Abstract
Alphaviruses are enveloped icosahedral viruses that mature by budding at the plasma membrane. According to a prevailing model maturation is driven by binding of membrane protein spikes to a preformed nucleocapsid (NC). The T = 4 geometry of the membrane is thought to be imposed by the NC through one-to-one interactions between spike protomers and capsid proteins (CPs). This model is challenged here by a Semliki Forest virus capsid gene mutant. Its CPs cannot assemble into NCs, or its intermediate structures, due to defective CP–CP interactions. Nevertheless, it can use its horizontal spike–spike interactions on membrane surface and vertical spike–CP interactions to make a particle with correct geometry and protein stoichiometry. Thus, our results highlight the direct role of membrane proteins in organizing the icosahedral conformation of alphaviruses.
Keywords: alphavirus/budding/glycoprotein/horizontal interactions
Introduction
Alphaviruses are simple enveloped viruses with a positive and single stranded RNA genome, a 42S RNA. They have been useful models to study the assembly, structure and functions of a viral envelope (Strauss and Strauss, 1994). Their structural proteins, i.e. the capsid protein (CP) and the (trans)membrane proteins p62 and E1, are translated as a polyprotein from the viral subgenome, a 26S RNA. The CP contains a serine protease activity in its C-terminal part, with which it cleaves itself off from the polyprotein, and a positively charged N-terminal part, with which it associates with 42S RNA when forming a nucleocapsid (NC). The membrane protein subunits are released from each other by signal peptidase in the endoplasmic reticulum. They oligomerize into p62–E1 heterodimers and are transported to the cell surface. When passing through the Golgi complex p62 is cleaved into the transmembrane E2 part and the small peripheral E3 subunit. The final maturation of the alphavirus occurs at the plasma membrane (PM) where the NC becomes enwrapped by a lipid envelope containing the viral membrane proteins (virus budding) (Acheson and Tamm, 1967; Murphy, 1980).
The overall structure of the mature alphavirus has been resolved by cryo-electron microscopic (cryo-EM) analyses and image processing (Vogel et al., 1986; Paredes et al., 1993; Cheng et al., 1995; Fuller et al., 1995; Mancini et al., 2000). Both the membrane proteins and the CPs are organized according to icosahedral symmetry with T = 4. The 240 spike protomers, i.e. the E1–E2 complexes, are grouped into 80 trimeric, corolla-like spikes at the 3- and quasi 3-fold axes. The 240 copies of CPs are clustered into 30 hexameric and 12 pentameric capsomeres, with distinct projections, at the 2- and 5-fold axes. On the internal side of the envelope the spike protomers are engaged in one-to-one interactions with CPs. These are so employed that the protomers of a spike at a 3-fold axis make contacts with CPs in three adjacent hexameric capsomeres and those of a spike at a quasi 3-fold axis contact CPs at adjacent hexameric and pentameric capsomeres. On the external surface of the lipid bilayer the spike protomers extend laterally, forming an enclosing protein shell with horizontal contacts between the spikes. In the NC the capsomere projections rest on an internal shell, which encloses the innermost part of the virion. This shell most likely provides a template for CP interactions that maintain the T = 4 organization of the capsomeres. The structure of the C-terminal, serine protease part of the CP has been resolved at atomic resolution and fits precisely into the EM density of the capsomere projection (Cheng et al., 1995). This suggests that the N-terminal part of the CP mediates the horizontal capsid protein interactions in the NC shell.
According to a prevailing model, alphavirus budding is driven by the binding of spikes to a preformed NC at the PM (Garoff and Simons, 1974; Brown, 1980; Murphy, 1980; Harrison, 1986; Lee et al., 1996). The T = 4 arrangement of the spikes in the viral envelope is thought to be a consequence of the one-to-one binding of spike protomers to CPs in the NC particle (Cheng et al., 1995). This ‘assembly-from-within’ model is supported by the facts that: (i) NC particles are abundant in infected cells; (ii) virus formation requires coexpression of both spike and capsid genes in cells; and (iii) budding can be abrogated by mutations of residues participating in the CP–spike interaction (Acheson and Tamm, 1967; Söderlund, 1973; Suomalainen and Garoff, 1992; Skoging et al., 1996). However, the realization that spikes are engaged in horizontal interactions on the virus surface raises the question about the role of these interactions in budding. Can they support or direct the formation of the T = 4 icosahedral alphavirus particle by ‘assembly-from-without’? This is an intriguing possibility for all icosahedral enveloped viruses.
Our approach to the study of this question has been to analyse the budding ability of Semliki Forest virus (SFV) mutants with defective NC assembly. We have already described two SFV mutants that were budding competent although they assembled only incomplete or intermediate (90S) forms of the NC in the cytoplasm (Forsell et al., 1996). This showed that the preformation of a complete NC (150S) is not required for budding, but it did not prove the role of spike–spike interactions in budding, as the incomplete NCs might still coordinate spikes according to the original model. In the present work we have characterized the assembly and structure of an SFV mutant with an extensive deletion in the N-terminal CP domain. The deletion resulted in the complete ‘knock out’ of all forms of cytoplasmic NC particles. Nevertheless, the mutant could still assemble at the PM and release particles with correct protein composition. These were subjected to structural analysis using cryo-EM. This showed that the mutant particle was organized according to T = 4 icosahedral symmetry. The spike layer appeared intact but the mutant core was lacking the NC shell, which provided horizontal CP interactions in wild-type SFV (SFV-wt). Thus, in the absence of the latter interactions, the spike proteins can still direct the formation of T = 4 particles via their horizontal interactions on the membrane surface and also transmit the corresponding regularity to the core via their vertical interactions with the CPs. This suggests that spike-mediated assembly ‘from without’ plays an important role in alphavirus budding.
Results
Design of mutant
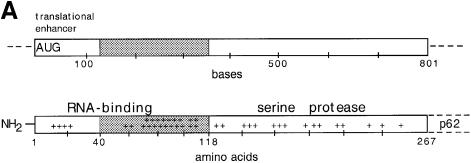
In order to find out whether horizontal spike interactions could drive particle formation we designed an SFV mutant which was expected to be defective in NC assembly but still retain the critical CP–spike interactions. A mutant with a deletion of the gene region encoding residues 40–118 of the CP appeared to be the most suitable. This mutant retained the 5′ coding region of the CP, which constitutes an effective translational enhancer on the viral 26S RNA (Frolov and Schlesinger, 1994; Sjöberg et al., 1994), and the region encoding the C-terminal domain of the CP (residues 119–267), which contains the serine protease required for CP–p62 cleavage during polyprotein synthesis, as well as the site for spike binding (Figure 1A). On the other hand, most of the region encoding the N-terminal domain of the capsid protein, which mediates binding of the CP to the viral RNA and probably also CP interactions in the NC shell, was deleted in the mutant. The deletion was made in the plasmid pSP6-SFV4 and the resulting mutant was called pSFV-CΔ40–118. For our expression analyses replication-competent SFV-CΔ40–118 RNA was transcribed in vitro from pSFV-CΔ40–118 and introduced by electroporation into BHK-21 cells. In some experiments the mutant RNA was introduced into cells by virus infection using SFV vectors.

