.T. and Baldoma,L. (1996) glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the Glc regulator protein. J. Bacteriol., 178, 2051–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Raman N. and DiRusso,C.C. (1995) Analysis of acyl coenzyme A binding to the transcription factor FadR and identification of amino acid residues in the carboxyl terminus required for ligand binding. J. Biol. Chem., 270, 1092–1097. [DOI] [PubMed] [Google Scholar]
- Raman N., Black,P.N. and DiRusso,C.C. (1997) Characterization of the fatty acid-responsive transcription factor FadR. J. Biol. Chem., 272, 30645–30650. [DOI] [PubMed] [Google Scholar]
- Rossbach S., Kulpa,D.A., Rossbach,U. and de Bruijn,F.J. (1994) Molecular and genetic characterization of the rhizopine catabolism (mocabrc) genes of Rhizobium meliloti l5-30. Mol. Gen. Genet., 245, 11–24. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander,C. and Schneider,R. (1994) PHD—an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci., 10, 53–60. [DOI] [PubMed] [Google Scholar]
- Sacchettini J.C., Gordon,J.I. and Banaszak,L.J. (1989) Refined apoprotein structure of rat intestinal fatty-acid binding protein produced in Escherichia coli. Proc. Natl Acad. Sci. USA, 86, 7736–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeck F. and Dahl,M.K. (1996) Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J. Bacteriol., 178, 4576–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S.C., Shields,G.C. and Steitz,T.A. (1991) Crystal structure of a CAP–DNA complex—the DNA is bent by 90°. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Schwartz T., Rould,M.A., Lowenhaupt,K., Herbert,A. and Rich,A. (1999) Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science, 284, 1841–1845. [DOI] [PubMed] [Google Scholar]
- Spector M.P., DiRusso,C.C., Pallen,M.J., del Portillo,F.G., Dougan,G. and Finlay,B.B. (1999) The medium-/long chain fatty acyl-CoA dehydrogenase (fadf) gene of Salmonella typhimurium is a phase 1 starvation-stress response locus. Microbiology, 145, 15–31. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL-W—improving the sensitivity of progressive multiple sequence aligment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L. and Rossmann,M.G. (1997) Rotation function calculation with glrf program. Methods Enzymol., 276, 594–611. [PubMed] [Google Scholar]
- van Aalten D.M.F., Knudsen,J., DiRusso,C.C., Kokko,T. and Wierenga,R.K. (2000) Crystallization and X-ray diffraction studies of the fatty acid responsive transcription factor FadR from Escherichia coli. Acta Crystallogr. D, 56, 469–471. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Winter N.S., Bratt,J.M. and Banaszak,L.J. (1993) Crystal structures of holo and apo-cellular retinol-binding protein. II. J. Mol. Biol., 230, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Yoshida K.-I., Fujita,Y. and Sarai,A. (1993) Missense mutations in the Bacillus subtilis gnt repressor that diminish operator binding ability. J. Mol. Biol., 231, 167–174. [DOI] [PubMed] [Google Scholar]
- Yoshida K.-I., Ohmori,H., Miwa,Y. and Fujita,Y. (1995) Bacillus subtilis gnt repressor mutants that diminish gluconate-binding ability. J. Bacteriol., 177, 4813–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Fraenkel,E., Pabo,C.O. and Pavletich,N.P. (1999) Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev., 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]