Fig. 1. The deletion in SFV-CΔ40–118 and expression analysis. (A) Upper and lower panels show functional and structural regions of the capsid gene and CP, respectively. The approximate location of residues with positively charged side chains are indicated (+) in lower panel. The deletion is indicated by grey shading. (B) SDS–PAGE analyses of cell-associated and released viral proteins. Two pairs of cell cultures were transfected with SFV-wt or SFV-CΔ40–118 RNA. The cells were incubated for 6.5 h, pulse-labelled with [35S]methionine for 30 min and then chased for 15 or 180 min. Chase media were collected and cells were lysed with NP-40. Samples of cell lysates (C), pelleted particles from the chase media (P), TCA precipitates of unfractionated media (total, T) and corresponding supernatants (S) were analysed on a 10% gel under reducing conditions. The molecular weight standards (St) were myosin (220 kDa), phosphorylase b (97 kDa), bovine serum albumin (66 kDa), ovalbumin (46 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (21 kDa) and lysozyme (14 kDa). The figure represents a fluorography of the gel.
Ineffective release of mutant virus despite efficient synthesis and processing of structural proteins
Cell cultures were transfected with SFV-wt and mutant RNA in parallel, pulse-labelled with [35S]methionine and the synthesis and fate of the viral proteins followed. The results showed that the mutant RNA directed synthesis of correctly sized membrane proteins E1 and p62 (Figure 1B, lanes 2 and 7). In addition, a 97 kDa protein was seen in both samples. This is an uncleaved cytoplasmic p62–E1 polyprotein that is produced as a by-product during viral protein synthesis in the cell (Garoff et al., 1978). The CP band that migrated as a 33 kDa protein in the SFV-wt sample was absent in the mutant. Instead, a smaller protein with an apparent mol. wt of 23 kDa was seen. This corresponded to a CP with a deletion of 78 amino acid residues, i.e. the CΔ40–118 protein. During chase of SFV-wt transfected cells, the p62 precursor protein was cleaved into E2 and E3 and labelled proteins appeared in virus particles that were released into the medium (Figure 1B, lanes 3–6). In cells transfected with the mutant RNA the p62 precursor protein also underwent maturation cleavage but very few labelled proteins were released as particles (Figure 1B, lanes 8–11). Note that the gel analyses shown in Figure 1B has been carried out under reducing conditions at which E1 and E2 comigrate. Separate identification of E2 was done using unreduced samples (data not shown). Also note that the host protein synthesis was virtually shut off in both SFV-wt and mutant RNA transfected cells and that mutant and SFV-wt proteins were labelled with a similar intensity. Altogether we conclude that the mutant RNA directed a high-level synthesis of structural proteins that underwent normal processing in the cells. However, the release of mutant virus was significantly compromised.
Isolation of SFV-CΔ40–118 particles from medium and analyses of protein content
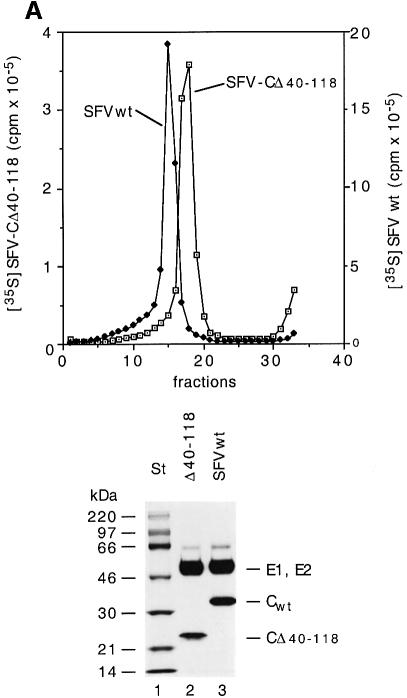
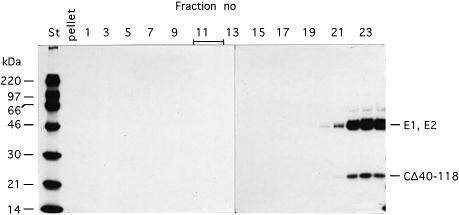
Our preliminary pulse–chase analyses suggested that some particles were released from cells transfected with the SFV-CΔ40–118 RNA. This was investigated thoroughly using larger cultures of BHK-21 cells. These were infected with SFV vectors carrying the mutant RNA and long-term labelled with [35S]methionine. Particles were recovered from the culture medium by pelleting and purified by sedimentation in a sucrose gradient. As a control, SFV-wt was produced and purified in parallel. The vector infected cells were found to produce particles that sedimented as a distinct band slightly more slowly than SFV-wt (Figure 2A, upper panel). The yield per infected cell was ∼5% of that of SFV-wt.
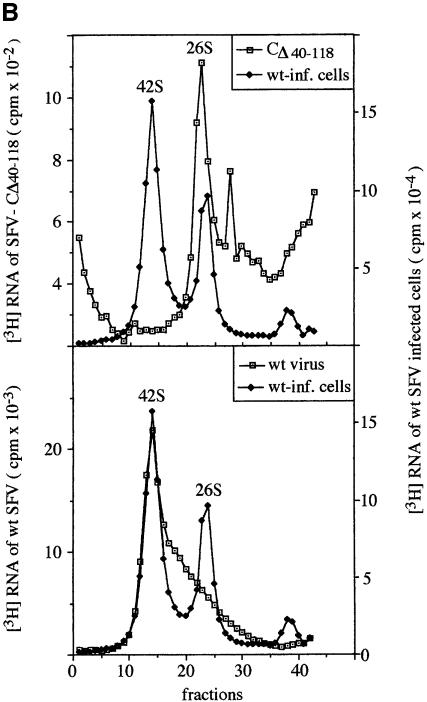
Fig. 2. Purification and composition of CΔ40–118 particles. (A) Sedimentation and protein composition. Four 162 cm2 cell cultures were infected with SFV vectors carrying mutant RNA (m.o.i. = 10) and one with SFV-wt at the same m.o.i. The cultures were labelled with [35S]methionine for 15 h. Particles were collected from the media by pelleting, resuspended and sedimentated in a linear 15–30% (w/w) sucrose gradient run at RCFmax = 150 000 g for 3 h (upper panel). The 35S radioactivity in each fraction was measured. Note that different scales are used for SFV-wt (filled squares, right scale) and SFV-CΔ40–118 (open squares, left scale). Fractions with peak radioactivity were analysed by SDS–PAGE (12%) under reducing conditions (lower panel). (B) RNA composition. Twenty 162 cm2 cell cultures were infected with SFV vectors carrying mutant RNA (m.o.i. = 10) and five cultures with SFV-wt at the same m.o.i. The cultures were labelled with [3H]uridine for 15 h. SFV-wt and SFV-CΔ40–118 particles were then purified by sedimentation in sucrose gradients as described above. A sample of each virus preparation was incubated with SDS and separated on a 15–30% (w/w) sucrose gradient for analyses of 3H-labelled RNA. Fractionation and quantification of radioactivity were as described above. Upper and lower panels show analyses of RNA from the mutant and SFV-wt, respectively. Included in both graphs are analyses of 3H-labelled RNA extracted from SFV-wt infected cells. The latter shows [3H]RNA peaks for the genomic 42S and the subgenomic 26S RNAs. The 3H c.p.m. scale for the control is on the right side in both panels.
The long-term labelling with [35S]methionine ensured a steady-state labelling of the viral proteins in the infected cells. It was therefore possible to estimate the stoichiometry of viral proteins in particles on the basis of their radioactivity. For this purpose the structural proteins of isolated SFV-CΔ40–118 particles and SFV-wt were separated by SDS–PAGE (Figure 2A, lower panel) and the radioactivity measured by a phosphoimager. The values were used to calculate the molar ratio of membrane proteins to CΔ40–118 or CPs in the mutant and the SFV-wt particle, respectively. The results from three experiments showed that the ratio was 1.1 ± 0.17 SD for the SFV-wt and 1.03 ± 0.06 SD for the mutant particle. This suggests that a full complement (240 copies) of CΔ40–118 protein is incorporated into the mutant particle.
The SFV-CΔ40–118 particle contains 26S mRNA
In the CΔ40–118 protein most of the RNA binding regions have been deleted. Therefore, an interesting question was whether the SFV-CΔ40–118 particles incorporated RNA and, in case they did, was it the 26S RNA subgenome, the 42S RNA genome or both? In order to produce mutant particles for RNA analyses, BHK-21 cell cultures were infected with SFV vectors carrying the mutant RNA and labelled with [3H]uridine. Particles were isolated as in the previous experiment, solubilized by SDS and the RNA analysed by sedimentation in a sucrose gradient. As controls, labelled RNA from SFV-wt particles and SFV-wt infected cells were run in parallel. The sedimentation of mutant RNA resulted in one major RNA species at the position of the 26S RNA and several smaller ones (Figure 2B, upper panel). Part of the latter might represent degradation products of the 26S RNA. It should be noted that the 42S RNA was not detected in the mutant virus sample. In contrast, RNA from SFV-wt particles migrated as one major species of RNA at the position of the 42S RNA (Figure 2B, lower panel). This 42S band was followed by a trailing edge of more slowly migrating RNA species, which might represent degraded 42S RNA. There was no distinct peak at the position of the 26S RNA.
It was important to know how much 26S RNA was incorporated per mutant virus particle. Thus, we separated the proteins in the 3H-labelled mutant virus by SDS–PAGE and stained the gel with Coomassie Blue. The amount of protein was estimated by comparison with a series of albumin samples with known protein concentration. The ratio of RNA (3H c.p.m.) to protein (µg) was calculated for the mutant virus and related to that of SFV-wt. The latter had been measured in the same way using the 3H-labelled SFV-wt preparation. The calculation showed that the mutant contained 0.33 times the RNA of SFV-wt (0.30 and 0.37 in two experiments). As the 42S RNA is approximately three times longer than the 26S RNA (Takkinen et al., 1991), our result suggests that on average one mutant particle contains one 26S RNA molecule. To confirm the low level of 42S viral genomes in the mutant virus we measured the specific infectivities of 35S-labelled mutant and SFV-wt preparations by plaque assay. The mutant virus and the SFV-wt preparation contained 1.3 and 1800 p.f.u. per 35S c.p.m., respectively.
Structure of the SFV-CΔ40–118 particle
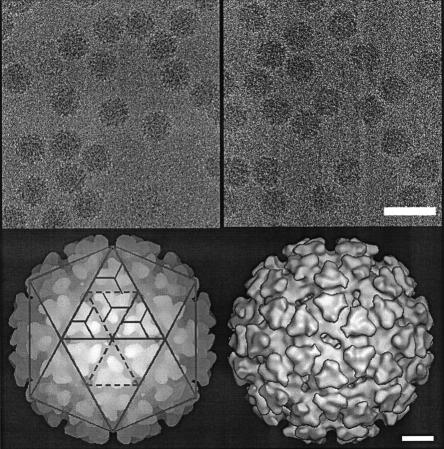
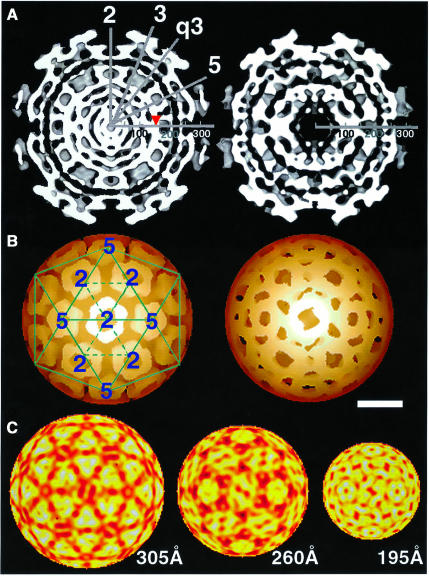
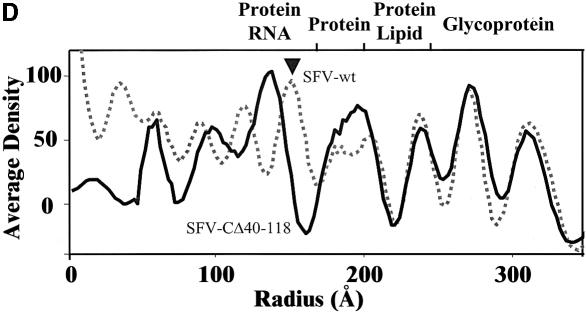
Samples of sucrose gradient purified mutant virus and SFV-wt were used for structural studies by cryo-EM (Figures 3 and 4). Vitrified mutant particles appeared intact without significant difference from SFV-wt particles (Figure 3, upper panel). The lower left panel shows a surface-shaded representation of SFV-wt along a 2-fold axis with the trimeric spikes organized in a T = 4 lattice as described (Cheng et al., 1995). An examination of the reconstructed mutant reveals a similar surface organization (Figure 3, lower right panel). Trimeric spikes can be localized at positions of icosahedral and local 3-fold axes. Like the spikes of SFV-wt, those of the mutant form corolla-like projections and horizontally extending shell domains that cover almost the whole particle surface. An equatorial cross-section of the SFV-wt reconstruction shows the densities of the two phospholipid headgroup layers in the lipid bilayer at radii 250 and 210 Å (Figure 4A, left panel). Underneath, the serine protease domains of the CPs are seen as radially distributed densities that project outward from a continuous dense layer at radius 170 Å. This is referred to as the NC shell. In the equatorial cross-section of the mutant particle the lipid bilayer can be seen in the same location as in SFV-wt (Figure 4A, right panel). However, neither the organized projection of the C proteins nor the supporting NC shell can be recognized underneath. Instead, there is a dense layer that merges with the inner phospholipid headgroup layer and a broad low-density region inside. The dense layer probably corresponds to the serine protease domains of the CΔ40–118 proteins, which contact the spike protomers below the lipid membrane. This was supported by similar mass estimates in the region between radii 170 and 205 Å in SFV-wt and mutant particles (data not shown). The low-density region inside replaces most of the NC shell region in the SFV-wt, thus suggesting that residues 40–118 of the CP contribute to this NC structure. Internal to the density gap in the mutant there is a bulky region of density that probably corresponds to the 26S RNA. Bridges of densities connecting the latter region with the serine protease layer below the lipid membrane are also evident. One-dimensional radial density profiles of the mutant and SFV-wt are shown in Figure 4D. From the particle periphery inwards the density peaks corresponding to the external spike glycoprotein and the lipid membrane are similar in SFV-wt and mutant particles. Further in, in the mutant, the peak corresponding to the inner phospholipid-headgroup layer and that of the protein layer inside (the serine protease domains) seem to have merged forming one wide peak. Moreover, a broad low-density region in the mutant replaces most of the protein/RNA peak corresponding to the NC shell in the SFV-wt, again indicating that residues 40–118 of the CP participate in formation of the shell structure. Figure 4B shows reconstructions of SFV-wt and mutant NCs. The hexameric and pentameric clustering of the SFV-wt CP projections are clearly seen at the 2- and 5-fold axes in SFV-wt (left panel). In the mutant, individual CΔ40–118 projections cannot be distinguished (right panel). Still, the general organization in hexameric and pentameric clusters is moderately preserved. When comparing the density distribution at different radii (Figure 4C), it is possible to follow how the spike protomers of the mutant particle separate from their trimeric clusters (305 Å) and re organize above the lipid bilayer (260 Å) in a way that imitates the arrangement of the CΔ40–118 proteins underneath (195 Å). This suggests that each spike protein interacts with one CΔ40–118 protein in the particle.
Fig. 3. Reconstructions of wt and mutant particles. Upper panel: cryo-micrographs of vitrified SFV-wt and SFV-CΔ40–118 particles. The images were recorded with an electron dose rate <10 e–/Å2/s and at 1 and 3 µm underfocus at the same area (only data set of 3 µm defocus is shown). The average radius of both particles is ∼345 Å. Lower panel: depth-cued and surface-shaded particle reconstructions of SFV-wt (left) and SFV-CΔ40–118 (right). The reconstructions are viewed along an icosahedral 2-fold axis. SFV-wt is overlaid with a T = 4 lattice to illustrate the spike positions relative to 5-, 3- and 2-fold symmetry axes. The facets represent positions of restricted and local 3-fold axes in two icosahedral faces. There are 80 trimeric spikes correspondingly seen in SFV-wt and SFV-CΔ40–118 particles. Enantio morphic information was assigned as described (Cheng et al., 1995).
Fig. 4. Comparative imaging of radial density distributions. (A) Equatorial cross-sections of SFV-wt (left) and SFV-CΔ40–118 (right) reconstructions along a 2-fold axis. High densities are white. Symmetry axes are indicated by numbered lines (e.g. ‘3’ and ‘q3’ are icosahedral and quasi 3-fold axes, respectively). The radial axis is marked in Å where the lipid membrane is located between radii 210 and 250 Å. The arrowhead pointed to the radial axis in SFV-wt indicates the NC shell. This supports the NC projections, seen as radially distributed densities between the NC shell and the lipid membrane. (B) Depth-cued representations of SFV-wt and mutant NC. The T = 4 lattice is superimposed on SFV-wt NC to illustrate the organization of the capsomeres. In the mutant, though compact capsomeres with distinct CP projections cannot be distinguished, the nature of T = 4 quasi-equivalence can still be uniquely observed with hexagonal and pentagonal clusters allocated at icosahedral 2-fold and 5-fold axes, respectively. Note that difference imaging with wt NC was carried out, but it gave, apart from the missing shell (seen in A), no additional meaningful information (data not shown). This can be explained by the less ordered positioning of the CΔ40–118 proteins in the mutant particle. Bar = 100 Å. (C) Projected density distributions at specific radii of the mutant to illustrate trimeric spike stems (305 Å) and the juxtaposition of spike protomers (260 Å) and CΔ40–118 proteins (195 Å) at each side of the lipid bilayer. High-to-low densities are indicated with a yellow-to-red scaled colour scheme. (D) One-dimensional density profile of SFV-wt (dotted line) and mutant (continuous line) particles. Major components at each radial bin are indicated for SFV-wt according to results from neutron scattering (Jacrot, 1987). Radial positions of the glycoprotein spikes and the phospholipid headgroups of inner and outer lipid bilayer leaflets agree well between the SFV-wt and the mutant. However, the mutant lacks density corresponding to the NC shell in SFV-wt (indicated by arrowhead).
The NC of the mutant virus is unstable without an envelope
A typical feature of SFV-wt is that its NC stays intact after solubilization of its membrane with a mild detergent like Nonidet P-40 (NP-40) (Kääriäinen et al., 1969). To find out whether this was also the case with the mutant virus we treated a sample with NP-40 and analysed the mixture for NC particles by sedimentation in a sucrose gradient. Surprisingly, we found that all CΔ40–118 proteins remained in the top fractions of the gradient together with the solubilized membrane proteins (Figure 5). This was also the case when virus solubilization and sedimentation were carried out in the presence of an RNase inhibitor (data not shown). These results suggested that the NC of the mutant virus is a labile structure that can only be maintained inside the particle, probably through effective CΔ40–118 protein–spike interactions.
Fig. 5. Stability analysis of the NC in the SFV-CΔ40–118. A sample of 35S-labelled SFV-CΔ40–118 particles, isolated as described in the legend to Figure 2A, was incubated in 1% NP-40 and then analysed by sedimentation in a 15–30% (w/w) sucrose gradient run at RCFmax = 288 000 g for 2 h. Gradient fractions were analysed by SDS–PAGE (12%) under reducing conditions. A sample of 35S-labelled SFV-wt was similarly treated and analysed. The migration of the NC of the SFV-wt is indicated by a bar at the top.
The CΔ40–118 proteins cannot assemble into NCs in cells
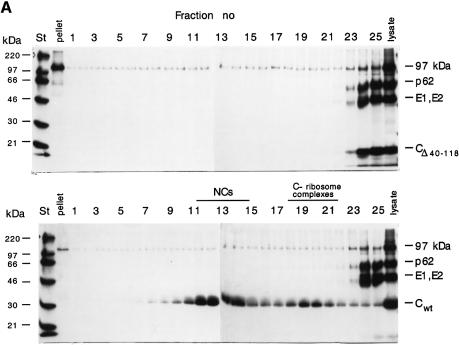
Newly synthesized CPs bind to the ribosomes (large subunit, 60S) before they assemble with 42S RNA into NCs (150S) (Söderlund and Ulmanen, 1977). When we analysed an NP-40 lysate of cells transfected with CΔ40–118 RNA by sedimentation in a sucrose gradient we could not detect any large complexes. Instead, all CΔ40–118 proteins remained at the top of the gradient with the membrane proteins (Figure 6A, upper panel). Control analysis of a lysate of cells transfected with SFV-wt RNA showed that most CP was present in NCs and some in ribosome complexes (Figure 6A, lower panel). The defective NC assembly of the mutant was confirmed by EM analysis of sections of infected cells. These did not reveal NC structures typically seen in SFV-wt infected cells (data not shown).
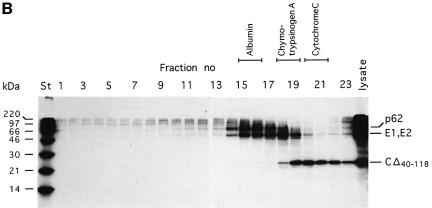
Fig. 6. Analysis of cell-associated CΔ40–118 proteins. (A) Sedimentation analysis of large complexes. Cells were transfected with SFV-CΔ40–118 or SFV-wt RNA, pulse-labelled with [35S]methionine for 30 min and chased for 15 min. NP-40 lysates were prepared and analysed in 15–30% (w/w) sucrose gradients run at RCFmax = 288 000 g for 2 h. A sample from each gradient fraction was analysed by SDS–PAGE (10%) under reducing conditions. Results with mutant and SFV-wt are shown in upper and lower panels, respectively. (B) Sedimentation analysis of small complexes. A lysate sample of cells transfected with mutant RNA was analysed in a 5–20% (w/w) sucrose gradient run at RCFmax = 260 000 g for 24 h. Samples from gradient fractions were analysed on a 12% gel under reducing conditions. The sedimentation of several monomeric standard proteins, run under identical conditions, is indicated at the top.
The CΔ40–118 proteins behave as monomers in cell and virus lysates
The CΔ40–118 protein could potentially form oligomers, e.g. capsomer-like structures with five or six monomers. The corresponding interaction sites are located in the serine protease domain, which is left intact in the CΔ40–118 protein. This possibility was studied in an NP-40 cell lysate by comparing the sedimentation of CΔ40–118 with that of several monomeric standard proteins. The results showed that the CΔ40–118 protein sedimented slightly more slowly than chymotrypsinogen A (25 kDa) (Figure 6B). This suggests that it is monomeric in an NP-40 cell lysate. A similar analysis was also done with NP-40 solubilized SFV-CΔ40–118 particles (data not shown). The result suggested that the CΔ40–118 proteins were released as monomers also in a virus lysate.
Discussion
The deletion in SFV-CΔ40–118 was designed to inactivate the horizontal CP interactions in the NC shell and thereby prevent assembly of NC particles in infected cells. The CP template for this interaction should reside in the N-terminal domain of CP (residues 1–118) as the rest of the CP, i.e. the C-terminal serine protease domain (residues 119–267) forms the CP projections (Cheng et al., 1995). The N-terminal CP domain contains two conserved regions spanning residues 39–53 and 104–118, which could be involved in such an interaction (Wengler et al., 1992; Tellinghuisen et al., 1999). Both of these regions have been deleted in the mutant. The first third of the N-terminal domain, i.e. the peptide spanning residues 1–39, was left intact in the mutant because the corresponding RNA sequence functions as an important translational enhancer in SFV gene expression (Sjöberg et al., 1994). This region could potentially participate in horizontal CP interactions. However, we considered it unlikely for two reasons. First, this region is not conserved among alphaviruses. Secondly, we have described an SFV mutant with a deletion of amino acid residues 11–29 of the CP and shown that it formed intracellular NCs although the expression level was significantly suppressed (Forsell et al., 1995). This shows that this region of the CP is not essential for CP interactions. Thus, it was expected that the CΔ40–118 proteins could not establish the horizontal interactions required for formation of an icosahedral NC particle. In agreement with this, our studies on SFV-CΔ40–118 assembly in infected cells showed that CΔ40–118 proteins were unable to assemble into intracellular NC particles. Indeed, the cell-associated CΔ40–118 proteins were found to be monomeric in an NP-40 lysate. Despite this the CΔ40–118 proteins were apparently able to bind to spike protomers at the PM and induce particle formation by budding. The binding of CΔ40–118 proteins to the spike protomers seemed to require simultaneous RNA interactions as all particles contained one 26S RNA molecule.
The structural analyses demonstrated that the SFV-CΔ40–118 particles contained an SFV-wt-like spike layer but a strikingly dissimilar interior. In particular, the mutant NC was lacking the shell region. As an apparent consequence of this, the serine protease units of the CΔ40–118 proteins could not organize themselves into the distinct pentameric and hexameric projections characteristic of SFV-wt, but formed a less-organized protein layer below the membrane. Nevertheless, it was still conceivable to localize the CΔ40–118 proteins in regular positions corres ponding to the tails of the spike protomers underneath the membrane. How is it possible for the CΔ40–118 proteins to maintain this order in the absence of horizontal interactions in the shell region? The most apparent interactions that can stabilize this regularity are the ones between spike tails and the serine protease domains of the CΔ40–118 proteins. It should also be possible for the latter domains to undergo capsomere subunit interactions. However, the facts that the CΔ40–118 proteins dissociate into monomers when the surrounding membrane of the particle is solubilized with a mild detergent, and that the deleted CPs cannot adopt the compact shape of the wild-type capsomeres suggest that the latter interactions are very weak or non-functional. In any case capsomere subunit interactions alone cannot explain the higher order structure of the mutant core. Neither can it be explained by interactions between CΔ40–118 proteins and the 26S RNA in the particle. Therefore, we conclude that the regularity of the mutant core is predominantly maintained by one-to-one interactions between the CΔ40–118 proteins and the spike protomers in the T = 4 icosahedral envelope.
The important question is how the mutant particle has been assembled. The apparent incapability of the CΔ40–118 protein to support icosahedral interactions rules out an NC-controlled budding model. Instead, one must assume that the assembly of the mutant particle must be governed by the horizontal interactions of the spike proteins. The role of the CΔ40–118 protein is probably restricted to triggering the spike protomers to establish these interactions. The triggering could mean that the spike protomers simply get a reduced dimensional mobility in the membrane as a result of CΔ40–118 binding, a change in conformation or both. Thus, budding of the mutant virus is most likely driven by the organization of the spikes into a T = 4 icosahedral lattice in the PM of the infected cell. The CΔ40–118 proteins, with the bound 26S RNA, will follow into the particle in a corresponding geometry via their one-to-one interactions with the spike protomers. Budding of SFV-CΔ40–118 according to this model is depicted in Figure 7, right panel. The fact that the production of SFV-CΔ40–118 was only 5% of that of SFV-wt suggests that budding of SFV-wt cannot be driven by horizontal spike protein interactions alone but, that in this case, they probably cooperate with spike–NC interactions. This would mean that particle formation in the alphavirus is secured by two overlapping activities of its structural proteins (Figure 7, left panel).
Fig. 7. Budding of SFV-wt and SFV-CΔ40–118. Budding of SFV-wt by vertical spike–NC and horizontal spike–spike interactions is shown in the left panel. The right panel shows budding of SFV-CΔ40–118 by horizontal spike–spike interactions only. In the latter case an NC particle cannot be formed and thus, not used as template for spike binding. The role of the CΔ40–118 protein in budding is restricted to triggering the spike to undergo horizontal interactions probably with the help of 26S RNA.
The finding that 26S RNA but not 42S RNA was incorporated into the SFV-CΔ40–118 particles is intriguing. SFV-wt incorporates only 42S RNA, supposedly via the specific recognition of the 42S RNA encapsidation signal by the CP (Weiss et al., 1989). The signal-binding region has been tentatively mapped to the C-terminal part of the N-terminal domain of the CP (Geigenmüller-Gnirke et al., 1993). Deletion of this region results in the loss of selectivity in RNA encapsidation, i.e. both 42 and 26S RNAs are incorporated (Owen and Kuhn, 1996). In SFV-CΔ40–118 the entire signal-binding region has been removed and one would therefore expect that both RNAs, and not only 26S RNA, are incorporated. Control experiments showed that the bias in RNA incorporation was not due to a distorted RNA ratio in the infected cell; here the two RNA species were approximately equally abundant. One possible explanation is that the remaining residues with positively charged side chains in the CΔ40–118 protein suffice for condensing a 26S RNA molecule into the mutant particle but not a 42S RNA. The CΔ40–118 protein contains only five out of the 30 residues with positively charged side chains that are present in the N-terminal domain of the SFV-wt CP.
In conclusion, our results highlight the direct role of membrane proteins in organizing an icosahedral virus. This assembly-from-without pathway contrasts the general view that the conformation of an icosahedral virus particle is determined mainly, with or without the aid of nucleic acid, through the horizontal interactions between CPs (assembly-from-within) (Rossmann and Johnson, 1989). A role of horizontal membrane protein interactions in alphavirus budding was originally suggested by von Bonsdorff et al. (1978). This was based on their observation of a hexagonal spike array in NC-free membrane sheets of Sindbis virus. Most likely assembly-from-without with membrane proteins is not going to be a mechanism specific only for alphaviruses but a general one, also used by other icosahedral enveloped viruses (e.g. von Bonsdorff and Pettersson, 1975). One can also speculate that there exist cellular transmembrane glycoproteins that can direct formation of intracellular membrane vesicles by making icosahedral interactions, similar to those of the alphavirus spikes. So far, there are only examples of cytoplasmic proteins that drive vesicle budding through the formation of a membrane-enclosing protein cage (Smith et al., 1998).
Materials and methods
Materials
Media and reagents for cell culture were purchased from Gibco (Paisley, UK). [35S]methionine (1000 Ci/mmol; SJ1515) was from Amersham International (Amersham, UK) and [3H]uridine (1000 µCi/mmol; NET-367) from Du Pont-NEN Products (Boston, MA). SDS was from Bio-Rad (Hercules, CA), NP-40 from Fluka Chemie AG (Buchs, Switzerland), sucrose was from J.T.Baker (Deventer, Holland) and actinomycin D from Sigma Biochemicals (St Louis, MO). [14C]methylated protein molecular weight markers (Rainbow™) were from Amersham International and rRNasin Ribonuclease Inhibitor from Promega (Madison, WI).
Cells and viruses
BHK-21 cells (clone 13), (American Type Culture Collection, Rockville, MD) were grown in BHK medium (Glasgow minimum essential medium, MEM) supplemented with 5% fetal calf serum (FCS), 10% tryptose phosphate broth, 2 mM glutamine and 20 mM HEPES. SFV-wt was derived from pSP6-SFV4 and was propagated in BHK-21 cells as described (Kääriäinen and Gomatos, 1969; Wahlberg et al., 1989; Liljeström and Garoff, 1991).
Construction of pSFV-CΔ40–118
The plasmid pSFV-CΔ40–118, which contains a deletion in the capsid gene encompassing codons for amino acids 40–118, was constructed by fusion–PCR using six different primers and an SFV engineering plasmid pGEM-7Z-SFV as template (H.Garoff, unpublished). First a 201 bp fragment containing the deletion in the capsid gene was synthesized. The primers used were: 5′-CGGCGGTCCTAGATTGGTG-3′ (nucleotides 751–769 in capsid gene encompassing a NarI site) and 5′-GTGTTTGACTTCGAAGATACA*GGCCTGGAAGTCGGGGACGAC-3′ (nucleotides 1188–1168 and 930–910 in capsid gene). The asterisk indicates the site of the deletion. Secondly a 356 bp fragment was made. The primers used were: 5′-TGTATCTTCGAAGTCAAACAC-3′ (nucleotides 1168–188 in capsid gene) and 5′-CCGCCAGGACGATAGC-3′ (nucleotides 1522–1507 in capsid gene encompassing a NheI site). The overlapping PCR fragments were joined and amplified in a third PCR using the primers 5′-CACCATGAATTACATCCCTAC-3′ and 5′-AGGTACTACTTGCTCGATTTCTTG-3′. The final PCR product was digested with NarI and NheI and used to replace the wt NarI–NheI fragment in the capsid gene of pSP6-SFV4. The construct was verified by DNA sequencing.
RNA transcription in vitro and transfection of cells
Replication and transcription competent SFV-wt RNA vectors were synthesized in vitro from SpeI-linearized plasmids using SP6 RNA polymerase and introduced into 107 BHK-21 cells by electroporation as described (Liljeström et al., 1991). Electroporated cells were suspended in 15 ml of BHK medium, plated in 35 mm dishes (Nunc A/S, Roskilde, Denmark) and further incubated. Packaging of SFV-CΔ40–118 RNA into infectious SFV vectors was done by cotransfection of BHK-21 cells with mutant and helper RNA (Helper 1) as described (Liljeström and Garoff, 1991).
Infection of cells
BHK-21 cell monolayers were infected with SFV-wt or SFV vectors at a m.o.i. of 10 in MEM. After 1 h absorption at 37°C the medium was replaced with fresh MEM and cells were further incubated for various lengths of time.
Metabolic labelling of cells
Cells were starved in methionine-free MEM for 30 min and pulsed in the same medium containing 100 µCi/ml of [35S]methionine for different times. After washing twice with MEM containing 750 mg/l methionine the cells were chased for various times with MEM containing 150 mg/l methionine. Alternatively, cells were long-term labelled with [35S]methionine (0.033 mCi/ml) in MEM containing 1.5 mg/l methionine or with [3H]uridine (3.3 µCi/ml) in Eagle’s minimal essential medium. Infected cultures were also incubated with [3H]uridine in the presence of actinomycin D (1 µg/ml) from 2.5 h post infection (p.i.) to 8 h p.i. to label specifically viral 26S and 42S RNAs in the cells.
Preparation of cell lysate
Cells were rinsed with ice-cold phosphate-buffered saline (PBS, with MgCl2 and CaCl2) and lysed in a buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 10 mM iodoacetamide, 2.5 mM phenylmethylsulfonyl fluoride, 2 µg aprotinin/ml, 5 µg antipain/ml, 0.5 µg leupeptin/ml and 0.7 µg pepstatin/ml. The lysate was cleared by centrifugation in an Eppendorf bench top centrifuge (6000 r.p.m. for 5 min at 4°C).
Recovery of virus particles from culture medium
Virus particles were recovered by pelleting either through 100 µl of 10% (w/w) sucrose cushion in TNE (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EDTA) in a JA 18.1 rotor (Beckman) run at 17 000 r.p.m. for 2 h at 4°C, or a 4 ml 10% (w/w) sucrose cushion in TNE using a SW28 rotor (Beckman, Palo Alto, CA) run at 25 000 r.p.m. for 90 min at 4°C. In the former case the virus pellet was directly taken up into gel-loading buffer and in the latter case it was resuspended in 100 µl of TNE for further purification of particles by velocity sedimentation (see below). The resuspension was done by incubating the mixture on ice for 10 h and then performing gentle pipetting.
Sedimentation analysis of virus and viral subcomponents
For detection of intracellular and viral NCs we used a 15–30% (w/w) linear sucrose gradient in TNE containing 0.1% NP-40, which was centrifuged in an SW41 (Beckman) rotor at 40 000 r.p.m. at 4°C for 2 h. In some experiments the gradient buffer also contained 6 U/ml recombinant ribonuclease inhibitor (rRNasin) and 5 mM dithiothreitol (DTT). The gradient was fractionated from the bottom into 500 µl fractions. To detect small protein oligomers we used a 5–20% (w/w) linear sucrose gradient in TNE containing 0.1% NP-40, which was run at 39 000 r.p.m. at 4°C in an SW41 rotor for 24 h. As standards we used albumin (67 kDa), chymotrypsinogen A (25 kDa) and cytochrome C (13 kDa). Virus particles (in suspension) were isolated by sedimentation in a 15–30% (w/w) linear sucrose gradient in TNE, which was run in an SW28.1 rotor at 28 000 r.p.m. at 4°C for 3 h.
RNA analysis
[3H]uridine-labelled virus was incubated in 2% SDS in TE (10 mM Tris–HCl pH 7.4 and 1 mM EDTA), heated for 2 min at 70°C, chilled to RT (23°C) and loaded directly on top of a 15–30% (w/w) sucrose gradient in 0.2% SDS. The gradient was run in an SW41 rotor at 40 000 r.p.m. and 23°C for 6.5 h and then fractionated from the bottom. The fractions were analysed for 3H radioactivity by scintillation counting. 3H-labeled viral 26S and 42S RNAs were isolated from infected cells using the TrizolTM system as described by the manufacturer (Gibco) and dissolved in 2% SDS.
Cryo-EM and image processing
Low dose images (<10 e–/Å2) were recorded at a magnification of 28 000× using a Philips CM120 electron microscope equipped with a Gatan 626DH cryo-transfer system. For the mutant particle, each specimen area was exposed twice as focal pair with 1 and 3 µm underfocus to facilitate the image selection and the orientation alignment (Cheng et al., 1992). The selected micrographs were digitized on Zeiss Phodis scanner with step size corresponding to 5 Å spacing per pixel at the specimen. The individual particle images were boxed out and analysed based on icosahedral symmetry (Cheng et al., 1994; Baker and Cheng, 1996). The data from the high defocus images were processed first to get a stable 3D density map with low resolution. The refinement was continued with the data from the close defocus images as described previously (Xing et al., 2000). To be free from model bias phase residues were evaluated across selected images. By minimizing this phase deviation, orientation parameters of individual images were further fine-tuned based on the consistency among the images. This assessed not only the quality of individual data images, but also the consistency of the phase, between the low and the high spatial frequency, when it was extended progressively at higher resolution (Cheng, 2000). Nevertheless, the real space comparison between raw images and corresponding model projections was used to exclude particles with significant size variation (Cheng et al., 1995). The resolution of refinement was extended to 24 Å, with reliability monitored by R-factors and Fourier-ring phase residues, and was probably limited by the less ordered positioning of the CΔ40–118 proteins. The best set of 89 and 54 images out of 427 (20%) wt and 324 (17%) mutant images, respectively, were combined to produce a 3D Fourier transform, low-pass-filtered with the same effective 1/26 Å–1 spatial frequency. The 3D density maps were produced by an inverse Fourier–Bessel method (Crowther et al., 1970). The completeness of the data sampling in the reconstruction was verified by eigenvalue spectra giving all eigenvalues >1 or 10. For the surface rendering, the 3D reconstruction was contoured at the value that represented the volume of the mass density (Xing et al., 1999). Far limited from the first zero-cross of microscope contrast transfer function, no significant difference of density distribution was observed after the attenuation function was systematically deconvoluted within a range of defocus values (Cheng et al., 1994).
Other methods
SDS–PAGE was performed as described (Wahlberg et al., 1989). The proteins on gels were visualized by staining with Coomassie Blue or processed for fluorography (Suomalainen et al., 1990). The radioactivity in protein bands was determined by a phosphoimager (Fuji, Bas 2000). For solubilization of viral membranes, 35S-labelled virus (∼1 µg) was incubated in a buffer containing 25 mM Tris–HCl pH 7.4, 50 mM NaCl, 0.25 mM EDTA and 1% NP-40 for 10 min on ice. In some experiments the solubilization buffer contained 0.6 U/µl rRNasin and 5 mM DTT in addition. Trichloroacetic acid (TCA) precipitation was carried out in 10% TCA for 2 h on ice.
Acknowledgments
Acknowledgements
We thank S.Kan and L.Haag for stimulating discussions and technical assistance, L.Rinnevuo for excellent cell cultures and technical assistance, M.Sjöberg for critical reading of the manuscript and I.Sigurdson for typing. The study was supported by grants from the EU (FMRX-CT-96-0004) and NFR (B-AA/BU 09353-312, -313, -314) to H.G. and by grants from the NFR (11691), MFR (12175) and Structural Biology Network in Sweden to R.H.C.
References
- Acheson N.H. and Tamm,I. (1967) Replication of Semliki Forest virus: an electron microscopic study. Virology, 32, 128–143. [DOI] [PubMed] [Google Scholar]
- Baker T.S. and Cheng,R.H. (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol., 116, 120–130. [DOI] [PubMed] [Google Scholar]
- Brown D. (1980) The togaviruses: biology, structure and replication. In Schlesinger,R.W. (ed.), The Assembly of Alphaviruses. Academic Press, New York, NY, pp. 473–501. [Google Scholar]
- Cheng R.H. (2000) Visualization of virus–host interactions. In Enquist,B., Johnsson,L. and Nieminen,R.M. (eds), Simulation and Visualization on the Grid. Springer-Verlag, New York, NY, pp. 141–154. [Google Scholar]
- Cheng R.H., Olson,N.H. and Baker,T.S. (1992) Cauliflower mosaic virus: A 420 subunit (T = 7) multilayer structure. Virology, 186, 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Reddy,V., Olson,N., Fisher,A., Baker,T. and Johnson,J. (1994) Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure, 2, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R.H., Kuhn,R.J., Olson,N.H., Rossmann,M.G., Choi,H.-K., Smith,T.J. and Baker,T.S. (1995) NC and glycoprotein organization in an enveloped virus. Cell, 80, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R.A., DeRosier,D.J. and Klug,A. (1970) The reconstruction of a 3D structure from projections and its application to electron microscopy. Proc. R. Soc. Lond. A, 317, 319–340. [Google Scholar]
- Forsell K., Suomalainen,M. and Garoff,H. (1995) Structure/function relation of the NH2-terminal domain of the Semliki Forest virus capsid protein. J. Virol., 69, 1556–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell K., Griffiths,G. and Garoff,H. (1996) Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J., 15, 6495–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I. and Schlesinger,S. (1994) Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol., 68, 8111–8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.D., Berriman,J.A., Butcher,S.J. and Gowen,B.E. (1995) Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell, 81, 715–725. [DOI] [PubMed] [Google Scholar]
- Garoff H. and Simons,K. (1974) Location of the spike glycoproteins in the Semliki Forest virus membrane. Proc. Natl Acad. Sci. USA, 71, 3988–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Simons,K. and Dobberstein,B. (1978) Assembly of Semliki Forest virus membrane glycoproteins in the membrane of the endoplasmic reticulum in vitro. J. Mol. Biol., 124, 587–600. [DOI] [PubMed] [Google Scholar]
- Geigenmüller-Gnirke U., Nitschko,H. and Schlesinger,S. (1993) Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J. Virol., 67, 1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. (1986) Alphavirus structure. In Schlesinger,S. and Schlesinger,M.J. (eds), The Togaviridae and Flaviviridae. Plenum Press, New York, NY, pp. 21–34. [Google Scholar]
- Jacrot B. (1987) Neutron scattering. In Nermut,M.V. and Steven,A.C. (eds), Animal Virus Structure. Elsevier Science Publishing, New York, NY, pp. 89–95. [Google Scholar]
- Johnson J.E. (1996) Functional implications of protein–protein interactions in icosahedral viruses. Proc. Natl Acad. Sci. USA, 93, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kääriäinen L. and Gomatos,P.J. (1969) A kinetic analysis of the synthesis in BHK-21 cells of RNAs specific for Semliki Forest virus. J. Gen. Virol., 5, 251–265. [DOI] [PubMed] [Google Scholar]
- Kääriäinen L., Simons,K. and von Bonsdorff,C.-H. (1969) Studies in subviral components of Semliki Forest virus. Ann. Med. Exp. Fenn., 47, 235–248. [PubMed] [Google Scholar]
- Lee S., Owen,K.E., Choi,H.-K., Lee,H., Lu,G., Wengler,G., Brown,D.T., Rossmann,M.G. and Kuhn,R.J. (1996) Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure, 4, 531–541. [DOI] [PubMed] [Google Scholar]
- Liljeström P. and Garoff,H. (1991) A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology, 9, 1356–1361. [DOI] [PubMed] [Google Scholar]
- Liljeström P., Lusa,S., Huylebroeck,D. and Garoff,H. (1991) In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol., 65, 4107–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E.J., Clarke,M., Gowen,B.E., Rutten,T. and Fuller,S.D. (2000) Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell, 5, 255–266. [DOI] [PubMed] [Google Scholar]
- Murphy F.A. (1980) The togaviruses: biology, structure, replication. In Schlesinger,R.W. (ed.), Togavirus Morphology and Morphogenesis. Academic Press, New York, NY, pp. 241–316. [Google Scholar]
- Owen K.E. and Kuhn,R.J. (1996) Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol., 70, 2757–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes A.M., Brown,D.T., Rothnagel,R., Chiu,W., Schoepp,R.J., Johnston,R.E. and Prasad,B.V.V. (1993) Thee-dimensional structure of a membrane-containing virus. Proc. Natl Acad. Sci. USA, 90, 9095–9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M.G. and Johnson,J.E. (1989) Icosahedral RNA virus structure. Annu. Rev. Biochem., 58, 533–573. [DOI] [PubMed] [Google Scholar]
- Sjöberg E.M., Suomalainen,M. and Garoff,H. (1994) A significantly improved Semliki Forest virus expression system based on translation enhancer segments from the viral capsid gene. Biotechnology, 12, 1127–1131. [DOI] [PubMed] [Google Scholar]
- Skoging U., Vihinen,M., Nilsson,L. and Liljeström,P. (1996) Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure, 4, 519–529. [DOI] [PubMed] [Google Scholar]
- Smith C., Grigorieff,N. and Pearse,B.M. (1998) Clathrin coats at 21 Å resolution: a cellular assembly designed to recycle multiple membrane receptors. EMBO J., 17, 4943–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderlund H. (1973) Kinetics of formation of the Semliki Forest virus nucleocapsid. Intervirology, 1, 354–361. [DOI] [PubMed] [Google Scholar]
- Söderlund H. and Ulmanen,I. (1977) Transient association of Semliki Forest virus capsid protein with ribosomes. J. Virol., 24, 907–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H. and Strauss,E.G. (1994) The alphaviruses: gene expression, replication and evolution. Microbiol. Rev., 58, 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M. and Garoff,H. (1992) Spike protein–nucleocapsid interactions drive the budding of alphaviruses. J. Virol., 66, 4737–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M., Garoff,H. and Baron,M. (1990) The E2 signal sequence of Rubella virus remains part of the capsid protein and confers membrane association in vitro. J. Virol., 64, 5500–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takkinen K., Peränen,J. and Kääriäinen,L. (1991) Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein. J. Gen. Virol., 72, 1627–1633. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen T.L., Hamburger,A.E., Fisher,B.R., Ostendorp,R. and Kuhn,R.J. (1999) In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli.J. Virol., 73, 5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel R.H., Provencher,S.W., von Bonsdorff,C.-H., Adrian,M. and Dubochet,J. (1986) Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature, 320, 533–535. [DOI] [PubMed] [Google Scholar]
- von Bonsdorff C.-H. and Pettersson,R. (1975) Surface structure of Uukuniemi virus. J. Virol., 95, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bonsdorff C.-H., Boere,W.A. and Garoff,H. (1978) Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol., 28, 578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg J.M., Boere,W.A. and Garoff,H. (1989) The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol., 63, 4991–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Nitschko,H., Ghattas,I., Wright,R. and Schlesinger,S. (1989) Evidence for specificity in the encapsidation of Sindbis virus RNAs. J. Virol., 63, 5310–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G., Würkner,D. and Wengler,G. (1992) Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology, 191, 880–888. [DOI] [PubMed] [Google Scholar]
- Xing L., Kato,K., Li,T., Takeda,N., Miyamura,T., Hammar,L. and Cheng,R.H. (1999) Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology, 265, 35–45. [DOI] [PubMed] [Google Scholar]
- Xing L., Tjärnlund,K., Lindqvist,B., Kaplan,G., Feigelstock,D., Cheng,R.H. and Casasnovas,J. (2000) Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J., 19, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]